Modulation of Amyloid β-Induced Microglia Activation and Neuronal Cell Death by Curcumin and Analogues
Abstract
1. Introduction
2. Results
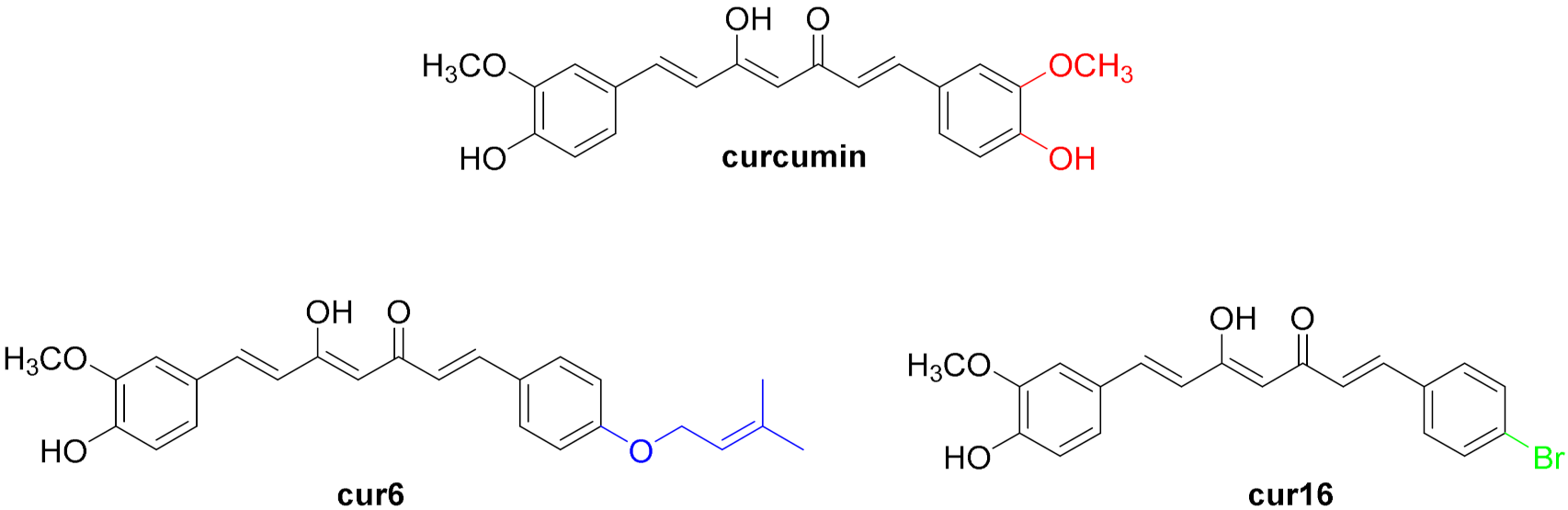
2.1. Rational Design and Synthesis of the Curcumin Analogues
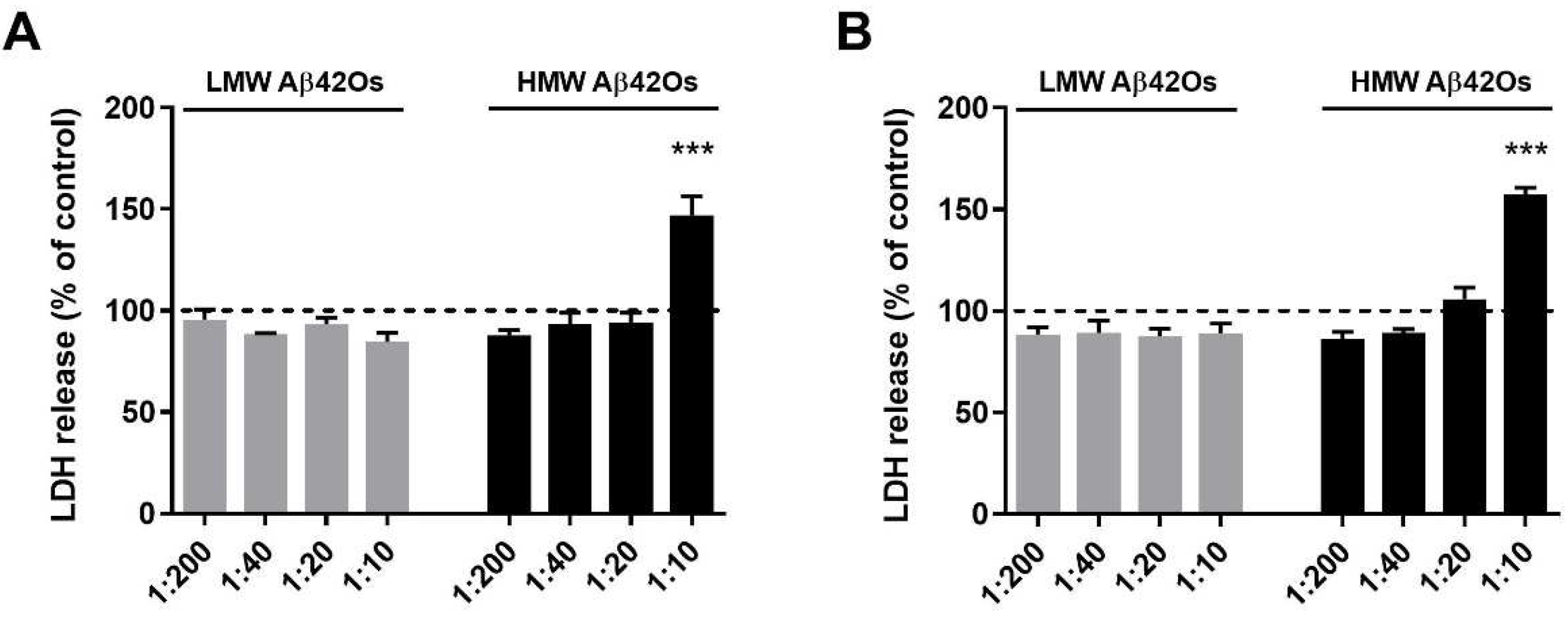
2.2. Effect of Aβ42 Oligomers on Microglia Cell Viability
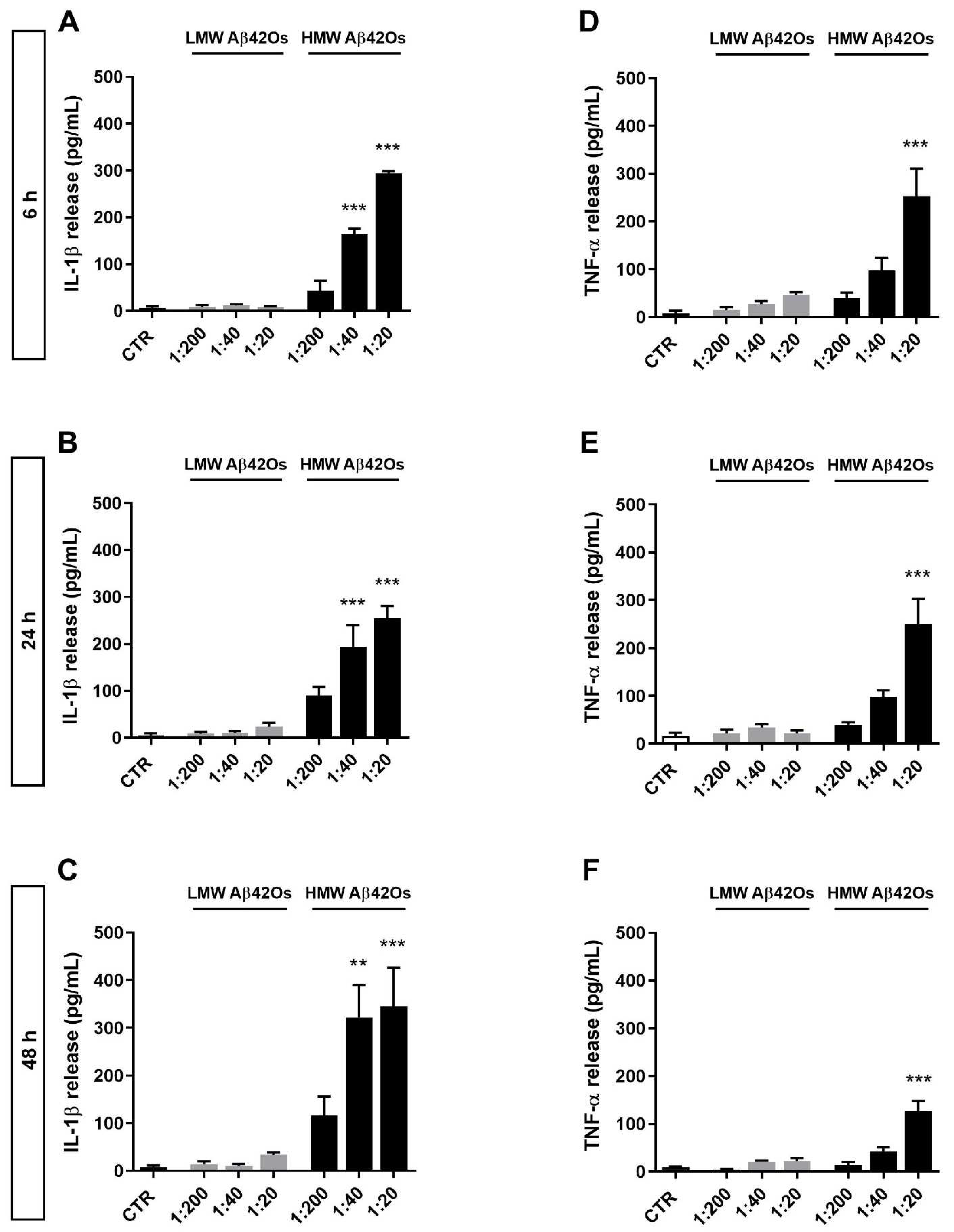
2.3. Effect of Aβ42 Oligomers on Proinflammatory Cytokine Release from Microglia
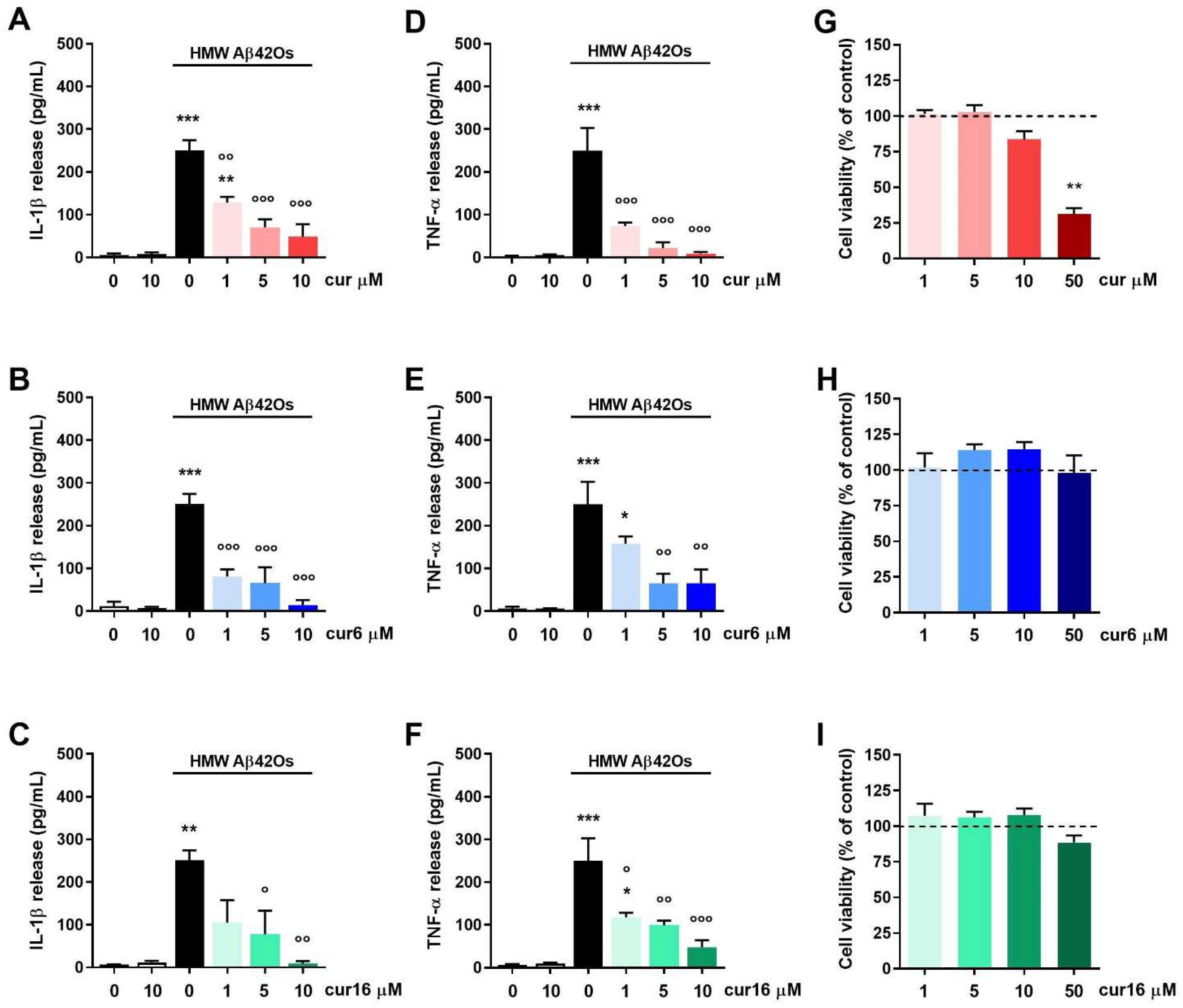
2.4. Effect of Curcumin and Analogues on Aβ42 Oligomer-Induced Microglia Inflammatory Response
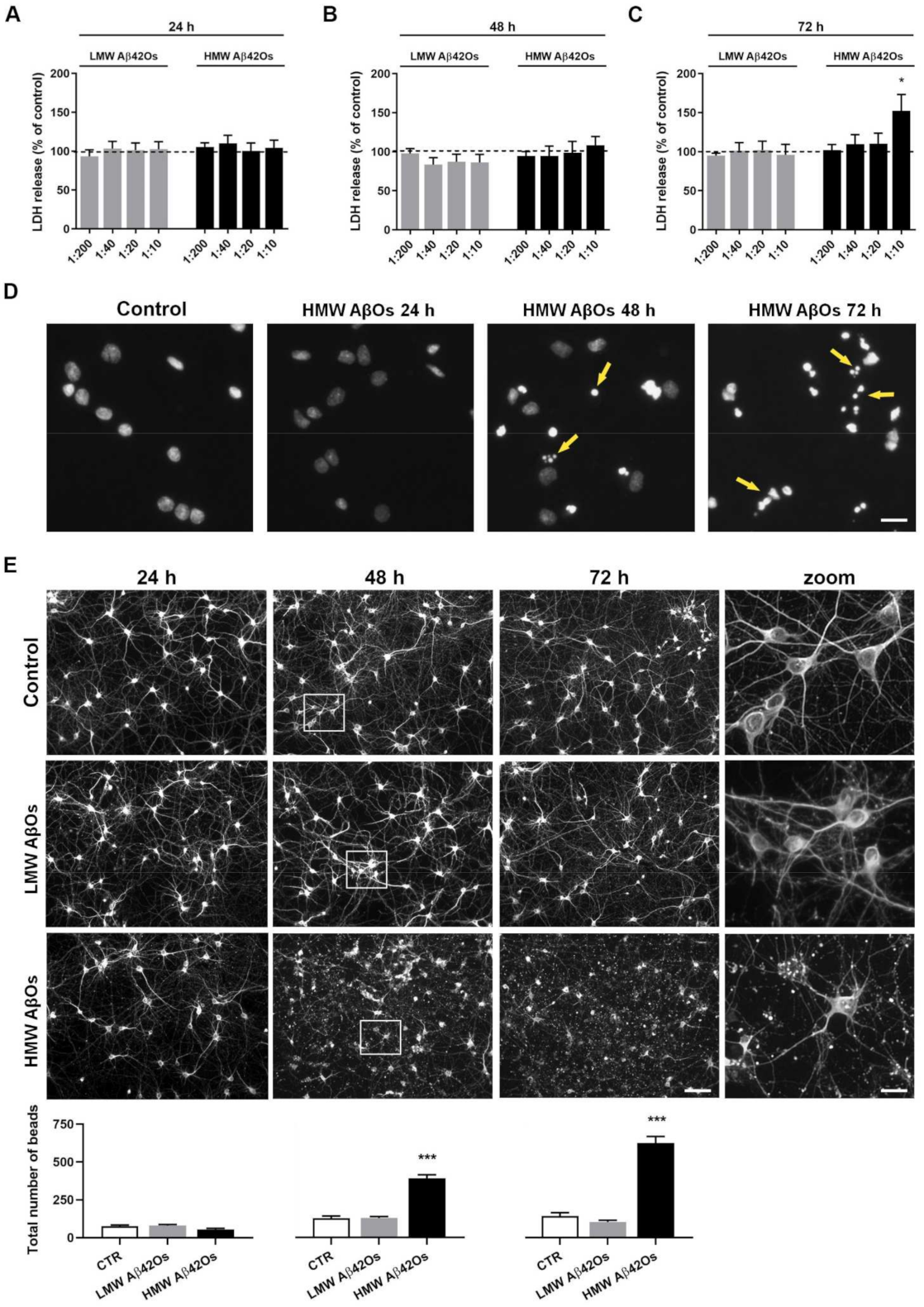
2.5. Effect of Aβ42 Oligomers on Neuronal Cell Toxicity
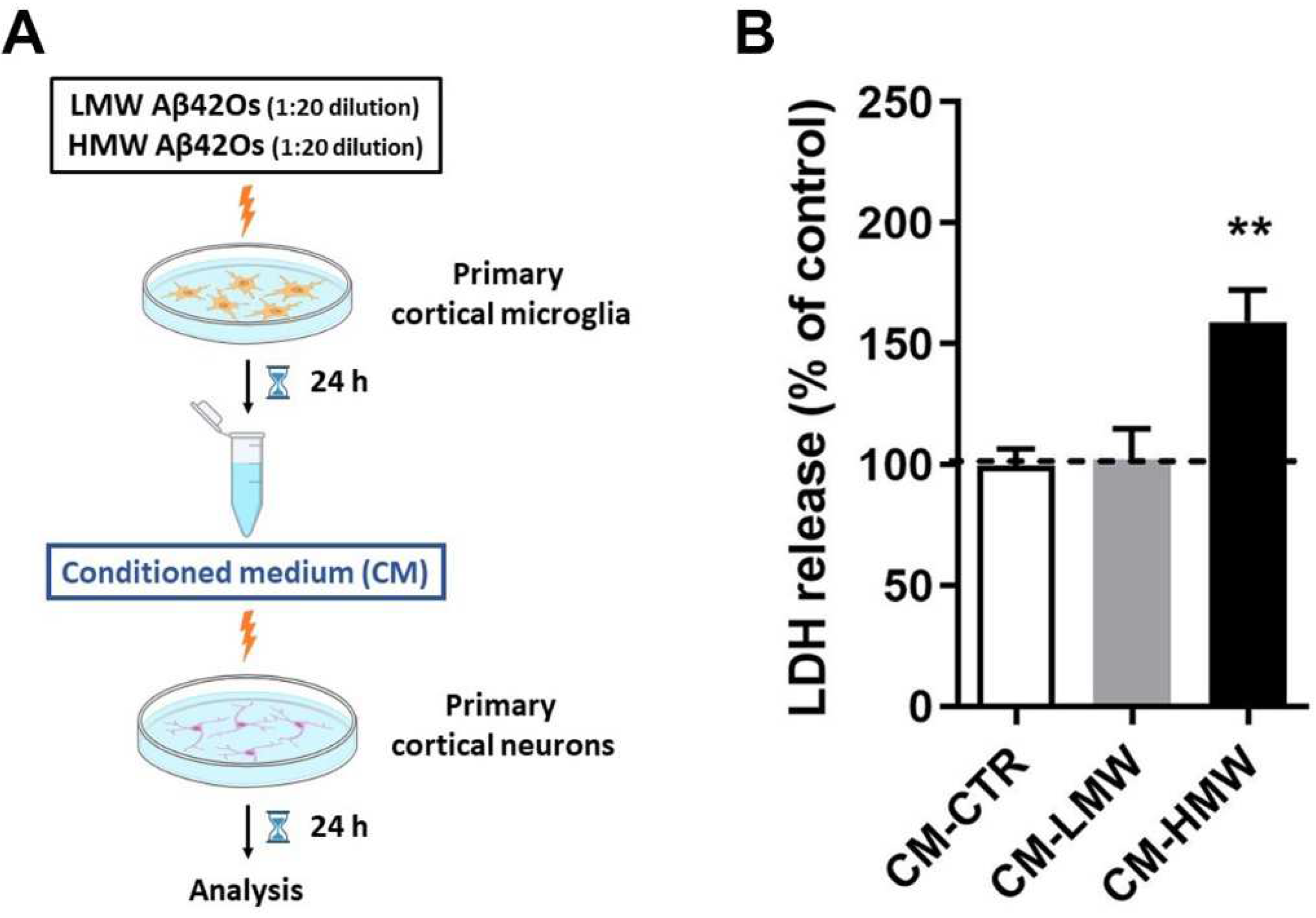
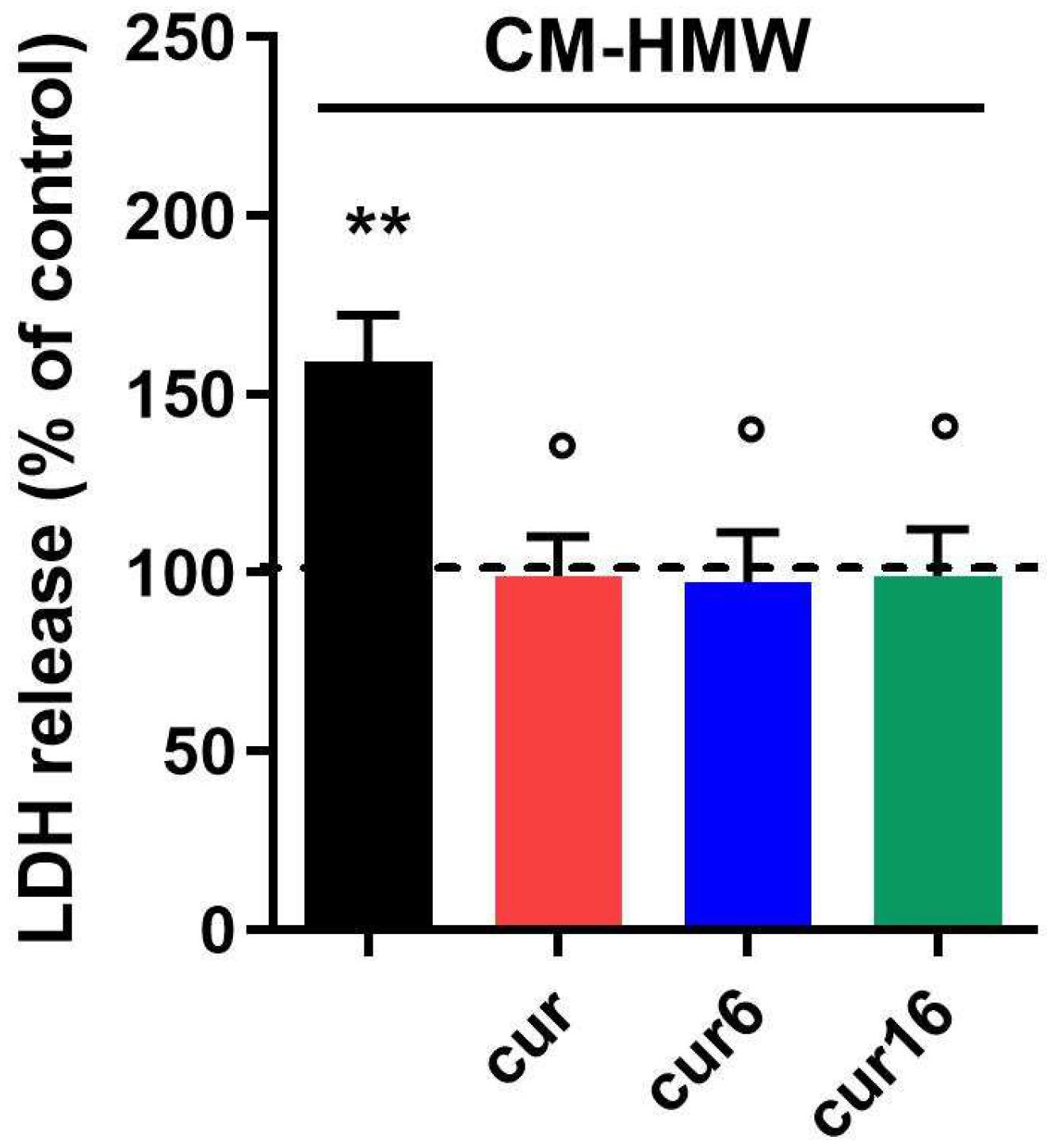
2.6. Effect of Curcumin and Analogues on Indirect Neuronal Cell Toxicity Induced by Aβ42 Oligomers
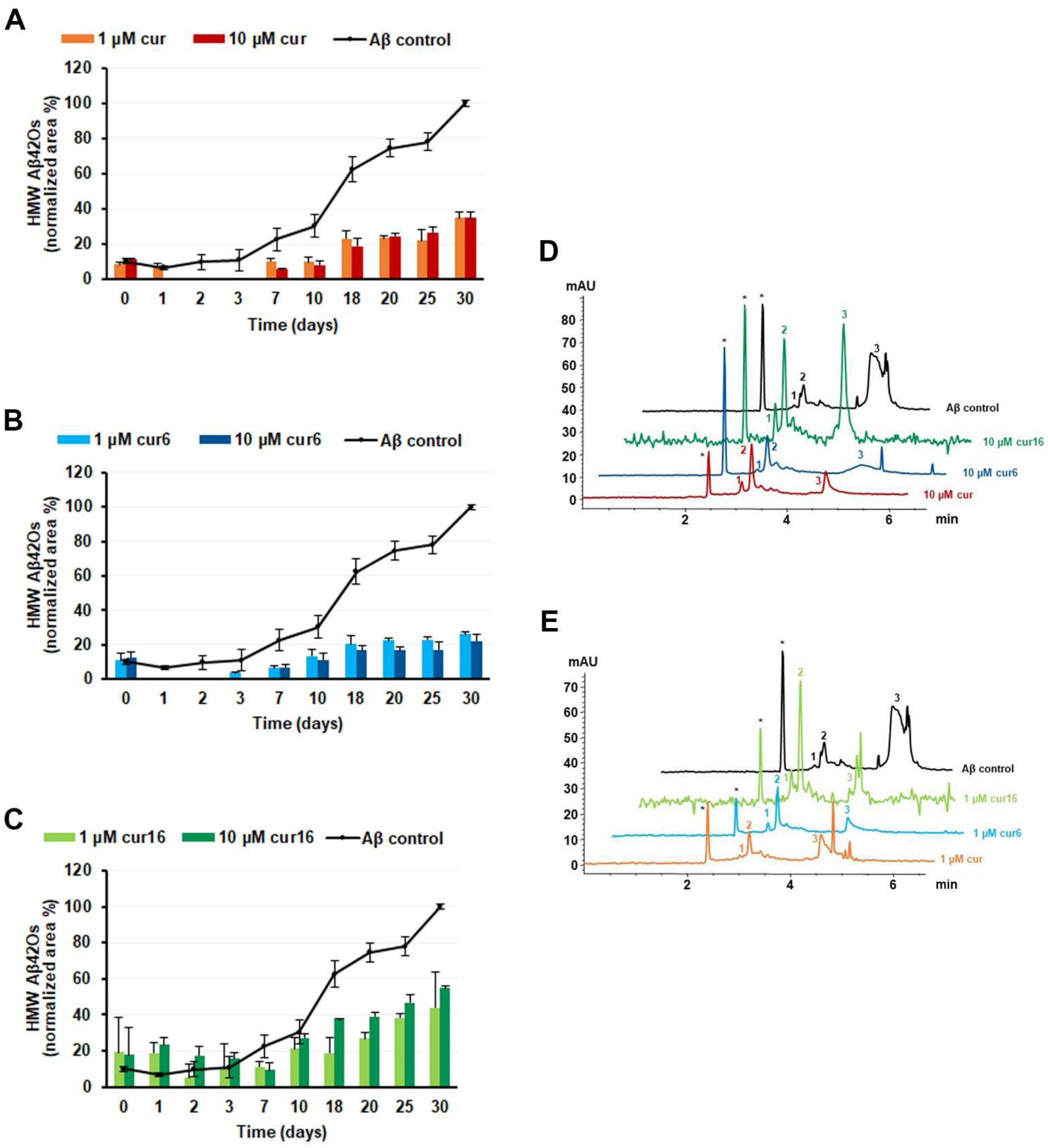
2.7. Effect of Curcumin and Analogues on Aβ42 Oligomerization
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. General Chemistry Procedures for cur6 and cur16 Synthesis
4.3. Synthesis of cur16 ((1E,4Z,6E)-1-(4-bromophenyl)-5-hydroxy-7-(4-hydroxy-3-methoxyphenyl)hepta-1,4,6-trien-3-one)
4.4. Preparation of Aβ42 Oligomers
4.5. Capillary Electrophoresis
4.6. Primary Cell Cultures
4.7. Preparation of Microglia-Conditioned Media
4.8. Lactate Dehydrogenase (LDH) Assay
4.9. MTT Assay
4.10. Cytokine Determination
4.11. Morphological Analysis of Apoptosis by DAPI Staining
4.12. Immunofluorescence
4.13. Quantification of Neuritic Beading
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Winblad, B.; Amouyel, P.; Andrieu, S.; Ballard, C.; Brayne, C.; Brodaty, H.; Cedazo-Minguez, A.; Dubois, B.; Edvardsson, D.; Feldman, H.; et al. Defeating Alzheimer’s disease and other dementias: A priority for European science and society. Lancet Neurol. 2016, 15, 455–532. [Google Scholar] [CrossRef]
- Querfurth, H.W.; LaFerla, F.M. Alzheimer’s disease. N. Engl. J. Med. 2010, 362, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Guo, Z. Alzheimer’s Aβ42 and Aβ40 peptides form interlaced amyloid fibrils. J. Neurochem. 2013, 126, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Goure, W.F.; Krafft, G.A.; Jerecic, J.; Hefti, F. Targeting the proper amyloid-beta neuronal toxins: A path forward for Alzheimer’s disease immunotherapeutics. Alzheimers Res. Ther. 2014, 6, 1–15. [Google Scholar] [CrossRef]
- Shankar, G.M.; Li, S.; Mehta, T.H.; Garcia-Munoz, A.; Shepardson, N.E.; Smith, I.; Brett, F.M.; Farrell, M.A.; Rowan, M.J.; Lemere, C.A.; et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008, 14, 837–842. [Google Scholar] [CrossRef]
- Roychaudhuri, R.; Yang, M.; Hoshi, M.M.; Teplow, D.B. Amyloid β-protein assembly and Alzheimer disease. J. Biol. Chem. 2009, 284, 4749–4753. [Google Scholar] [CrossRef]
- Ondrejcak, T.; Klyubin, I.; Hu, N.W.; Barry, A.E.; Cullen, W.K.; Rowan, M.J. Alzheimer’s disease amyloid β-protein and synaptic function. Neuromol. Med. 2010, 12, 13–26. [Google Scholar] [CrossRef]
- Sakono, M.; Zako, T. Amyloid oligomers: Formation and toxicity of Aβ oligomers. FEBS J. 2010, 277, 1348–1358. [Google Scholar] [CrossRef]
- Ferreira, S.T.; Klein, W.L. The Aβ oligomer hypothesis for synapse failure and memory loss in Alzheimer’s disease. Neurobiol. Learn Mem. 2011, 96, 529–543. [Google Scholar] [CrossRef]
- Li, S.; Jin, M.; Koeglsperger, T.; Shepardson, N.E.; Shankar, G.M.; Selkoe, D.J. Soluble Abeta oligomers inhibit long-term potentiation through a mechanism involving excessive activation of extrasynaptic NR2B-containing NMDA receptors. J. Neurosci. 2011, 31, 6627–6638. [Google Scholar] [CrossRef] [PubMed]
- Benilova, I.; Karran, E.; De Strooper, B. The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Forloni, G.; Artuso, V.; La Vitola, P.; Balducci, C. Oligomeropathies and pathogenesis of Alzheimer and Parkinson’s diseases. Mov. Disord. 2016, 31, 771–881. [Google Scholar] [CrossRef] [PubMed]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef]
- Varnum, M.M.; Ikezu, T. The classification of microglial activation phenotypes on neurodegeneration and regeneration in Alzheimer’s disease brain. Arch. Immunol. Ther. Exp. 2012, 60, 251–266. [Google Scholar] [CrossRef]
- Hanisch, U.K.; Kettenmann, H. Microglia: Active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007, 10, 1387–1394. [Google Scholar] [CrossRef]
- Parkhurst, C.N.; Yang, G.; Ninan, I.; Savas, J.N.; Yates, J.R., 3rd; Lafaille, J.J.; Hempstead, B.L.; Littman, D.R.; Gan, W.B. Microglia promote learning dependent synapse formation through brain-derived neurotrophic factor. Cell 2013, 155, 1596–1609. [Google Scholar] [CrossRef]
- Hickman, S.E.; Allison, E.K.; El Khoury, J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J. Neurosci. 2008, 28, 8354–8360. [Google Scholar] [CrossRef]
- Doens, D.; Fernández, P.L. Microglia receptors and their implications in the response to amyloid β for Alzheimer’s disease pathogenesis. J. Neuroinflamm. 2014, 11, 1–14. [Google Scholar] [CrossRef]
- Calsolaro, V.; Edison, P. Neuroinflamm. in Alzheimer’s disease: Current evidence and future directions. Alzheimers Dement. 2016, 12, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Morphy, R.; Rankovic, Z. Designed multiple ligands. An emerging drug discovery paradigm. J. Med. Chem. 2005, 48, 6523–6543. [Google Scholar] [CrossRef] [PubMed]
- Albertini, C.; Salerno, A.; de Sena Murteira Pinheiro, P.; Bolognesi, M.L. From combinations to multitarget-directed ligands: A continuum in Alzheimer’s disease polypharmacology. Med. Res. Rev. 2021, 41, 2606–2633. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lim, G.P.; Begum, A.N.; Ubeda, O.J.; Simmons, M.R.; Ambegaokar, S.S.; Chen, P.P.; Kayed, R.; Glabe, C.G.; Frautschy, S.A.; et al. Curcumin inhibits formation of amyloid beta oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 2005, 280, 5892–5901. [Google Scholar] [CrossRef]
- Mercanti, G.; Ragazzi, E.; Toffano, G.; Giusti, P.; Zusso, M. Phosphatidylserine and curcumin act synergistically to down-regulate release of interleukin-1β from lipopolysaccharide-stimulated cortical primary microglial cells. CNS Neurol. Disord. Drug Targets 2014, 13, 792–800. [Google Scholar] [CrossRef]
- Prasad, S.; Gupta, S.C.; Tyagi, A.K.; Aggarwal, B.B. Curcumin, a component of golden spice: From bedside to bench and back. Biotechnol. Adv. 2014, 32, 1053–1064. [Google Scholar] [CrossRef]
- Di Martino, R.M.C.; Bisi, A.; Rampa, A.; Gobbi, S.; Belluti, F. Recent progress on curcumin-based therapeutics: A patent review (2012–2016). Part II: Curcumin derivatives in cancer and neurodegeneration. Expert Opin Ther. Pat. 2017, 27, 953–965. [Google Scholar] [CrossRef]
- Zusso, M.; Mercanti, G.; Belluti, F.; Di Martino, R.M.C.; Pagetta, A.; Marinelli, C.; Brun, P.; Ragazzi, E.; Lo, R.; Stifani, S.; et al. Phenolic 1,3-diketones attenuate lipopolysaccharide-induced inflammatory response by an alternative magnesium-mediated mechanism. Br. J. Pharmacol. 2017, 174, 1090–1103. [Google Scholar] [CrossRef]
- Sorrenti, V.; Contarini, G.; Sut, S.; Dall’Acqua, S.; Confortin, F.; Pagetta, A.; Giusti, P.; Zusso, M. Curcumin prevents acute neuroinflammation and long-term memory impairment induced by systemic lipopolysaccharide in mice. Front. Pharmacol. 2018, 9, 183. [Google Scholar] [CrossRef]
- Bisceglia, F.; Seghetti, F.; Serra, M.; Zusso, M.; Gervasoni, S.; Verga, L.; Vistoli, G.; Lanni, C.; Catanzaro, M.; De Lorenzi, E.; et al. Prenylated curcumin analogues as multipotent tools to tackle Alzheimer’s disease. ACS Chem. Neurosci. 2019, 10, 1420–1433. [Google Scholar] [CrossRef]
- Caruso, G.; Torrisi, S.A.; Mogavero, M.P.; Currenti, W.; Castellano, S.; Godos, J.; Ferri, R.; Galvano, F.; Leggio, G.M.; Grosso, G.; et al. Polyphenols and neuroprotection: Therapeutic implications for cognitive decline. Pharmacol. Ther. 2022, 232, 108013. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Stohs, S.J.; Chen, O.; Ray, S.D.; Ji, J.; Bucci, L.R.; Preuss, H.G. Highly bioavailable forms of curcumin and promising avenues for curcumin-based research and application: A review. Molecules 2020, 25, 1397. [Google Scholar] [CrossRef]
- Anand, P.; Thomas, S.G.; Kunnumakkara, A.B.; Sundaram, C.; Harikumar, K.B.; Sung, B.; Tharakan, S.T.; Misra, K.; Priyadarsini, I.K.; Rajasekharan, K.N.; et al. Biological activities of curcumin and its analogues (Congeners) made by man and Mother Nature. Biochem. Pharmacol. 2008, 76, 1590–1611. [Google Scholar] [CrossRef]
- Di Martino, R.M.C.; De Simone, A.; Andrisano, V.; Bisignano, P.; Bisi, A.; Gobbi, S.; Rampa, A.; Fato, R.; Bergamini, C.; Perez, D.; et al. Versatility of the curcumin scaffold: Discovery of potent and balanced dual BACE-1 and GSK-3 beta inhibitors. J. Med. Chem. 2016, 59, 531–544. [Google Scholar] [CrossRef]
- Di Martino, R.M.C.; Pruccoli, L.; Bisi, A.; Gobbi, S.; Rampa, A.; Martinez, A.; Pérez, C.; Martinez-Gonzalez, L.; Paglione, M.; Di Schiavi, E.; et al. Novel curcumin-diethyl fumarate hybrid as a dualistic GSK-3β inhibitor/Nrf2 inducer for the treatment of Parkinson’s disease. ACS Chem. Neurosci. 2020, 11, 2728–2740. [Google Scholar] [CrossRef]
- Seghetti, F.; Di Martino, R.M.C.; Catanzaro, E.; Bisi, A.; Gobbi, S.; Rampa, A.; Canonico, B.; Montanari, M.; Krysko, D.V.; Papa, S.; et al. Curcumin-1,2,3-triazole conjugation for targeting the cancer apoptosis machinery. Molecules 2020, 25, 3066. [Google Scholar] [CrossRef]
- Reinke, A.A.; Gestwicki, J.E. Structure-activity relationships of amyloid beta-aggregation inhibitors based on curcumin: Influence of linker length and flexibility. Chem. Biol. Drug Des. 2007, 70, 206–215. [Google Scholar] [CrossRef]
- Heidrich, J.; Sperl, L.E.; Boeckler, F.M. Embracing the diversity of halogen bonding motifs in fragment-based drug discovery-construction of a diversity-optimized halogen-enriched fragment library. Front. Chem. 2019, 7, 9. [Google Scholar] [CrossRef]
- Wu, Q.; Huang, M.; Li, T.; Jiao, L.; Tu, Y.; Xu, X.; Ma, X.; Tian, H.; Qiao, Y. Crystal and electronic structure of poly-halogenated lanthanide Schiff base complex: Insights into halogen bond from structural and theoretical analysis. J. Mol. Struct. 2021, 1225, 129054. [Google Scholar] [CrossRef]
- Chan, F.K.; Moriwaki, K.; De Rosa, M.J. Detection of necrosis by release of lactate dehydrogenase activity. Methods Mol. Biol. 2013, 979, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.M.; Lieven, C.J.; Levin, L.A. Simultaneous labeling of projecting neurons and apoptotic state. J. Neurosci. Methods. 2007, 161, 281–284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, H.Y.; Hudry, E.; Hashimoto, T.; Kuchibhotla, K.; Rozkalne, A.; Fan, Z.; Spires-Jones, T.; Xie, H.; Arbel-Ornath, M.; Grosskreutz, C.; et al. Amyloid beta induces the morphological neurodegenerative triad of spine loss, dendritic simplification, and neuritic dystrophies through calcineurin activation. J. Neurosci. 2010, 30, 2636–2649. [Google Scholar] [CrossRef] [PubMed]
- Sadleir, K.R.; Kandalepas, P.C.; Buggia-Prévot, V.; Nicholson, D.A.; Thinakaran, G.; Vassar, R. Presynaptic dystrophic neurites surrounding amyloid plaques are sites of microtubule disruption, BACE1 elevation, and increased Aβ generation in Alzheimer’s disease. Acta Neuropathol. 2016, 132, 235–256. [Google Scholar] [CrossRef]
- Combs, C.K.; Karlo, J.C.; Kao, S.C.; Landreth, G.E. beta-Amyloid stimulation of microglia and monocytes results in TNFalpha-dependent expression of inducible nitric oxide synthase and neuronal apoptosis. J. Neurosci. 2001, 21, 1179–1188. [Google Scholar] [CrossRef]
- Floden, A.M.; Li, S.; Combs, C.K. Beta-amyloid-stimulated microglia induce neuron death via synergistic stimulation of tumor necrosis factor alpha and NMDA receptors. J. Neurosci. 2005, 25, 2566–2575. [Google Scholar] [CrossRef]
- Shukla, S.M.; Sharma, S.K. Sinomenine inhibits microglial activation by Aβ and confers neuroprotection. J. Neuroinflamm. 2011, 8, 117. [Google Scholar] [CrossRef]
- Bisceglia, F.; Natalello, A.; Serafini, M.M.; Colombo, R.; Verga, L.; Lanni, C.; De Lorenzi, E. An integrated strategy to correlate aggregation state, structure and toxicity of Aß 1–42 oligomers. Talanta 2018, 188, 17–26. [Google Scholar] [CrossRef]
- Sabella, S.; Quaglia, M.; Lanni, C.; Racchi, M.; Govoni, S.; Caccialanza, G.; Calligaro, A.; Bellotti, V.; De Lorenzi, E. Capillary electrophoresis studies on the aggregation process of beta-amyloid 1-42 and 1-40 peptides. Electrophoresis 2004, 25, 3186–3194. [Google Scholar] [CrossRef]
- Lee, S.J.; Nam, E.; Lee, H.J.; Savelieff, M.G.; Lim, M.H. Towards an understanding of amyloid-beta oligomers: Characterization, toxicity mechanisms, and inhibitors. Chem. Soc. Rev. 2017, 46, 310–323. [Google Scholar] [CrossRef]
- Matuszyk, M.M.; Garwood, C.J.; Ferraiuolo, L.; Simpson, J.E.; Staniforth, R.A.; Wharton, S.B. Biological and methodological complexities of beta-amyloid peptide: Implications for Alzheimer’s disease research. J. Neurochem. 2021, 160, 434–453. [Google Scholar] [CrossRef] [PubMed]
- Rambaldi, D.C.; Zattoni, A.; Reschiglian, P.; Colombo, R.; De Lorenzi, E. In vitro amyloid Aβ1-42 peptide aggregation monitoring by asymmetrical flow field-flow fractionation with multi-angle light scattering detection. Anal. Bioanal. Chem. 2009, 394, 2145–2149. [Google Scholar] [CrossRef] [PubMed]
- Nichols, M.R.; Colvin, B.A.; Hood, E.A.; Paranjape, G.S.; Osborn, D.C.; Terrill-Usery, S.E. Biophysical comparison of soluble Amyloid-β(1–42) protofibrils, oligomers, and protofilaments. Biochemistry 2015, 54, 2193–2204. [Google Scholar] [CrossRef] [PubMed]
- Colombo, R.; Carotti, A.; Catto, M.; Racchi, M.; Lanni, C.; Verga, L.; Caccialanza, G.; De Lorenzi, E. CE can identify small molecules that selectively target soluble oligomers of amyloid beta protein and display antifibrillogenic activity. Electrophoresis 2009, 30, 1418–1429. [Google Scholar] [CrossRef]
- Butini, S.; Brindisi, M.; Brogi, S.; Maramai, S.; Guarino, E.; Panico, A.; Saxena, A.; Chauhan, V.; Colombo, R.; Verga, L.; et al. Multifunctional cholinesterase and amyloid Beta fibrillization modulators. Synthesis and biological investigation. ACS Med. Chem. Lett. 2013, 4, 1178–1182. [Google Scholar] [CrossRef][Green Version]
- Brogi, S.; Butini, S.; Maramai, S.; Colombo, R.; Verga, L.; Lanni, C.; De Lorenzi, E.; Lamponi, S.; Andreassi, M.; Bartolini, M.; et al. Disease-modifying anti-Alzheimer’s drugs: Inhibitors of human cholinesterases interfering with β-amyloid aggregation. CNS Neurosci. Ther. 2014, 20, 624–632. [Google Scholar] [CrossRef]
- Kaffy, J.; Brinet, D.; Soulier, J.-L.; Correia, I.; Tonali, N.; Fera, K.F.; Iacone, Y.; Hoffmann, A.R.F.; Khemtémourian, L.; Crousse, B.; et al. Designed glycopeptidomimetics disrupt protein-protein interactions mediating amyloid β-peptide aggregation and restore neuroblastoma cell viability. J. Med. Chem. 2016, 59, 2025–2040. [Google Scholar] [CrossRef]
- Brinet, D.; Gaie-Levrel, F.; Delatour, V.; Kaffy, J.; Ongeri, S.; Taverna, M. In vitro monitoring of amyloid β-peptide oligomerization by Electrospray differential mobility analysis: An alternative tool to evaluate Alzheimer’s disease drug candidates. Talanta 2017, 165, 84–91. [Google Scholar] [CrossRef]
- Kaffy, J.; Berardet, C.; Mathieu, L.; Legrand, B.; Taverna, M.; Halgand, F.; Van Der Rest, G.; Maillard, L.T.; Ongeri, S. Helical γ-peptide foldamers as dual inhibitors of amyloid-β peptide and islet amyloid polypeptide oligomerization and fibrillization. Chemistry 2020, 26, 14612–14622. [Google Scholar] [CrossRef]
- Huang, Y.R.; Liu, R.-T. The toxicity and polymorphism of amyloid oligomers. Int. J. Mol. Sci. 2020, 21, 4477. [Google Scholar] [CrossRef]
- Li, S.; Selkoe, D.J. A mechanistic hypothesis for the impairment of synaptic plasticity by soluble Aβ oligomers from Alzheimer’s brain. J. Neurochem. 2020, 154, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Shea, D.; Hsu, C.-C.; Bi, T.M.; Paranjapye, N.; Childers, M.C.; Cochran, J.; Tomberlin, C.P.; Wang, L.; Paris, D.; Zonderman, J.; et al. α-Sheet secondary structure in amyloid β-peptide drives aggregation and toxicity in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2019, 116, 8895–8900. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Li, S.; Xu, H.; Walsh, D.M.; Selkoe, D.J. Large soluble oligomers of amyloid β-protein from Alzheimer brain are far less neuroactive than the smaller oligomers to which they dissociate. J. Neurosci. 2017, 37, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Sondag, C.M.; Dhawan, G.; Combs, C.K. Beta amyloid oligomers and fibrils stimulate differential activation of primary microglia. J. Neuroinflamm. 2009, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Álvarez, M.; Santos-Lozano, A.; Sanchis-Gomar, F.; Fiuza-Luces, C.; Pareja-Galeano, H.; Garatachea, N.; Lucia, A. Non-steroidal anti-inflammatory drugs as a treatment for Alzheimer’s disease: A systematic review and meta-analysis of treatment effect. Drugs Aging 2015, 32, 139–147. [Google Scholar] [CrossRef]
- Atamna, H.; Boyle, K. Amyloid-beta peptide binds with heme to form a peroxidase: Relationship to the cytopathologies of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2006, 103, 3381–3386. [Google Scholar] [CrossRef]
- Garcia-Alloza, M.; Borrelli, L.A.; Rozkalne, A.; Hyman, B.T.; Bacskai, B.J. Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. J. Neurochem. 2007, 102, 1095–1104. [Google Scholar] [CrossRef]
- Reddy, P.H.; Manczak, M.; Yin, X.; Grady, M.C.; Mitchell, A.; Tonk, S.; Kuruva, C.S.; Bhatti, J.S.; Kandimalla, R.; Vijayan, M.; et al. Protective effects of Indian spice curcumin against amyloid-β in Alzheimer’s disease. J. Alzheimer’s Dis. 2018, 61, 843–866. [Google Scholar] [CrossRef]
- Simon, E.; Obst, J.; Gomez-Nicola, D. The evolving dialogue of microglia and neurons in Alzheimer’s disease: Microglia as necessary transducers of pathology. Neuroscience 2019, 405, 24–34. [Google Scholar] [CrossRef]
- Greenwood, S.M.; Mizielinska, S.M.; Frenguelli, B.G.; Harvey, J.; Connolly, C.N. Mitochondrial dysfunction and dendritic beading during neuronal toxicity. J. Biol. Chem. 2007, 282, 26235–26244. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.N.; Chiu, S.W.; Benoit, J.; Chew, L.Y.; Mu, Y. The effect of curcumin on the stability of Aβ dimers. J. Phys. Chem. B 2012, 116, 7428–7435. [Google Scholar] [CrossRef] [PubMed]
- Nedumpully-Govindan, P.; Kakinen, A.; Pilkington, E.H.; Dsvis, T.P.; Chun Ke, P.; Ding, F. Stabilizing off-pathway oligomers by polyphenol nanoassemblies for IAPP aggregation inhibition. Sci. Rep. 2016, 6, 1–12. [Google Scholar]
- Facci, L.; Barbierato, M.; Zusso, M.; Skaper, S.D.; Giusti, P. Serum amyloid A primes microglia for ATP-dependent interleukin-1β release. J. Neuroinflamm. 2018, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Facci, L.; Skaper, S.D. Culture of rodent cortical, hippocampal, and striatal neurons. Methods Mol. Biol. 2018, 1727, 39–47. [Google Scholar] [CrossRef]
- Facci, L.; Skaper, S.D. Amyloid β-peptide neurotoxicity assay using cultured rat cortical neurons. Methods Mol. Biol. 2012, 846, 57–65. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and citotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Lorenzi, E.; Franceschini, D.; Contardi, C.; Di Martino, R.M.C.; Seghetti, F.; Serra, M.; Bisceglia, F.; Pagetta, A.; Zusso, M.; Belluti, F. Modulation of Amyloid β-Induced Microglia Activation and Neuronal Cell Death by Curcumin and Analogues. Int. J. Mol. Sci. 2022, 23, 4381. https://doi.org/10.3390/ijms23084381
De Lorenzi E, Franceschini D, Contardi C, Di Martino RMC, Seghetti F, Serra M, Bisceglia F, Pagetta A, Zusso M, Belluti F. Modulation of Amyloid β-Induced Microglia Activation and Neuronal Cell Death by Curcumin and Analogues. International Journal of Molecular Sciences. 2022; 23(8):4381. https://doi.org/10.3390/ijms23084381
Chicago/Turabian StyleDe Lorenzi, Ersilia, Davide Franceschini, Cecilia Contardi, Rita Maria Concetta Di Martino, Francesca Seghetti, Massimo Serra, Federica Bisceglia, Andrea Pagetta, Morena Zusso, and Federica Belluti. 2022. "Modulation of Amyloid β-Induced Microglia Activation and Neuronal Cell Death by Curcumin and Analogues" International Journal of Molecular Sciences 23, no. 8: 4381. https://doi.org/10.3390/ijms23084381
APA StyleDe Lorenzi, E., Franceschini, D., Contardi, C., Di Martino, R. M. C., Seghetti, F., Serra, M., Bisceglia, F., Pagetta, A., Zusso, M., & Belluti, F. (2022). Modulation of Amyloid β-Induced Microglia Activation and Neuronal Cell Death by Curcumin and Analogues. International Journal of Molecular Sciences, 23(8), 4381. https://doi.org/10.3390/ijms23084381







