Slc38a9 Deficiency Induces Apoptosis and Metabolic Dysregulation and Leads to Premature Death in Zebrafish
Abstract
1. Introduction
2. Results
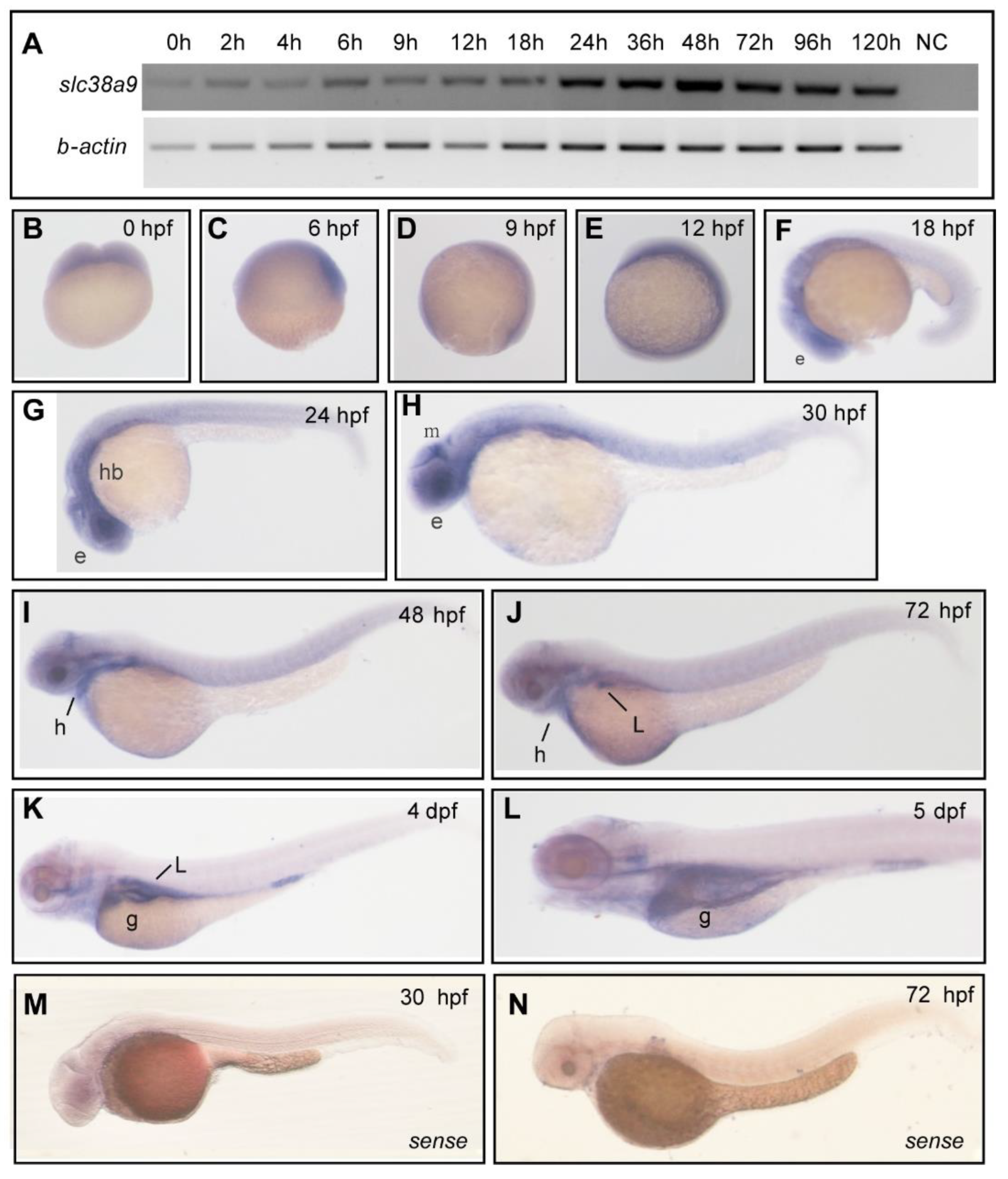
2.1. Spatiotemporal Expression Pattern of slc38a9 in Zebrafish
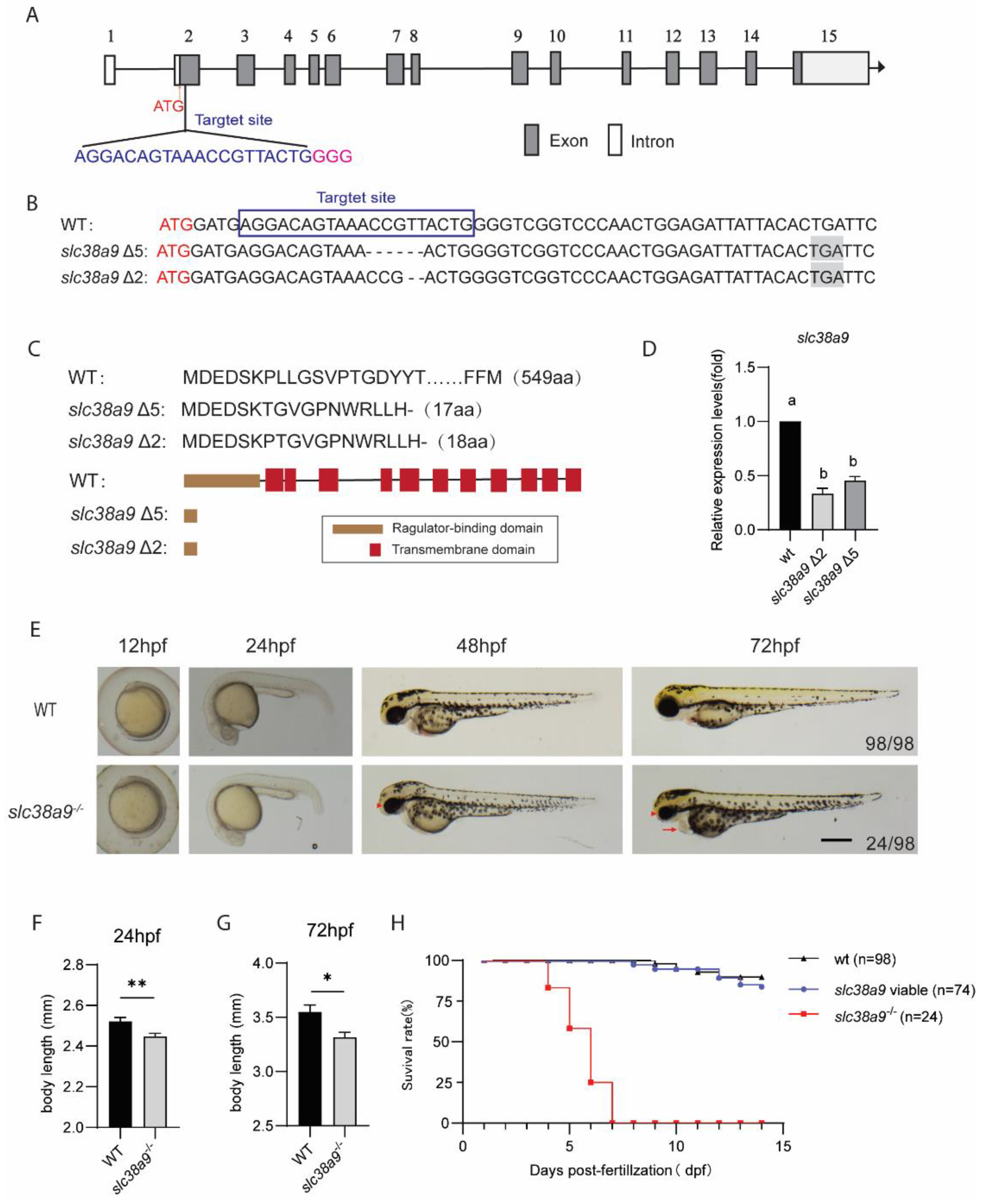
2.2. Knockout slc38a9 Causes Developmental Defects and Premature Death in Zebrafish
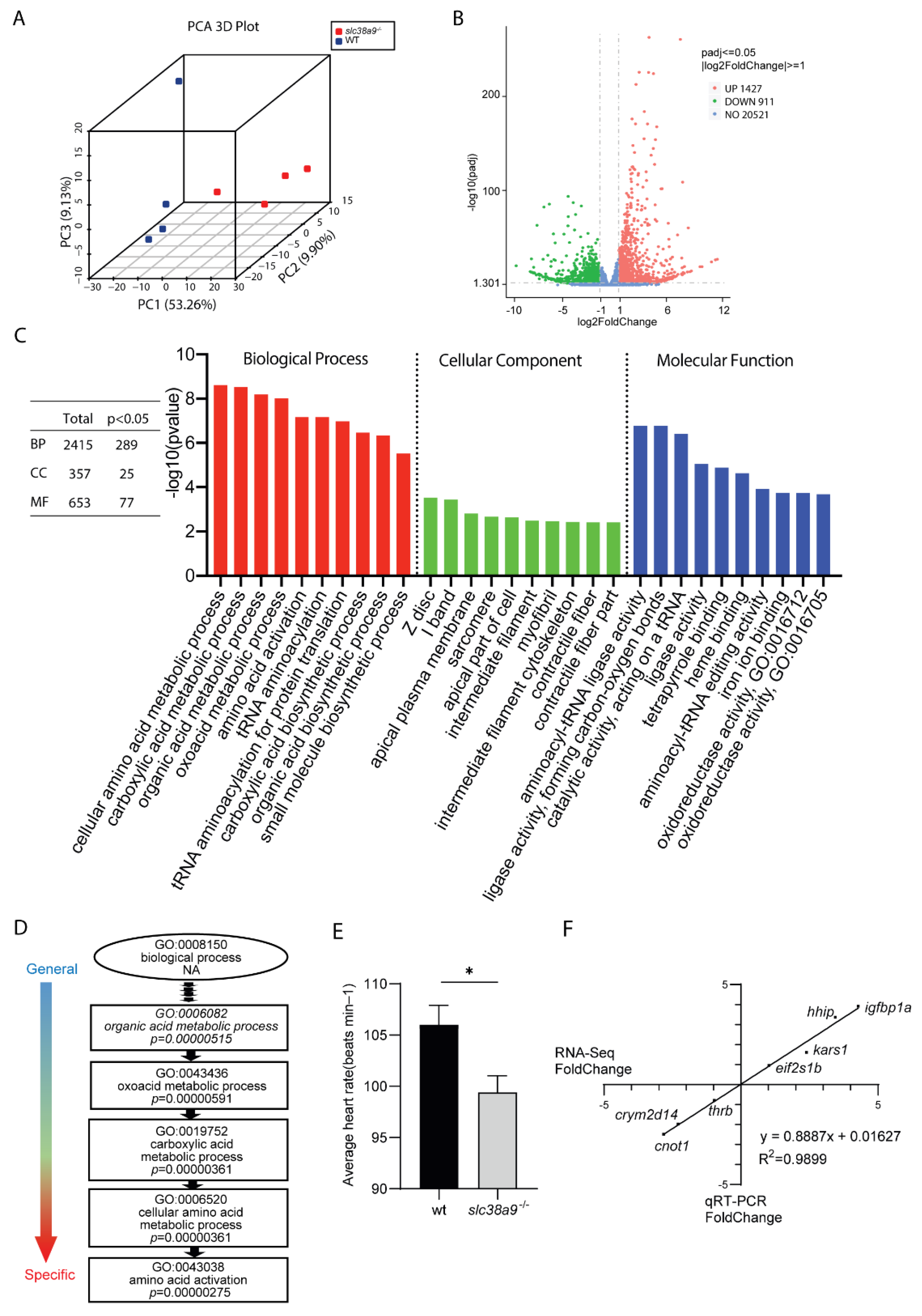
2.3. Transcriptional Profiling of slc38a9−/− Mutant
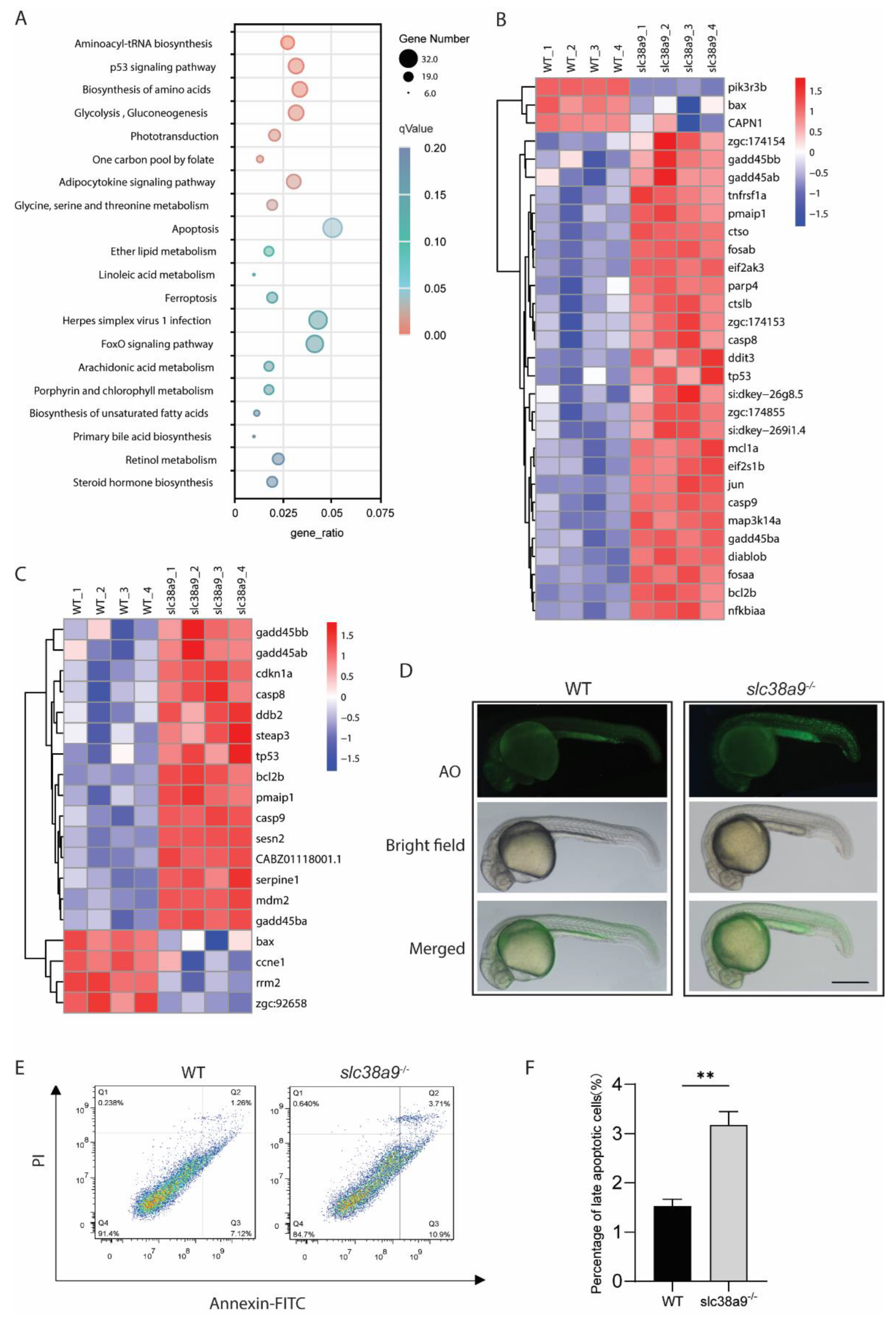
2.4. Slc38a9 Deficiency Induces Apoptosis and Alters Multiple Metabolic Pathways
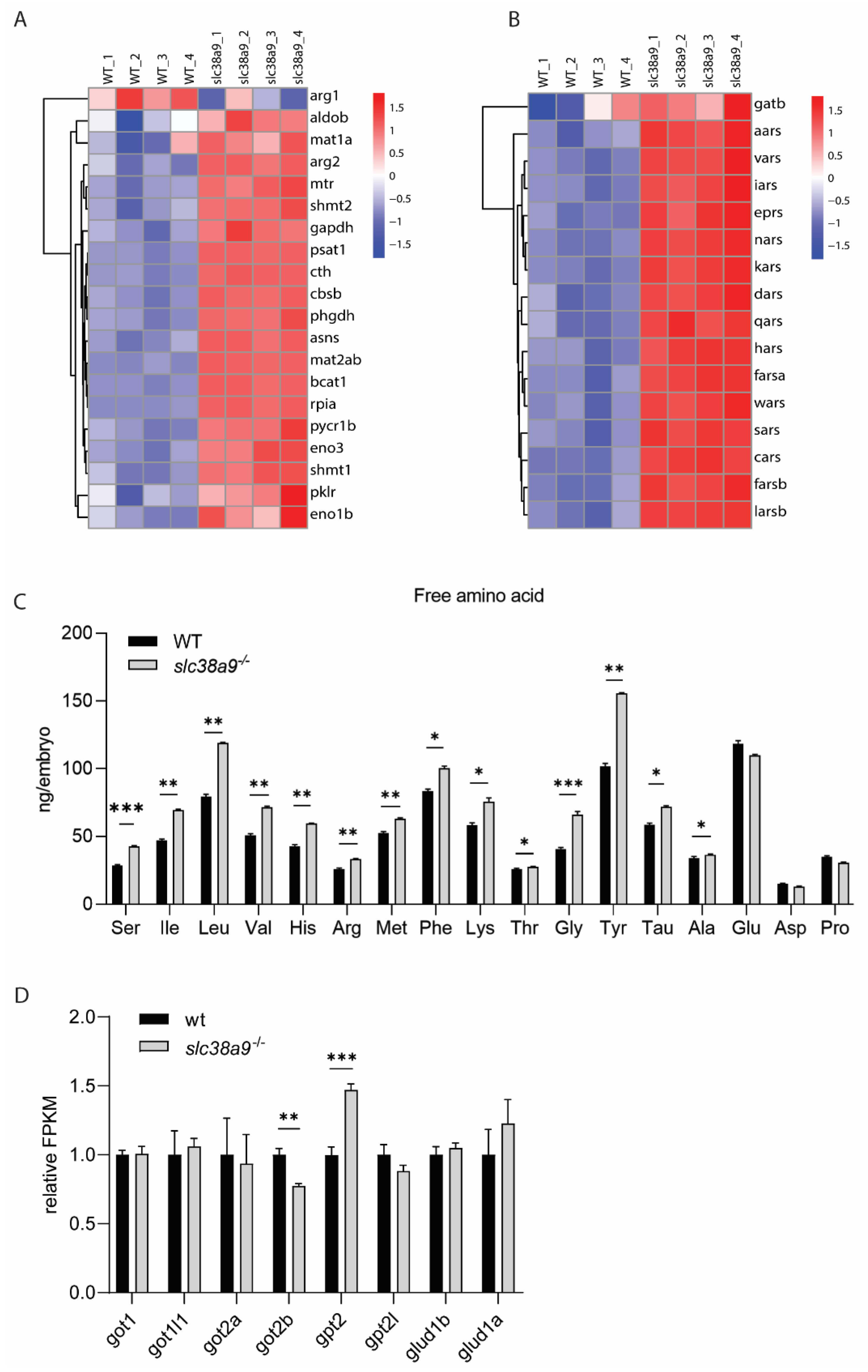
2.5. Biosynthesis of Amino Acids and Aminoacyl-tRNA Were Enhanced in slc38a9−/− Embryos
2.6. Slc38a9 Regulates Glycolysis and Gluconeogenesis in Zebrafish Embryo
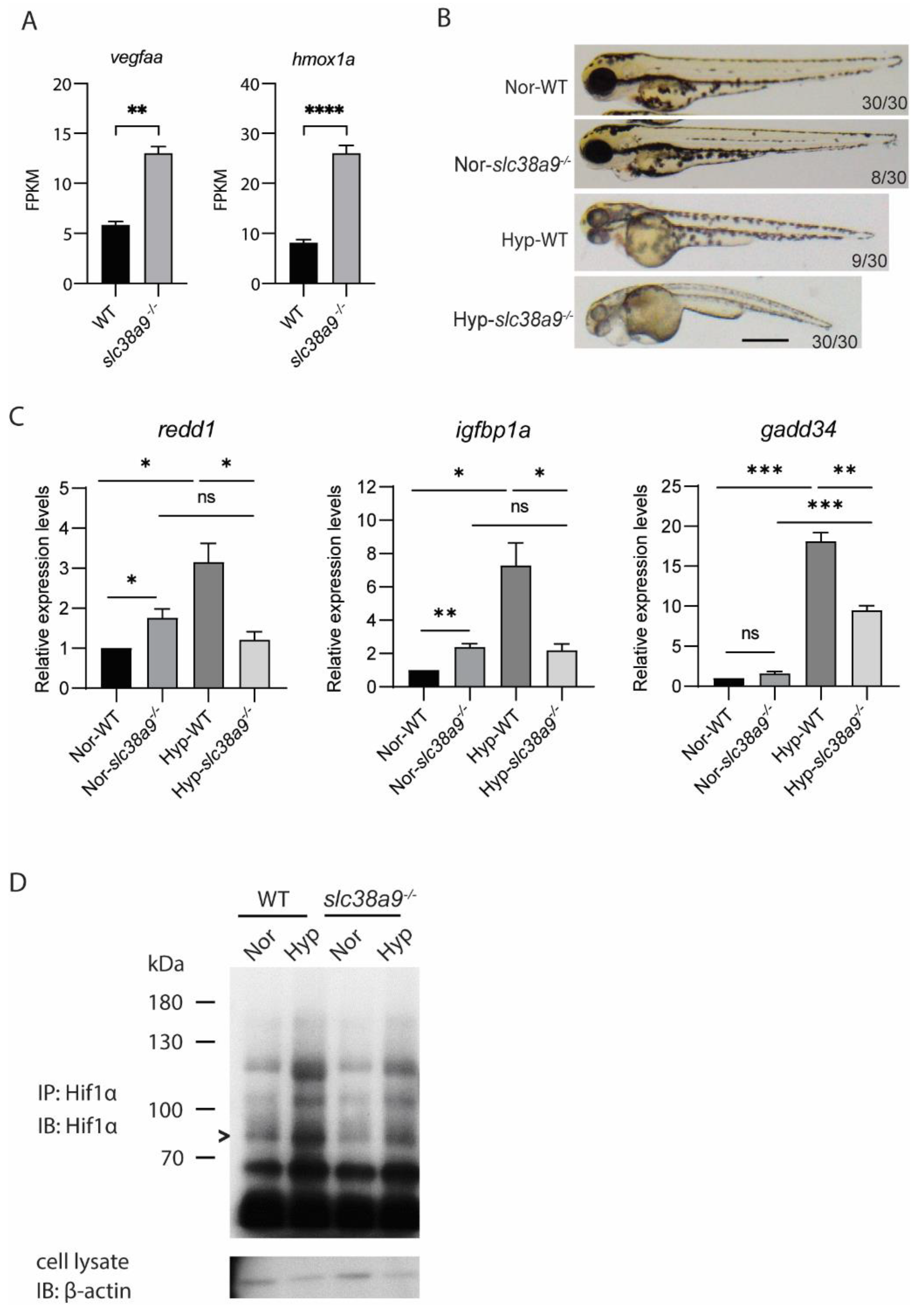
2.7. Slc38a9 Deficiency Leads to Dysregulated Hypoxia Response
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. slc38a9 Knockout by CRISPR/Cas9 System
4.3. Hypoxia Treatment
4.4. Quantitative RT-PCR (qRT-PCR)
4.5. Whole-Mount In Situ Hybridization
4.6. High-Throughput RNA Sequencing
4.7. Bioinformatics Analysis of RNA Sequence Data
4.8. AO Staining
4.9. Apoptosis Analysis by Annexin V-FITC/PI Staining
4.10. Free Amino Acid Analysis
4.11. Mass Spectrometry
4.12. Western Blot and Immunoprecipitation
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velazquez, M.A. Impact of maternal malnutrition during the periconceptional period on mammalian preimplantation embryo development. Domest. Anim. Endocrinol. 2015, 51, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Budanov, A.V.; Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef]
- Chen, J.; Ou, Y.; Luo, R.; Wang, J.; Wang, D.; Guan, J.; Li, Y.; Xia, P.; Chen, P.R.; Liu, Y. SAR1B senses leucine levels to regulate mTORC1 signalling. Nature 2021, 596, 281–284. [Google Scholar] [CrossRef]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef]
- Wang, S.; Tsun, Z.-Y.; Wolfson, R.L.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; de Araujo, M.E.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef]
- Sundberg, B.E.; Wååg, E.; Jacobsson, J.A.; Stephansson, O.; Rumaks, J.; Svirskis, S.; Alsiö, J.; Roman, E.; Ebendal, T.; Klusa, V.; et al. The evolutionary history and tissue mapping of amino acid transporters belonging to solute carrier families SLC32, SLC36, and SLC38. J. Mol. Neurosci. 2008, 35, 179–193. [Google Scholar] [CrossRef]
- Mackenzie, B.; Erickson, J.D. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflug. Arch. 2004, 447, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Hosseini, K.; Arapi, V.; Fredriksson, R.; Bagchi, S. SLC38A10 (SNAT10) is Located in ER and Golgi Compartments and Has a Role in Regulating Nascent Protein Synthesis. Int. J. Mol. Sci. 2019, 20, 6265. [Google Scholar] [CrossRef] [PubMed]
- Chapel, A.; Kieffer-Jaquinod, S.; Sagné, C.; Verdon, Q.; Ivaldi, C.; Mellal, M.; Thirion, J.; Jadot, M.; Bruley, C.; Garin, J.; et al. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell Proteom. 2013, 12, 1572–1588. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Sabatini, D.M. Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc. Natl. Acad. Sci. USA 2018, 115, 9545–9550. [Google Scholar] [CrossRef] [PubMed]
- Castellano, B.M.; Thelen, A.M.; Moldavski, O.; Feltes, M.; Van Der Welle, R.E.; Mydock-McGrane, L.; Jiang, X.; Van Eijkeren, R.J.; Davis, O.B.; Louie, S.M. Lysosomal cholesterol activates mTORC1 via an SLC38A9–Niemann-Pick C1 signaling complex. Science 2017, 355, 1306–1311. [Google Scholar] [CrossRef]
- Januchowski, R.; Zawierucha, P.; Andrzejewska, M.; Ruciński, M.; Zabel, M. Microarray-based detection and expression analysis of ABC and SLC transporters in drug-resistant ovarian cancer cell lines. Biomed. Pharm. 2013, 67, 240–245. [Google Scholar] [CrossRef]
- Jia, J.; Abudu, Y.P.; Claude-Taupin, A.; Gu, Y.; Kumar, S.; Choi, S.W.; Peters, R.; Mudd, M.H.; Allers, L.; Salemi, M.; et al. Galectins Control mTOR in Response to Endomembrane Damage. Mol. Cell 2018, 70, 120.e8–135.e8. [Google Scholar] [CrossRef]
- Lei, H.T.; Mu, X.; Hattne, J.; Gonen, T. A conformational change in the N terminus of SLC38A9 signals mTORC1 activation. Structure 2021, 29, 426.e8–432.e8. [Google Scholar] [CrossRef]
- Garcia-Alvarez, M.; Marik, P.; Bellomo, R. Stress hyperlactataemia: Present understanding and controversy. Lancet Diabetes Endocrinol 2014, 2, 339–347. [Google Scholar] [CrossRef]
- Lei, H.-T.; Ma, J.; Martinez, S.S.; Gonen, T. Crystal structure of arginine-bound lysosomal transporter SLC38A9 in the cytosol-open state. Nat. Struct. Mol. Biol. 2018, 25, 522–527. [Google Scholar] [CrossRef]
- Wyant, G.A.; Abu-Remaileh, M.; Wolfson, R.L.; Chen, W.W.; Freinkman, E.; Danai, L.V.; Vander Heiden, M.G.; Sabatini, D.M. mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell 2017, 171, 642.e12–654.e12. [Google Scholar] [CrossRef] [PubMed]
- Sekine, Y.; Nishibori, Y.; Akimoto, Y.; Kudo, A.; Ito, N.; Fukuhara, D.; Kurayama, R.; Higashihara, E.; Babu, E.; Kanai, Y.; et al. Amino acid transporter LAT3 is required for podocyte development and function. J. Am. Soc. Nephrol. 2009, 20, 1586–1596. [Google Scholar] [CrossRef] [PubMed]
- Narumanchi, S.; Wang, H.; Perttunen, S.; Tikkanen, I.; Lakkisto, P.; Paavola, J. Zebrafish Heart Failure Models. Front Cell Dev. Biol. 2021, 9, 662583. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Wu, S.Q.; Guo, S.Y.; Yang, H.; Xia, B.; Li, P.; Li, C.Q. A Zebrafish Heart Failure Model for Assessing Therapeutic Agents. Zebrafish 2018, 15, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Cory, S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018, 25, 27–36. [Google Scholar] [CrossRef]
- He, K.; Zheng, X.; Li, M.; Zhang, L.; Yu, J. mTOR inhibitors induce apoptosis in colon cancer cells via CHOP-dependent DR5 induction on 4E-BP1 dephosphorylation. Oncogene 2016, 35, 148–157. [Google Scholar] [CrossRef]
- Gangloff, Y.G.; Mueller, M.; Dann, S.G.; Svoboda, P.; Sticker, M.; Spetz, J.F.; Um, S.H.; Brown, E.J.; Cereghini, S.; Thomas, G.; et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell Biol. 2004, 24, 9508–9516. [Google Scholar] [CrossRef]
- Yilmaz, O.; Patinote, A.; Com, E.; Pineau, C.; Bobe, J. Knock out of specific maternal vitellogenins in zebrafish (Danio rerio) evokes vital changes in egg proteomic profiles that resemble the phenotype of poor quality eggs. BMC Genom. 2021, 22, 308. [Google Scholar] [CrossRef]
- Moss, J.J.; Wirth, M.; Tooze, S.A.; Lane, J.D.; Hammond, C.L. Autophagy coordinates chondrocyte development and early joint formation in zebrafish. FASEB J. 2021, 35, e22002. [Google Scholar] [CrossRef]
- Pei, W.; Williams, P.H.; Clark, M.D.; Stemple, D.L.; Feldman, B. Environmental and genetic modifiers of squint penetrance during zebrafish embryogenesis. Dev. Biol. 2007, 308, 368–378. [Google Scholar] [CrossRef]
- Zanon, A.; Pramstaller, P.P.; Hicks, A.A.; Pichler, I. Environmental and Genetic Variables Influencing Mitochondrial Health and Parkinson’s Disease Penetrance. Parkinsons Dis. 2018, 2018, 8684906. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xin, Y.; Bai, Y.; Lewin, G.; He, G.; Mai, K.; Duan, C. Ca(2+) concentration-dependent premature death of igfbp5a(−/−) fish reveals a critical role of IGF signaling in adaptive epithelial growth. Sci. Signal 2018, 11, eaat2231. [Google Scholar] [CrossRef] [PubMed]
- Sofer, A.; Lei, K.; Johannessen, C.M.; Ellisen, L.W. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell Biol. 2005, 25, 5834–5845. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hu, L.; Wu, F.; Zou, L.; He, T. Poor prognosis of hexokinase 2 overexpression in solid tumors of digestive system: A meta-analysis. Oncotarget 2017, 8, 32332–32344. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.M.; Zhang, Y.M. Targeting FBPase is an emerging novel approach for cancer therapy. Cancer Cell Int. 2018, 18, 36. [Google Scholar] [CrossRef]
- Chou, H.Y.; Lin, Y.H.; Shiu, G.L.; Tang, H.Y.; Cheng, M.L.; Shiao, M.S.; Pai, L.M. ADI1, a methionine salvage pathway enzyme, is required for Drosophila fecundity. J. Biomed. Sci. 2014, 21, 64. [Google Scholar] [CrossRef]
- Bousquet, F.; Chauvel, I.; Flaven-Pouchon, J.; Farine, J.P.; Ferveur, J.F. Dietary rescue of altered metabolism gene reveals unexpected Drosophila mating cues. J. Lipid. Res. 2016, 57, 443–450. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Liu, C.; Dai, W.; Bai, Y.; Chi, C.; Xin, Y.; He, G.; Mai, K.; Duan, C. Development of a Whole Organism Platform for Phenotype-Based Analysis of IGF1R-PI3K-Akt-Tor Action. Sci. Rep. 2017, 7, 1994. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Chen, J.; Liu, C.; Wang, X.; Zhou, H.; Mai, K.; He, G. Slc38a9 Deficiency Induces Apoptosis and Metabolic Dysregulation and Leads to Premature Death in Zebrafish. Int. J. Mol. Sci. 2022, 23, 4200. https://doi.org/10.3390/ijms23084200
Wu X, Chen J, Liu C, Wang X, Zhou H, Mai K, He G. Slc38a9 Deficiency Induces Apoptosis and Metabolic Dysregulation and Leads to Premature Death in Zebrafish. International Journal of Molecular Sciences. 2022; 23(8):4200. https://doi.org/10.3390/ijms23084200
Chicago/Turabian StyleWu, Xiya, Jianyang Chen, Chengdong Liu, Xuan Wang, Huihui Zhou, Kangsen Mai, and Gen He. 2022. "Slc38a9 Deficiency Induces Apoptosis and Metabolic Dysregulation and Leads to Premature Death in Zebrafish" International Journal of Molecular Sciences 23, no. 8: 4200. https://doi.org/10.3390/ijms23084200
APA StyleWu, X., Chen, J., Liu, C., Wang, X., Zhou, H., Mai, K., & He, G. (2022). Slc38a9 Deficiency Induces Apoptosis and Metabolic Dysregulation and Leads to Premature Death in Zebrafish. International Journal of Molecular Sciences, 23(8), 4200. https://doi.org/10.3390/ijms23084200




