Identification of the Wheat (Triticum aestivum) IQD Gene Family and an Expression Analysis of Candidate Genes Associated with Seed Dormancy and Germination
Abstract
:1. Introduction
2. Results
2.1. Identification of IQD Gene Family Members in Wheat
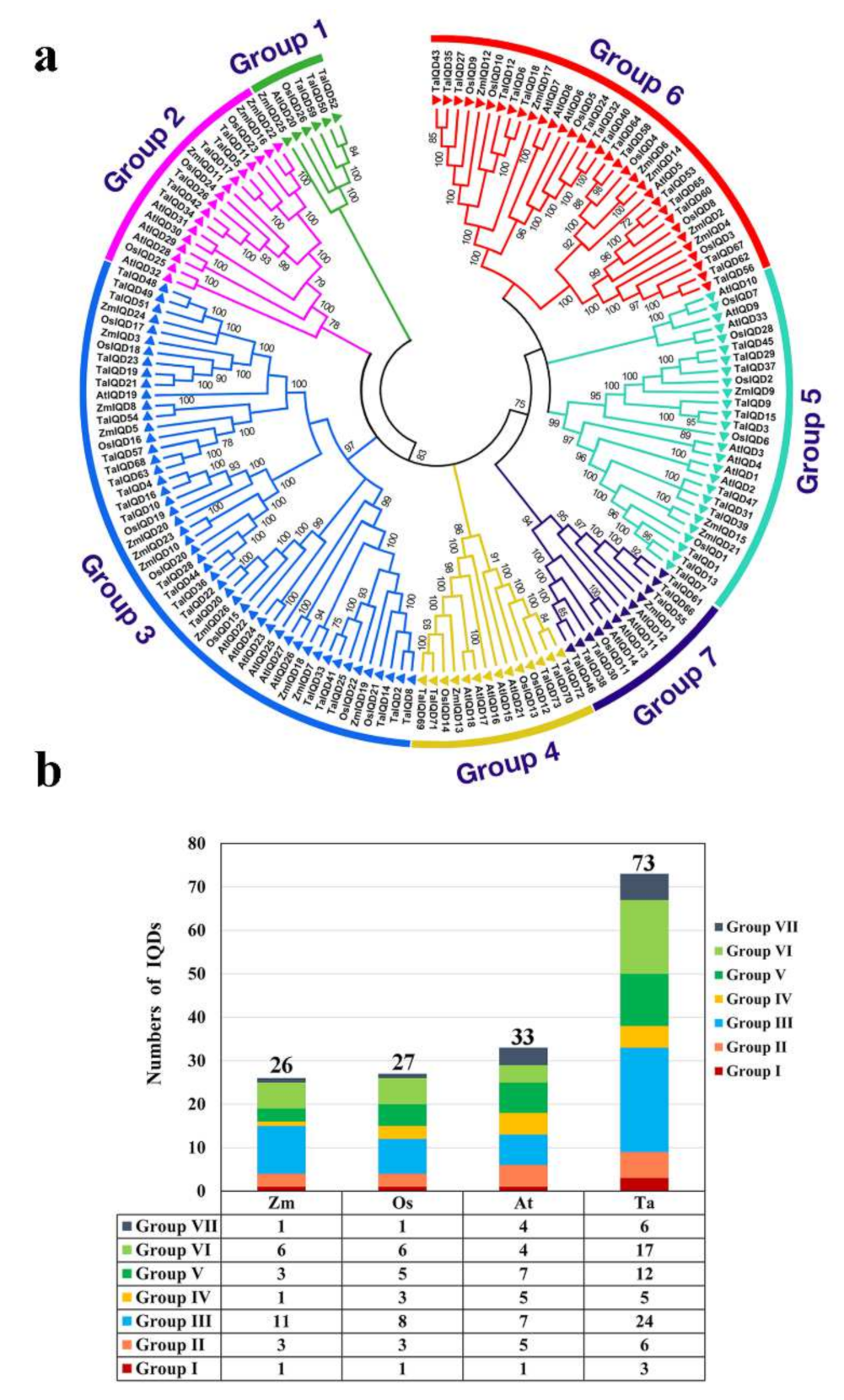
2.2. Phylogenetic Analysis and Conserved Sequence Alignment
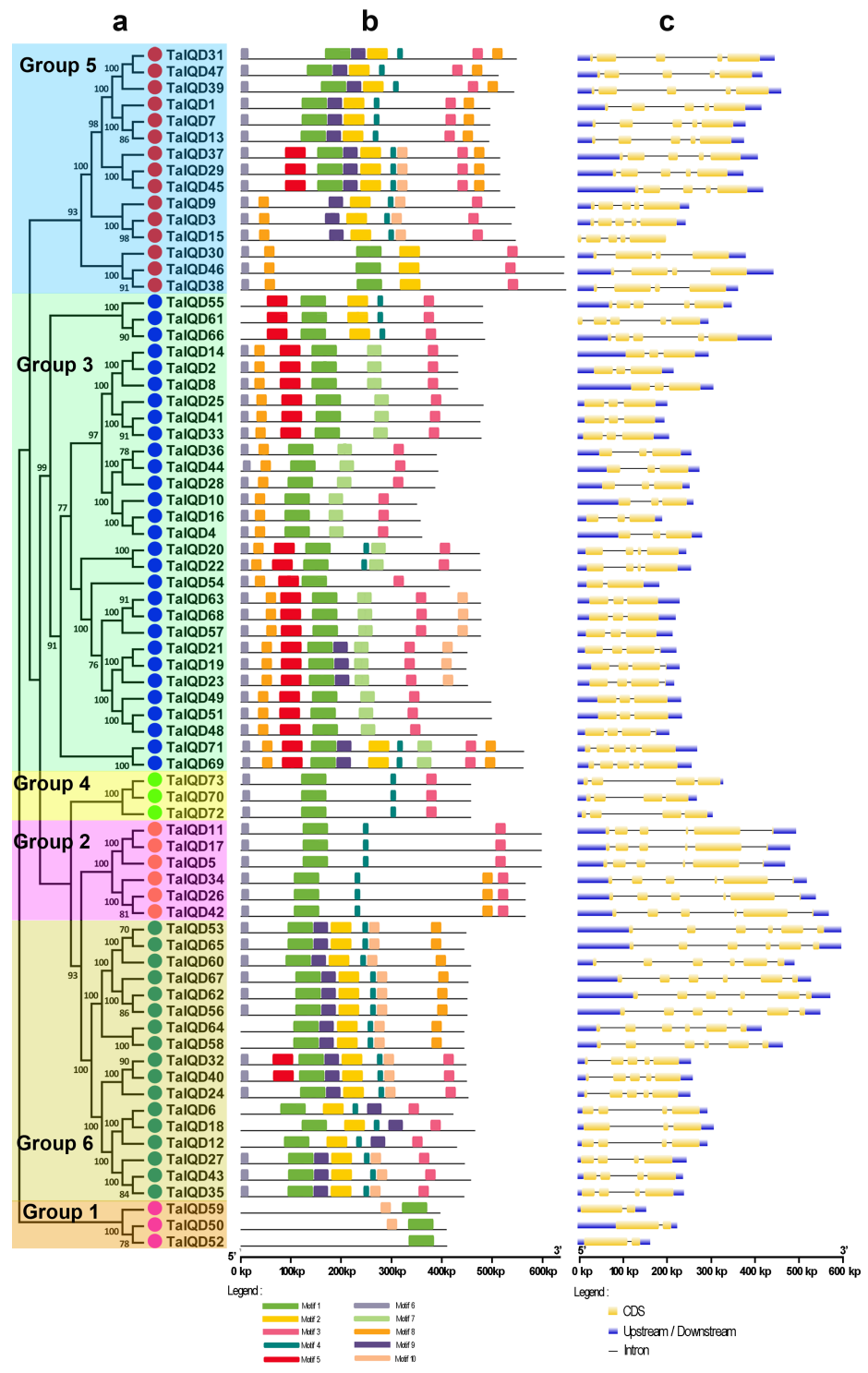
2.3. Gene Structure and Motif Composition Analysis
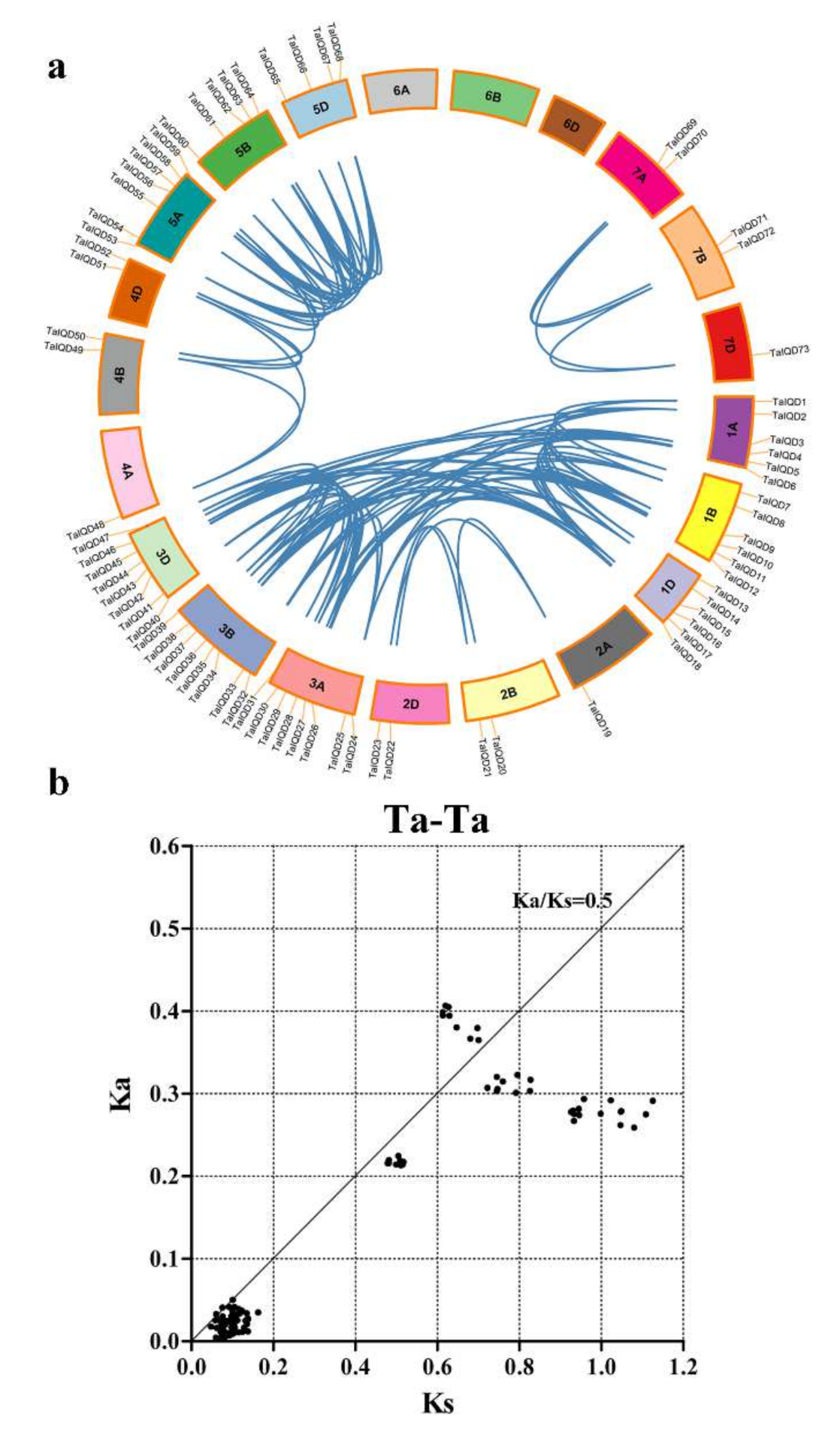
2.4. Analysis of the Chromosomal Locations and Duplications of TaIQD Genes
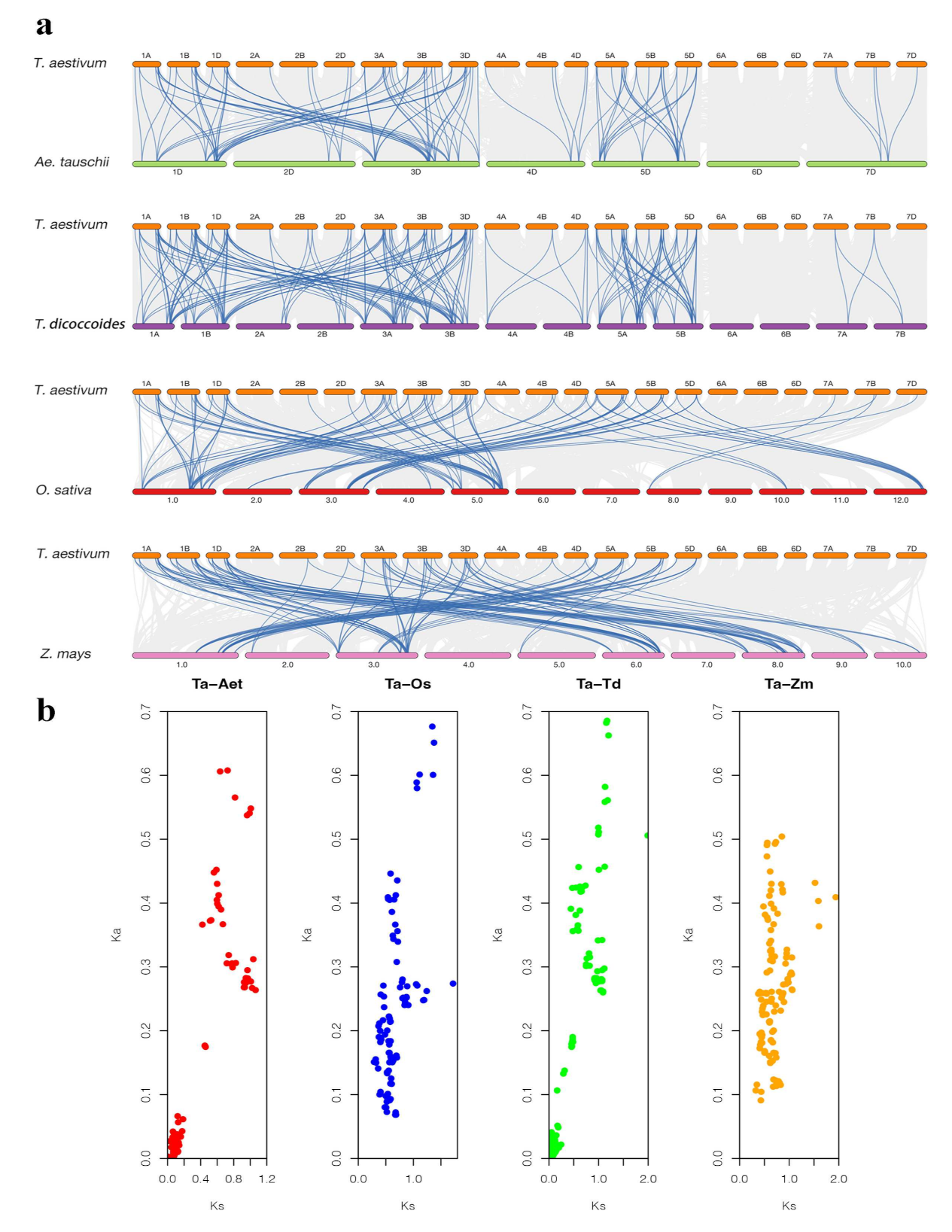
2.5. Micro-Collinearity Analysis
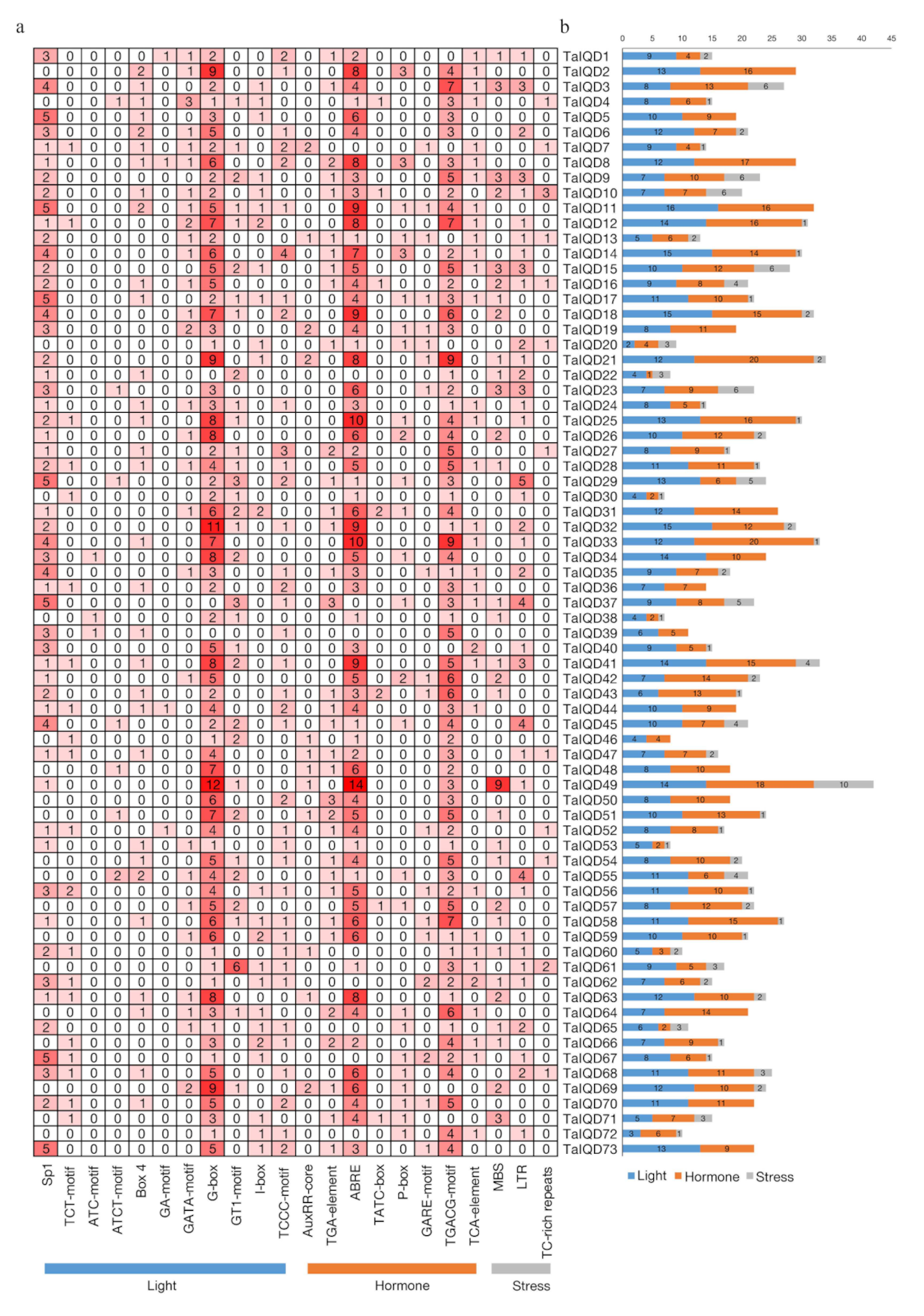
2.6. Analysis of Cis Elements in TaIQD Promoters
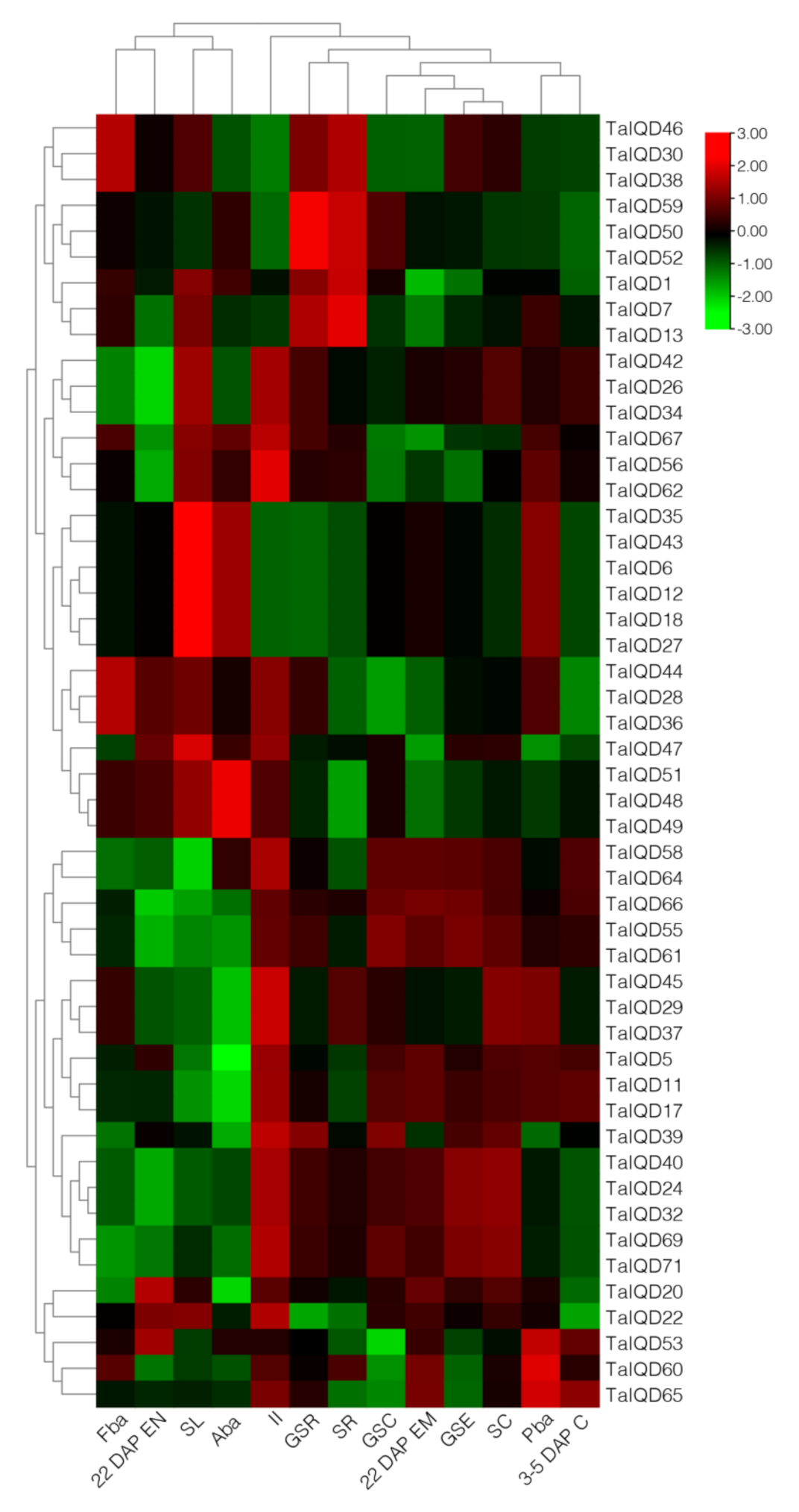
2.7. Expression Profiles of TaIQD Genes in Different Tissues
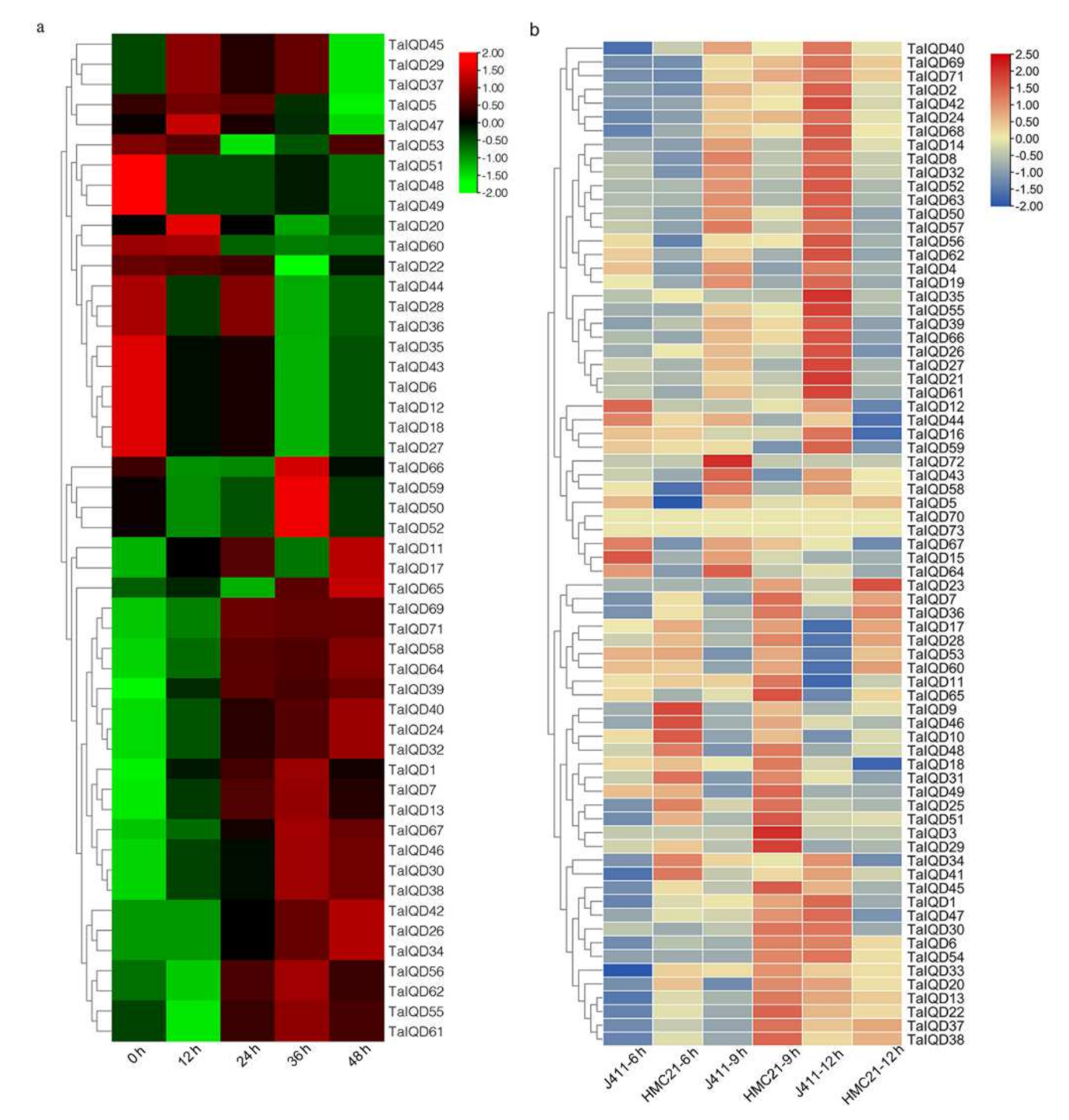
2.8. Expression Pattern Analysis of TaIQDs in the Transcriptome
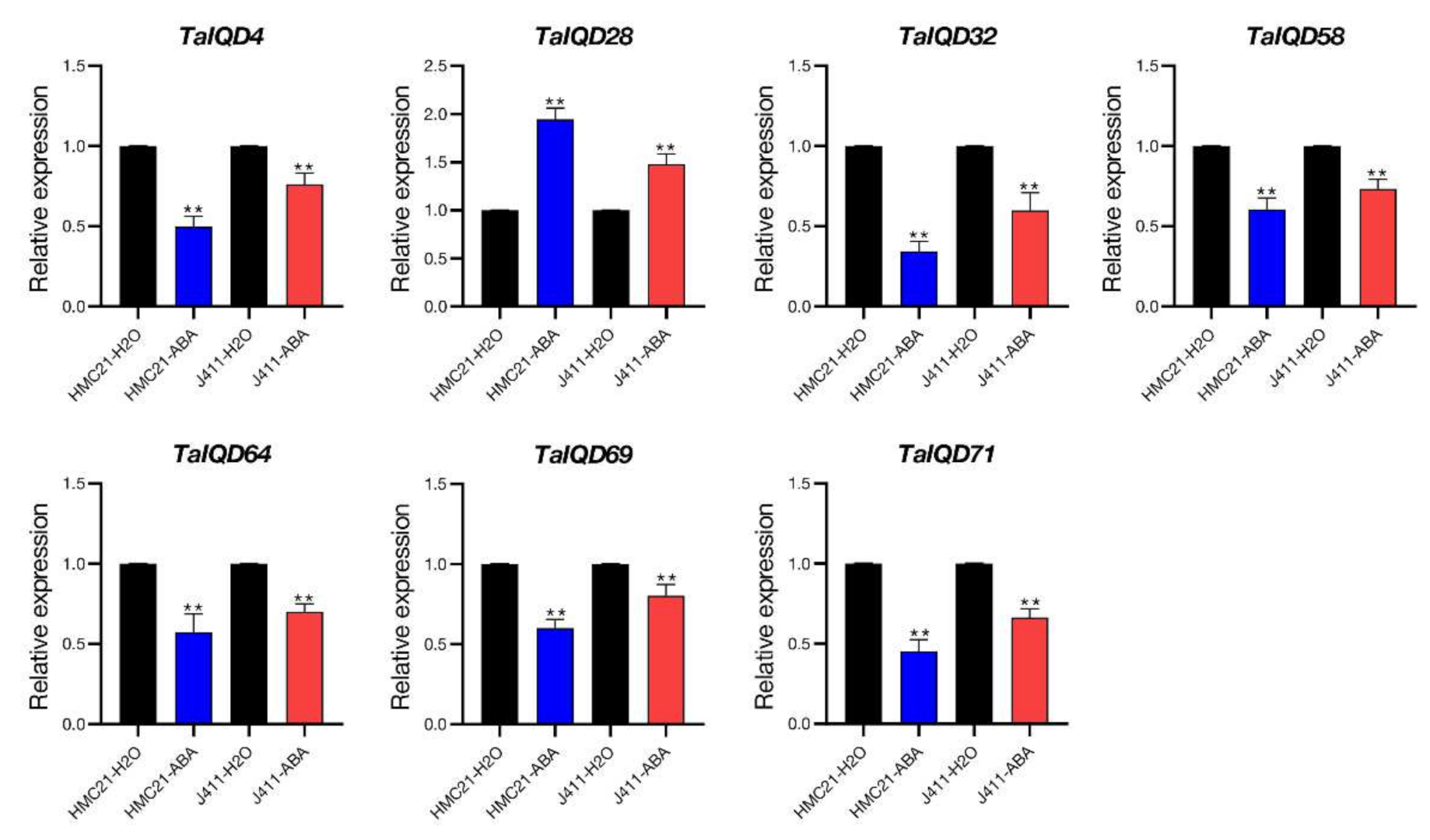
2.9. TaIQDs Expression Analysis by qRT-PCR
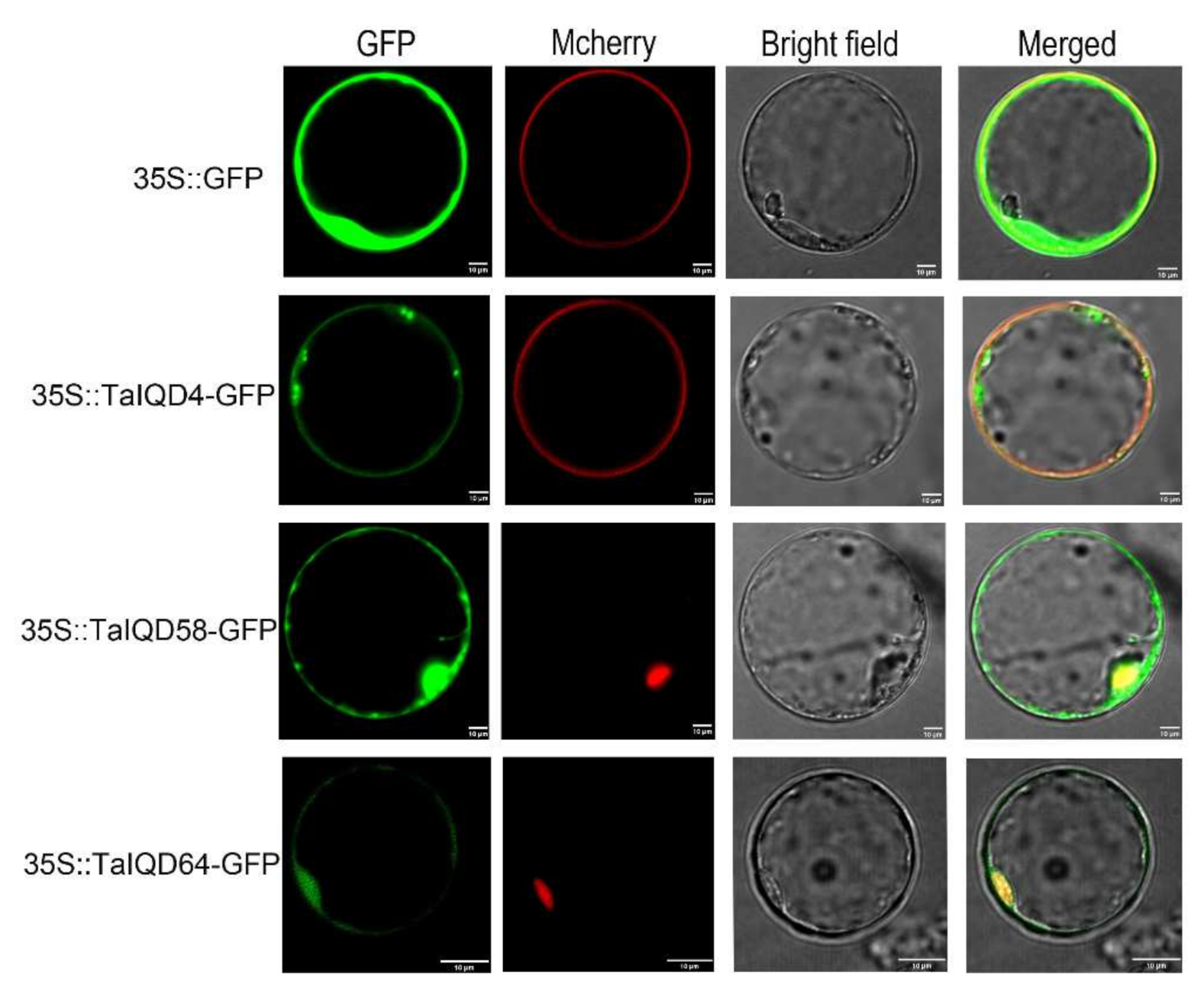
2.10. Subcellular Localization Analysis
3. Discussion
4. Materials and Methods
4.1. Identification of IQD Genes in Wheat
4.2. Multiple Sequence Alignments and Phylogenetic Analysis
4.3. Gene Structure, Conserved Motif, and Cis-Acting Element Analysis
4.4. Chromosomal Location and Gene Duplication
4.5. Expression Pattern Analysis
4.6. Plant Materials and Stress Treatments
4.7. RNA Isolation and qRT-PCR Analysis
4.8. Subcellular Localization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| qRT-PCR | quantitative real-time PCR |
| AI | aliphatic indices |
| MW | molecular weight |
| GRAVY | grand average of hydropathicity |
| PI | isoelectric point |
| bp | base pair |
| aa | amino acids |
| Da | Dalton |
| PHS | pre-harvest sprouting |
References
- Nonogaki, H.; Bassel, G.W.; Bewley, J.D. Germination—Still a mystery. Plant Sci. 2010, 179, 574–581. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular Aspects of Seed Dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Sehgal, S.K.; Li, J.; Lin, M.; Trick, H.N.; Yu, J.; Gill, B.S.; Bai, G. Cloning and Characterization of a Critical Regulator for Preharvest Sprouting in Wheat. Genetics 2013, 195, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, M.; Bewley, J.; Halmer, P. The Encyclopedia of Seeds: Science, Technology and Uses; CABI: Cambridge, UK, 2006. [Google Scholar]
- Wei, H.U. Characteristics of Pre-harvest Sprouting in Sterile Lines in Hybrid Rice Seeds Production. Acta Agron. Sin. 2003, 29, 441–446. [Google Scholar]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’amore, R.; Allen, A.M.; Mckenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Zhu, Y.; Ma, D.; Xu, W.; Yin, J. Screening, Identification, and Optimization of Fermentation Conditions of an Antagonistic Endophyte to Wheat Head Blight. Agronomy 2019, 9, 476. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Cai, S.; Graybosch, R.; Chen, C.; Bai, G. Quantitative trait loci for resistance to pre-harvest sprouting in US hard white winter wheat Rio Blanco. Theor. Appl. Genet. 2008, 117, 691–699. [Google Scholar] [CrossRef]
- Cheng, X.; Cao, J.; Gao, C.; Gao, W.; Yan, S.; Yao, H.; Xu, K.; Liu, X.; Xu, D.; Pan, X. Identification of the wheat C3H gene family and expression analysis of candidates associated with seed dormancy and germination. Plant Physiol. Biochem. 2020, 156, 524–537. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, X.L.; Xia, L.Q.; Chen, X.M.; Xia, X.C.; Yu, Z.; Röder, Z.H.H. Development and validation of a Viviparous-1 STS marker for pre-harvest sprouting tolerance in Chinese wheats. Theor. Appl. Genet. 2007, 115, 971–980. [Google Scholar] [CrossRef]
- Nakamura, S.; Abe, F.; Kawahigashi, H.; Kou, N.; Tagiri, A.; Matsumoto, T.; Utsugi, S.; Ogawa, T.; Handa, H.; Ishida, H. A Wheat Homolog of Mother of FT and TFL1 Acts in the Regulation of Germination. Plant Cell 2011, 23, 3215–3229. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Miao, X.; Xia, X.; He, Z. Cloning of seed dormancy genes (TaSdr) associated with tolerance to pre-harvest sprouting in common wheat and development of a functional marker. Theor. Appl. Genet. 2014, 127, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, X.; He, Z. The seed dormancy allele TaSdr-A1a associated with pre-harvest sprouting tolerance is mainly present in Chinese wheat landraces. Theor. Appl. Genet. 2017, 130, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Torada, A.; Koike, M.; Ogawa, T.; Takenouchi, Y.; Tadamura, K.; Wu, J.; Matsumoto, T.; Kawaura, K.; Ogihara, Y. A Causal Gene for Seed Dormancy on Wheat Chromosome 4A Encodes a MAP Kinase Kinase. Curr. Biol. 2016, 26, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Min, X.; Shan, S.; Jiang, H.; Chang, C. Isolation and characterization of TaQsd1 genes for period of dormancy in common wheat (Triticum aestivum L.). Mol. Breed. 2019, 39, 150. [Google Scholar] [CrossRef]
- Dodd, A.N.; Kudla, J.; Sanders, D. The Language of Calcium Signalling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with Stresses: Roles of Calcium- and Calcium/Calmodulin-Regulated Gene Expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [Green Version]
- White, P.J. Calcium channels in higher plants. Biochim. Biophys. Acta Biomembr. 2000, 1465, 171–189. [Google Scholar] [CrossRef] [Green Version]
- Nagamune, K.; Xiong, L.; Chini, E.; Sibley, L.D. Plants, endosymbionts and parasites: Abscisic acid and calcium signaling. Commun. Integr. Biol. 2008, 1, 62–65. [Google Scholar] [CrossRef]
- Asano, T.; Hayashi, N.; Kikuchi, S.; Ohsugi, R. CDPK-mediated abiotic stress signaling. Plant Signal. Behav. 2012, 7, 817–821. [Google Scholar] [CrossRef] [Green Version]
- Ranty, B.; Aldon, D.; Galaud, J.P. Plant calmodulins and calmodulin-related proteins: Multifaceted relays to decode calcium signals. Plant Signal. Behav. 2006, 1, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Nomura, S. Calcium Signaling in Plant Endosymbiotic Organelles: Mechanism and Role in Physiology. Mol. Plant 2014, 7, 1094–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snedden, W.A.; Fromm, H. Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 2010, 151, 35–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Day, D.H. CaMBOT: Profiling and characterizing calmodulin-binding proteins. Cell. Signal. 2003, 15, 347–354. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 2010, 425, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Abel, S.; Bürstenbinder, K.; Müller, J. The emerging function of IQD proteins as scaffolds in cellular signaling and trafficking. Plant Signal. Behav. 2013, 8, e24369. [Google Scholar] [CrossRef] [Green Version]
- Bürstenbinder, K.; Möller, B.; Plötner, R.; Stamm, G.; Hause, G.; Mitra, D.; Abel, S. The IQD Family of Calmodulin-Binding Proteins Links Calcium Signaling to Microtubules, Membrane Subdomains, and the Nucleus. Plant Physiol. 2017, 173, 1692–1708. [Google Scholar] [CrossRef] [Green Version]
- Bahler, M.; Rhoads, A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002, 513, 107–113. [Google Scholar] [CrossRef]
- Choi, J.Y.; Lee, S.H.; Park, C.Y.; Heo, W.D.; Kim, J.C.; Kim, M.C.; Chung, W.S.; Moon, B.C.; Cheong, Y.H.; Kim, C.Y.; et al. Identification of calmodulin isoform-specific binding peptides from a phage-displayed random 22-mer peptide library. J. Biol. Chem. 2002, 277, 21630–21638. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.H.; Zhang, C.S.; Zhao, Y.; Zhu, K.J.; Wang, Y.F.; Jiang, H.Y.; Xiang, Y.; Cheng, B.J. Genome-wide analysis of the IQD gene family in maize. Mol. Genet. Genom. 2016, 291, 543–558. [Google Scholar] [CrossRef]
- Abel, S.; Savchenko, T.; Levy, M. Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol. Biol. 2005, 5, 72. [Google Scholar] [CrossRef] [Green Version]
- Filiz, E.; Tombuloglu, H.; Ozyigit, I.I. Genome-wide analysis of IQ67 domain (IQD) gene families in Brachypodium distachyon. Plant Omics 2013, 6, 425–432. [Google Scholar]
- Wu, M.; Li, Y.; Chen, D.M.; Liu, H.L.; Zhu, D.Y.; Xiang, Y. Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2016, 6, 24520. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.P.; Liu, T.K.; Yu, Z.H.; Li, Y.; Ren, H.B.; Hou, X.L.; Li, Y. Genome-wide analysis of the Chinese cabbage IQD gene family and the response of BrIQD5 in drought resistance. Plant Mol. Biol. 2019, 99, 603–620. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Haider, M.S.; Khan, N.; Fang, J.G. Comprehensive Sequence Analysis of IQD Gene Family and their Expression Profiling in Grapevine (Vitis vinifera). Genes 2020, 11, 235. [Google Scholar] [CrossRef] [Green Version]
- Mei, C.; Liu, Y.W.; Dong, X.; Song, Q.N.; Wang, H.J.; Shi, H.W.; Feng, R.Y. Genome-Wide Identification and Characterization of the Potato IQD Family during Development and Stress. Front. Genet. 2021, 12, 693936. [Google Scholar] [CrossRef]
- Ma, H.; Feng, L.; Chen, Z.; Chen, X.; Zhao, H.L.; Xiang, Y. Genome-wide identification and expression analysis of the IQD gene family in Populus trichocarpa. Plant Sci. 2014, 229, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Zhang, Y.; Martinez, P.; Rasmussen, C.G.; Xu, T.D.; Yang, Z.B. The Microtubule-Associated Protein IQ67 DOMAIN5 Modulates Microtubule Dynamics and Pavement Cell Shape. Plant Physiol. 2018, 177, 1555–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef]
- Bi, L.L.; Weng, L.; Jiang, Z.Y.; Xiao, H. The tomato IQD gene SUN24 regulates seed germination through ABA signaling pathway. Planta 2018, 248, 919–931. [Google Scholar] [CrossRef]
- Song, S.W.; Hao, L.Y.; Zhao, P.; Xu, Y.; Zhong, N.Q.; Zhang, H.J.; Liu, N. Genome-wide Identification, Expression Profiling and Evolutionary Analysis of Auxin Response Factor Gene Family in Potato (Solanum tuberosum Group Phureja). Sci. Rep. 2019, 9, 1755. [Google Scholar] [CrossRef]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; Olsen, O.A.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Fatima, F.; Haider, M.S.; Shazadee, H.; Liu, Z.J.; Zheng, T.; Fang, J.G. Genome-Wide Identification and Expression Profiling of the Polygalacturonase (PG) and Pectin Methylesterase (PME) Genes in Grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 3180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, A.; Peng, Z.; Li, H.G.; Qin, G.Y.; Jia, Y.H.; Pan, Z.; He, S.P.; Qayyum, A.; Du, X.M. Genome wide analysis of IQD gene family in diploid and tetraploid species of cotton (Gossypium spp.). Int. J. Biol. Macromol. 2021, 184, 1035–1061. [Google Scholar] [CrossRef]
- Huang, B.; Huang, Z.; Ma, R.; Ramakrishnan, M.; Yrjl, K. Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis). BMC Plant Biol. 2021, 21, 296. [Google Scholar] [CrossRef] [PubMed]
- Walker-Simmons, M. ABA Levels and Sensitivity in Developing Wheat Embryos of Sprouting Resistant and Susceptible Cultivars. Plant Physiol. 1987, 84, 61–66. [Google Scholar] [CrossRef] [Green Version]
- He, H.Z.; Vidigal, D.D.; Snoek, L.B.; Schnabel, S.; Nijveen, H.; Hilhorst, H.; Bentsink, L. Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J. Exp. Bot. 2014, 65, 6603–6615. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Chen, F.Y.; Wang, Z.; Cao, H.; Li, X.Y.; Deng, X.; Soppe, W.J.J.; Li, Y.; Liu, Y.X. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012, 193, 605–616. [Google Scholar] [CrossRef]
- Zhang, M.T.; Cui, G.B.; Bai, X.C.; Ye, Z.; Zhang, S.M.; Xie, K.L.; Sun, F.L.; Zhang, C.; Xi, Y.J. Regulatory Network of Preharvest Sprouting Resistance Revealed by Integrative Analysis of mRNA, Noncoding RNA, and DNA Methylation in Wheat. J. Agric. Food Chem. 2021, 69, 4018–4035. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Liu, Y.; Ma, L.; Wang, X.; Ma, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Gao, Y.M.; Liu, H.L.; Zhang, K.M.; Li, F.; Wu, M.; Xiang, Y. A moso bamboo transcription factor, Phehdz1, positively regulates the drought stress response of transgenic rice. Plant Cell Rep. 2021, 40, 187–204. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, Q.Q.; Zhao, Y.; Han, G.M.; Zhu, S.W. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and Diversification of the Populus R2R3-MYB Family of Transcription Factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bryfczynski, S.; Pargas, R.P. GraphPad: A Graph Creation Tool for CS2/CS7. SIGCSE Bull. 2009, 41, 389. [Google Scholar] [CrossRef]

| Name | Gene ID | Location | Open Reading Frame Length (bp) | Size (Amino Acid) | Molecular Weight (Da) | Isoelectric Points | Exons | Instability Index | Aliphatic Indices | Grand Averages of Hydropathicity |
|---|---|---|---|---|---|---|---|---|---|---|
| TaIQD1 | TraesCS1A02G064100 | Chr1A:45793606-45797826 | 1443 | 480 | 52,361.54 | 10.15 | 5 | Unstable | 71.17 | −0.71 |
| TaIQD2 | TraesCS1A02G122900 | Chr1A:140960645-14096280 | 1257 | 418 | 44,891.95 | 10.46 | 3 | Unstable | 65.67 | −0.47 |
| TaIQD3 | TraesCS1A02G288100 | Chr1A:485337870-48534035 | 1566 | 521 | 56,328.25 | 10.55 | 5 | Unstable | 48.27 | −0.938 |
| TaIQD4 | TraesCS1A02G341900 | Chr1A:531281089-53128394 | 1050 | 349 | 37,937.17 | 10.66 | 3 | Unstable | 65.56 | −0.672 |
| TaIQD5 | TraesCS1A02G357500 | Chr1A:540047888-54005224 | 1740 | 579 | 64,209.32 | 9.75 | 6 | Unstable | 66.94 | −0.816 |
| TaIQD6 | TraesCS1A02G362400 | Chr1A:543060516-54306349 | 1230 | 409 | 44,715.09 | 10.71 | 4 | Unstable | 61.69 | −0.721 |
| TaIQD7 | TraesCS1B02G082100 | Chr1B:65547252-65551108 | 1443 | 480 | 52,439.53 | 10.07 | 5 | Unstable | 68.71 | −0.746 |
| TaIQD8 | TraesCS1B02G142100 | Chr1B:190629188-19063230 | 1257 | 418 | 44,925.92 | 10.46 | 3 | Unstable | 64.5 | −0.488 |
| TaIQD9 | TraesCS1B02G297500 | Chr1B:517901878-51790443 | 1587 | 528 | 56,591.61 | 10.63 | 5 | Unstable | 52.27 | −0.863 |
| TaIQD10 | TraesCS1B02G354600 | Chr1B:584324452-58432711 | 1020 | 339 | 37,096.08 | 10.95 | 3 | Unstable | 64.07 | −0.755 |
| TaIQD11 | TraesCS1B02G374100 | Chr1B:604548650-60455366 | 1740 | 579 | 63,871.93 | 9.56 | 6 | Unstable | 68 | −0.778 |
| TaIQD12 | TraesCS1B02G379600 | Chr1B:612863317-61286629 | 1251 | 416 | 45,415.84 | 10.66 | 4 | Unstable | 61.35 | −0.736 |
| TaIQD13 | TraesCS1D02G064700 | Chr1D:46181934-46185754 | 1437 | 478 | 52,189.3 | 10.07 | 5 | Unstable | 69.44 | −0.726 |
| TaIQD14 | TraesCS1D02G123800 | Chr1D:126471856-12647475 | 1257 | 418 | 44,775.75 | 10.46 | 3 | Unstable | 64.98 | −0.484 |
| TaIQD15 | TraesCS1D02G287100 | Chr1D:385615809-38561783 | 1590 | 529 | 56,884.84 | 10.54 | 5 | Unstable | 51.1 | −0.914 |
| TaIQD16 | TraesCS1D02G344200 | Chr1D:432807534-43280947 | 1041 | 346 | 37,654.85 | 10.88 | 3 | Unstable | 64.45 | −0.703 |
| TaIQD17 | TraesCS1D02G361800 | Chr1D:444361818-44436669 | 1740 | 579 | 64,058.13 | 9.69 | 6 | Unstable | 67.13 | −0.805 |
| TaIQD18 | TraesCS1D02G367300 | Chr1D:447254643-44725776 | 1356 | 451 | 49,366.40 | 10.42 | 3 | Unstable | 65.23 | −0.623 |
| TaIQD19 | TraesCS2A02G470100 | Chr2A:712980746-71298308 | 1305 | 434 | 47,480.54 | 10.08 | 4 | Unstable | 61.59 | −0.658 |
| TaIQD20 | TraesCS2B02G418900 | Chr2B:600440674-60044316 | 1383 | 460 | 49,548.51 | 10.55 | 4 | Unstable | 51.78 | −0.78 |
| TaIQD21 | TraesCS2B02G492900 | Chr2B:690907263-69090953 | 1311 | 436 | 47,526.50 | 10.20 | 4 | Unstable | 58.85 | −0.663 |
| TaIQD22 | TraesCS2D02G398300 | Chr2D:511437009-51143961 | 1389 | 462 | 49,645.65 | 10.56 | 4 | Unstable | 53.7 | −0.747 |
| TaIQD23 | TraesCS2D02G470000 | Chr2D:574631280-57463349 | 1314 | 437 | 47,638.78 | 10.22 | 4 | Unstable | 60.07 | −0.655 |
| TaIQD24 | TraesCS3A02G105600 | Chr3A:69504313-69506901 | 1317 | 438 | 47,911.68 | 10.56 | 5 | Unstable | 70.75 | −0.609 |
| TaIQD25 | TraesCS3A02G108100 | Chr3A:73512378-73514437 | 1404 | 467 | 49,931.39 | 10.16 | 3 | Unstable | 64.84 | −0.518 |
| TaIQD26 | TraesCS3A02G244100 | Chr3A:457460875-45746634 | 1647 | 548 | 60,598.78 | 9.78 | 6 | Unstable | 75.88 | −0.694 |
| TaIQD27 | TraesCS3A02G251400 | Chr3A:471278391-47128089 | 1296 | 431 | 47,074.55 | 10.71 | 4 | Unstable | 62.09 | −0.741 |
| TaIQD28 | TraesCS3A02G275900 | Chr3A:505523600-50552617 | 1125 | 374 | 41,766.59 | 10.81 | 3 | Unstable | 61.5 | −0.794 |
| TaIQD29 | TraesCS3A02G352500 | Chr3A:600339974-60034378 | 1500 | 499 | 55,768.28 | 10.27 | 5 | Unstable | 61.86 | −0.833 |
| TaIQD30 | TraesCS3A02G398900 | Chr3A:645637395-64564125 | 1872 | 623 | 69,213.26 | 11.35 | 4 | Unstable | 50.14 | −0.906 |
| TaIQD31 | TraesCS3A02G538100 | Chr3A:749341062-74934558 | 1596 | 531 | 56,655.68 | 10.23 | 5 | Unstable | 64.29 | −0.645 |
| TaIQD32 | TraesCS3B02G124000 | Chr3B:97076190-97078793 | 1305 | 434 | 47,645.28 | 10.56 | 5 | Unstable | 70.05 | −0.65 |
| TaIQD33 | TraesCS3B02G127100 | Chr3B:104970381-10497248 | 1392 | 463 | 49,945.46 | 10.17 | 3 | Unstable | 64.73 | −0.541 |
| TaIQD34 | TraesCS3B02G276000 | Chr3B:445602528-44560775 | 1647 | 548 | 60,603.81 | 9.85 | 6 | Unstable | 78.18 | −0.692 |
| TaIQD35 | TraesCS3B02G280900 | Chr3B:451530157-45153259 | 1293 | 430 | 46,916.43 | 10.65 | 4 | Unstable | 63.37 | −0.718 |
| TaIQD36 | TraesCS3B02G309600 | Chr3B:498325074-49832768 | 1134 | 377 | 42,099.99 | 11.00 | 3 | Unstable | 63.58 | −0.78 |
| TaIQD37 | TraesCS3B02G385100 | Chr3B:605267059-60527119 | 1500 | 499 | 55,811.31 | 10.27 | 5 | Unstable | 61.86 | −0.832 |
| TaIQD38 | TraesCS3B02G431200 | Chr3B:670023177-67002686 | 1881 | 626 | 69,491.65 | 11.41 | 4 | Unstable | 49.44 | −0.907 |
| TaIQD39 | TraesCS3B02G603500 | Chr3B:822903185-82290786 | 1581 | 526 | 56,241.10 | 10.31 | 5 | Unstable | 64.35 | −0.671 |
| TaIQD40 | TraesCS3D02G107700 | Chr3D:61311367-61314011 | 1308 | 435 | 47,677.38 | 10.56 | 5 | Unstable | 69.89 | −0.632 |
| TaIQD41 | TraesCS3D02G109900 | Chr3D:63778901-63780895 | 1386 | 461 | 49,561.05 | 10.24 | 3 | Unstable | 65.86 | −0.508 |
| TaIQD42 | TraesCS3D02G247300 | Chr3D:346354612-34636037 | 1647 | 548 | 60,756.97 | 9.84 | 6 | Unstable | 75.33 | −0.734 |
| TaIQD43 | TraesCS3D02G251800 | Chr3D:352612821-35261524 | 1332 | 443 | 48,523.32 | 10.68 | 4 | Unstable | 63.27 | −0.728 |
| TaIQD44 | TraesCS3D02G275900 | Chr3D:382629319-38263211 | 1143 | 380 | 42,344.28 | 10.94 | 3 | Unstable | 63.87 | −0.766 |
| TaIQD45 | TraesCS3D02G346500 | Chr3D:457554464-45755872 | 1500 | 499 | 55,768.28 | 10.27 | 5 | Unstable | 62.06 | −0.829 |
| TaIQD46 | TraesCS3D02G392900 | Chr3D:508079513-50808400 | 1869 | 622 | 69,151.24 | 11.35 | 4 | Unstable | 50.39 | −0.899 |
| TaIQD47 | TraesCS3D02G543500 | Chr3D:613703281-61370752 | 1491 | 496 | 53,101.63 | 10.14 | 5 | Unstable | 61.96 | −0.702 |
| TaIQD48 | TraesCS4A02G028700 | Chr4A:21051778-21053883 | 1368 | 455 | 48,711.53 | 11.15 | 4 | Unstable | 58.31 | −0.647 |
| TaIQD49 | TraesCS4B02G277100 | Chr4B:558725978-55872825 | 1449 | 482 | 52,255.78 | 11.16 | 3 | Unstable | 63.94 | −0.547 |
| TaIQD50 | TraesCS4B02G330700 | Chr4B:621671148-62167343 | 1191 | 396 | 40,618.63 | 11.33 | 2 | Unstable | 72.5 | −0.2 |
| TaIQD51 | TraesCS4D02G275700 | Chr4D:446651522-44665391 | 1452 | 483 | 52,450.89 | 11.24 | 3 | Unstable | 62.57 | −0.606 |
| TaIQD52 | TraesCS4D02G327500 | Chr4D:486907798-48690946 | 1194 | 397 | 40,630.77 | 11.47 | 2 | Unstable | 70.88 | −0.206 |
| TaIQD53 | TraesCS5A02G029600 | Chr5A:25698699-25704674 | 1305 | 434 | 46,543.00 | 10.25 | 6 | Unstable | 60.88 | −0.961 |
| TaIQD54 | TraesCS5A02G055100 | Chr5A:51736382-51738258 | 1209 | 402 | 42,506.48 | 10.86 | 2 | Unstable | 58.81 | −0.65 |
| TaIQD55 | TraesCS5A02G163000 | Chr5A:348717683-34872122 | 1401 | 466 | 51,850.83 | 10.25 | 5 | Unstable | 65.9 | −0.7 |
| TaIQD56 | TraesCS5A02G373000 | Chr5A:571149994-57115556 | 1311 | 436 | 47,752.33 | 9.69 | 6 | Unstable | 67.43 | −0.869 |
| TaIQD57 | TraesCS5A02G377500 | Chr5A:574931819-57493400 | 1389 | 462 | 49,373.67 | 10.33 | 3 | Unstable | 62.97 | −0.513 |
| TaIQD58 | TraesCS5A02G425800 | Chr5A:611132861-61113757 | 1293 | 430 | 47,774.71 | 10.00 | 6 | Unstable | 68.81 | −0.805 |
| TaIQD59 | TraesCS5A02G502100 | Chr5A:667123853-66712542 | 1155 | 384 | 39,565.30 | 11.39 | 2 | Unstable | 71.69 | −0.267 |
| TaIQD60 | TraesCS5B02G028200 | Chr5B:27294672-27299651 | 1332 | 443 | 47,698.23 | 10.02 | 6 | Unstable | 60.47 | −0.953 |
| TaIQD61 | TraesCS5B02G160400 | Chr5B:295380499-29538350 | 1401 | 466 | 50,900.00 | 10.72 | 5 | Unstable | 64.87 | −0.649 |
| TaIQD62 | TraesCS5B02G375000 | Chr5B:552482573-55248837 | 1311 | 436 | 48,004.64 | 9.64 | 6 | Unstable | 68.33 | −0.855 |
| TaIQD63 | TraesCS5B02G381100 | Chr5B:558870075-55887241 | 1389 | 462 | 49,424.76 | 10.44 | 3 | Unstable | 62.34 | −0.525 |
| TaIQD64 | TraesCS5B02G427800 | Chr5B:603883827-60388805 | 1293 | 430 | 47,671.63 | 9.89 | 6 | Unstable | 70.44 | −0.817 |
| TaIQD65 | TraesCS5D02G037300 | Chr5D:36476089-36482142 | 1293 | 430 | 46,336.84 | 10.17 | 6 | Unstable | 62.56 | −0.955 |
| TaIQD66 | TraesCS5D02G168000 | Chr5D:262838385-26284284 | 1413 | 470 | 52,167.06 | 10.16 | 5 | Unstable | 65.74 | −0.704 |
| TaIQD67 | TraesCS5D02G382500 | Chr5D:452235938-45224129 | 1317 | 438 | 48,184.85 | 9.72 | 6 | Unstable | 65.8 | −0.896 |
| TaIQD68 | TraesCS5D02G387500 | Chr5D:457006913-45700916 | 1392 | 463 | 49,554.10 | 10.47 | 3 | Unstable | 63.05 | −0.506 |
| TaIQD69 | TraesCS7A02G317400 | Chr7A:457568641-45757125 | 1635 | 544 | 59,224.93 | 10.73 | 5 | Unstable | 54.56 | −0.787 |
| TaIQD70 | TraesCS7A02G332500 | Chr7A:485743582-48574631 | 1332 | 443 | 48,287.92 | 10.41 | 4 | Unstable | 58.8 | −0.729 |
| TaIQD71 | TraesCS7B02G218800 | Chr7B:408567032-40856977 | 1638 | 545 | 59,385.10 | 10.78 | 5 | Unstable | 54.46 | −0.799 |
| TaIQD72 | TraesCS7B02G244900 | Chr7B:453334625-45333772 | 1332 | 443 | 48,516.26 | 10.4 | 4 | Unstable | 59.23 | −0.757 |
| TaIQD73 | TraesCS7D02G341000 | Chr7D:436877993-43688133 | 1332 | 443 | 48,511.23 | 10.55 | 4 | Unstable | 58.13 | −0.774 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Wang, Z.; Wang, C.; Pan, X.; Gao, W.; Yan, S.; Cao, J.; Lu, J.; Chang, C.; Ma, C.; et al. Identification of the Wheat (Triticum aestivum) IQD Gene Family and an Expression Analysis of Candidate Genes Associated with Seed Dormancy and Germination. Int. J. Mol. Sci. 2022, 23, 4093. https://doi.org/10.3390/ijms23084093
Liu M, Wang Z, Wang C, Pan X, Gao W, Yan S, Cao J, Lu J, Chang C, Ma C, et al. Identification of the Wheat (Triticum aestivum) IQD Gene Family and an Expression Analysis of Candidate Genes Associated with Seed Dormancy and Germination. International Journal of Molecular Sciences. 2022; 23(8):4093. https://doi.org/10.3390/ijms23084093
Chicago/Turabian StyleLiu, Mingli, Zhuofan Wang, Chenchen Wang, Xu Pan, Wei Gao, Shengnan Yan, Jiajia Cao, Jie Lu, Cheng Chang, Chuanxi Ma, and et al. 2022. "Identification of the Wheat (Triticum aestivum) IQD Gene Family and an Expression Analysis of Candidate Genes Associated with Seed Dormancy and Germination" International Journal of Molecular Sciences 23, no. 8: 4093. https://doi.org/10.3390/ijms23084093
APA StyleLiu, M., Wang, Z., Wang, C., Pan, X., Gao, W., Yan, S., Cao, J., Lu, J., Chang, C., Ma, C., & Zhang, H. (2022). Identification of the Wheat (Triticum aestivum) IQD Gene Family and an Expression Analysis of Candidate Genes Associated with Seed Dormancy and Germination. International Journal of Molecular Sciences, 23(8), 4093. https://doi.org/10.3390/ijms23084093






