Comparative Proteomic Profiling of Secreted Extracellular Vesicles from Breast Fibroadenoma and Malignant Lesions: A Pilot Study
Abstract
:1. Introduction
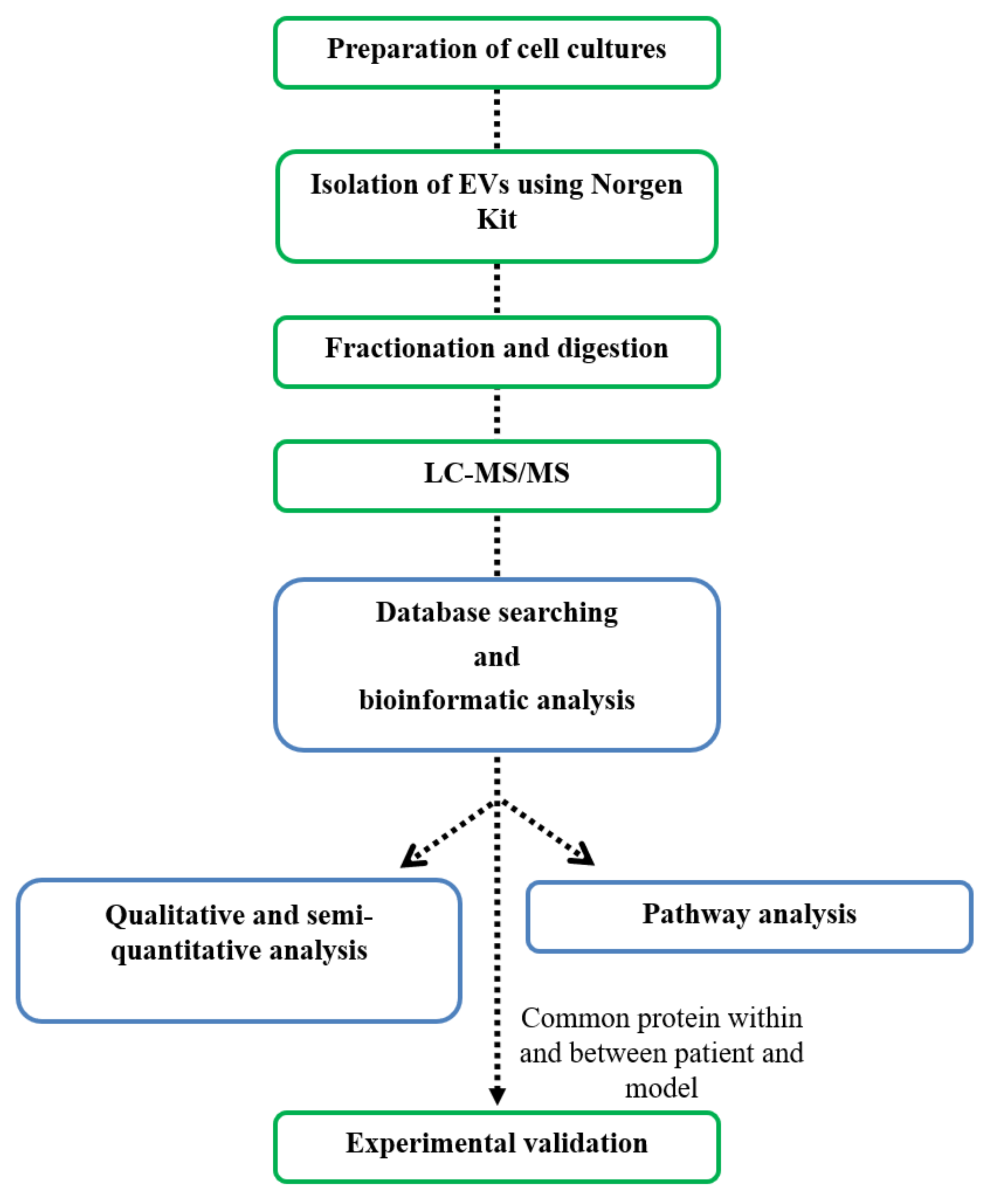
2. Materials and Methods
2.1. Cell Lines and Primary Cultures
2.2. Purification of Extracellular Vesicles
2.3. Dynamic Light Scattering (DLS) Measurements
2.4. Scanning Electron Microscopy (SEM)
2.5. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
2.6. Database Searches and Bioinformatics Analyses
2.7. Western Blot Analysis
3. Results
3.1. Study Design and Sample Description
3.2. Characterization of EVs from Primary and IBCC Lines
| Patient | Sample #37 | Sample #46 | Sample #72 | Sample #44 | Sample #170 | Sample #148 |
|---|---|---|---|---|---|---|
| Median age at diagnosis (years) | 49.5 | 43 | 53.5 | |||
| Age at diagnosis (years) | 66 | 33 | 44 | 42 | 56 | 51 |
| Histological type | Ductal infiltrating carcinoma (NOS) | Ductal infiltrating carcinoma high grade | Fibroadenoma | Fibroadenoma with adenosi | Ductal infiltrating carcinoma (NOS) CK19 (+++) | Lobular infiltrating carcinoma, poorly differentiated with 10% of lobular neoplasia in situ with high grade, E-cadherin negative |
| Tumor stage | pT1c | pT2 | _ | _ | pT2 | pT2 |
| Grade | pG3 | pG3 | _ | _ | pG3 | pG3 |
| Lymph node | pN1a | pN1a | _ | _ | 0 | 0 |
| ER/PR/HER2 status (positivity) | ER − (0)/PR + (<5)/HER2 + (2+) | ER +(10)/PR + a/HER2 + (3+) | _ | _ | ER − (0)/PR − (0)/HER2 − (0) | ER − (0)/PR − (<5)/HER2 − (0) |
| ki 67 status (positivity) | High (30) | High (50) | _ | _ | High (80) | High (45) |
| Subtype | HER2+ BC | HER2+ BC | FAD | FAD | TNBC | TNBC |
| Corresponding cell model | BT-549 | MCF10-A | MDA-MB-231 | |||
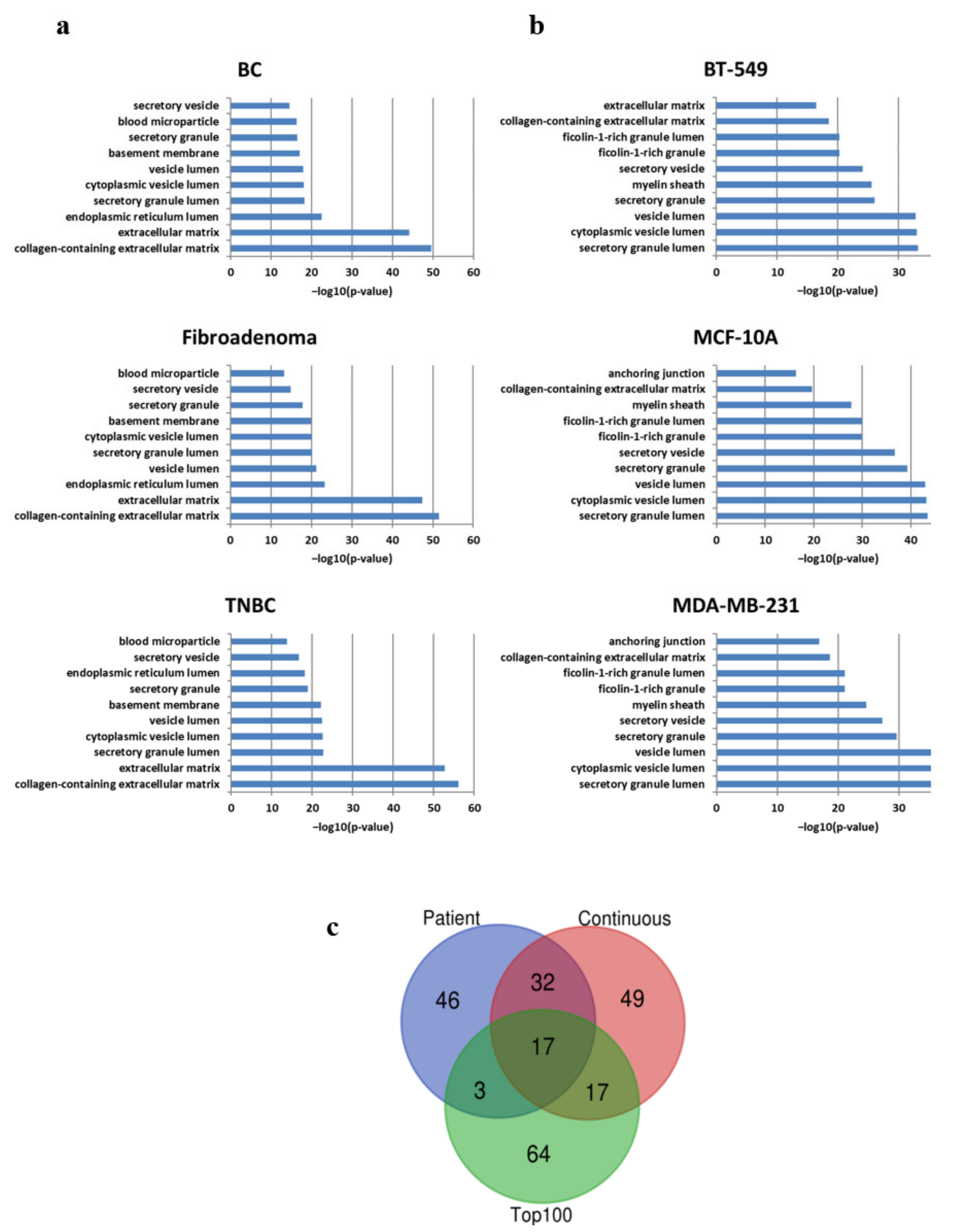
3.3. Qualitative and Semi-Quantitative MS-Based Proteomic Profiling
3.4. Pathway Analysis
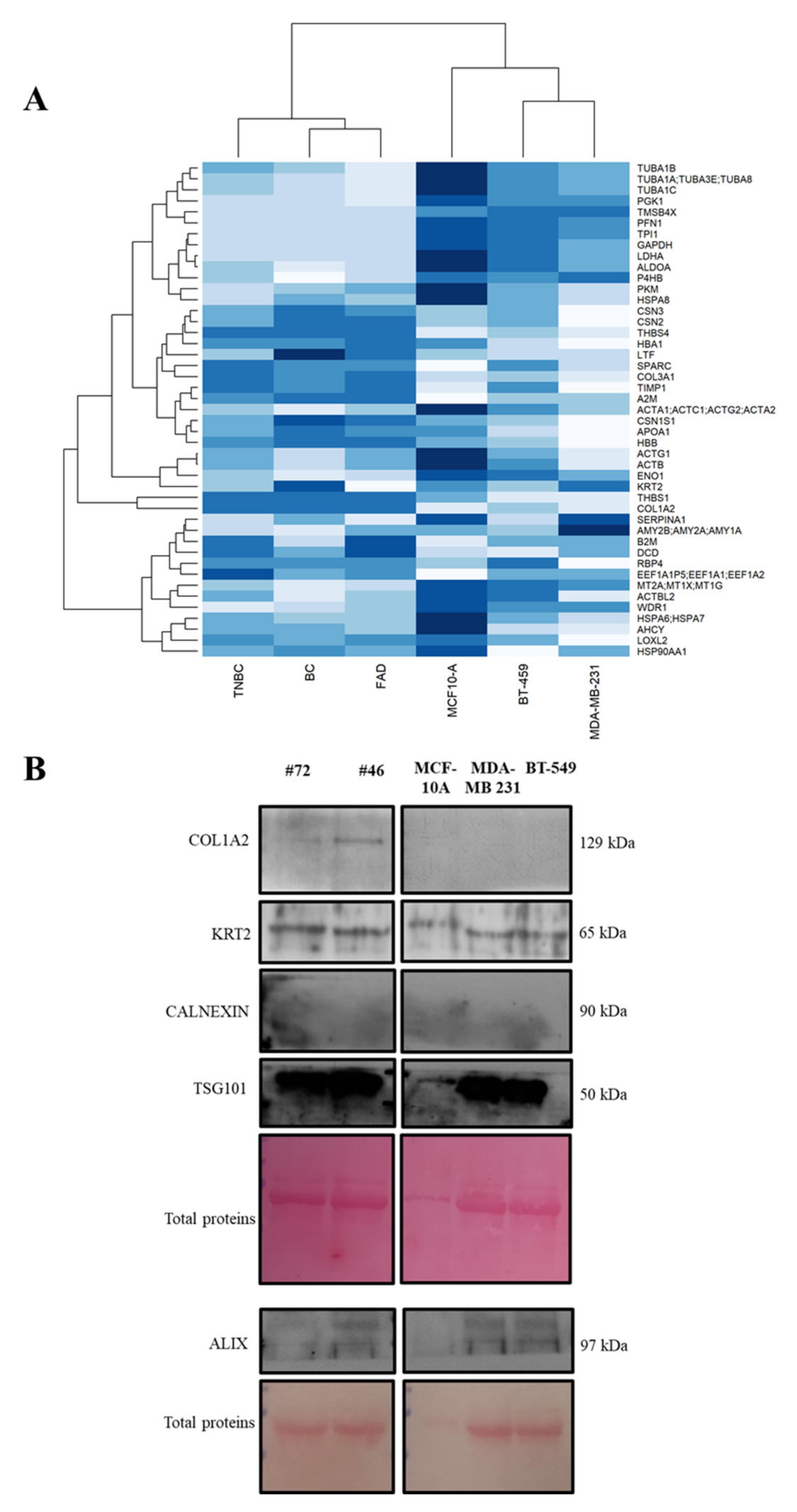
3.5. Protein Abundance Patterns and In Vitro Investigation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | breast cancer |
| DDM | n-dodecyl β-D-maltoside |
| DLS | dynamic light scattering |
| ESI | electrospray ionization |
| ER | estrogen receptor |
| EVs | extracellular vesicles |
| FAD | fibroadenoma |
| FDR | false discovery rate |
| HER2 | human epidermal growth factor receptor 2 |
| IBCC | immortalized breast cell culture |
| PR | progesterone receptor |
| SED | secondary electron detector |
| SEM | scanning electron microscopy |
| TNBC | triple–negative breast cancer |
References
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primer. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Irwig, L.; Macaskill, P.; Houssami, N. Evidence Relevant to the Investigation of Breast Symptoms: The Triple Test. Breast 2002, 11, 215–220. [Google Scholar] [CrossRef]
- Ingenito, F.; Roscigno, G.; Affinito, A.; Nuzzo, S.; Scognamiglio, I.; Quintavalle, C.; Condorelli, G. The Role of Exo-MiRNAs in Cancer: A Focus on Therapeutic and Diagnostic Applications. Int. J. Mol. Sci. 2019, 20, 4687. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H. The Clinical Relevance of Circulating, Exosomal MiRNAs as Biomarkers for Cancer. Expert Rev. Mol. Diagn. 2015, 15, 1159–1169. [Google Scholar] [CrossRef]
- Sandim, V.; Monteiro, R.Q. Extracellular Vesicle Fingerprinting: The next Generation for Cancer Diagnosis? Signal Transduct. Target. Ther. 2020, 5, 263. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key Players in Cancer and Potential Therapeutic Strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass Spectrometry-Based Proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- Shteynberg, D.; Nesvizhskii, A.I.; Moritz, R.L.; Deutsch, E.W. Combining Results of Multiple Search Engines in Proteomics. Mol. Cell. Proteomics MCP 2013, 12, 2383–2393. [Google Scholar] [CrossRef] [Green Version]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic Profiling of Extracellular Vesicles Allows for Human Breast Cancer Subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinik, Y.; Ortega, F.G.; Mills, G.B.; Lu, Y.; Jurkowicz, M.; Halperin, S.; Aharoni, M.; Gutman, M.; Lev, S. Proteomic Analysis of Circulating Extracellular Vesicles Identifies Potential Markers of Breast Cancer Progression, Recurrence, and Response. Sci. Adv. 2020, 6, eaba5714. [Google Scholar] [CrossRef] [PubMed]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-Associated Fibroblasts Release Exosomal MicroRNAs That Dictate an Aggressive Phenotype in Breast Cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallo, E.; Diaferia, C.; Balasco, N.; Sibillano, T.; Roviello, V.; Giannini, C.; Vitagliano, L.; Morelli, G.; Accardo, A. Fabrication of Fluorescent Nanospheres by Heating PEGylated Tetratyrosine Nanofibers. Sci. Rep. 2021, 11, 2470. [Google Scholar] [CrossRef]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The Proteomic Analysis of Breast Cell Line Exosomes Reveals Disease Patterns and Potential Biomarkers. Sci. Rep. 2020, 10, 13572. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Bandu, R.; Oh, J.W.; Kim, K.P. Mass Spectrometry-Based Proteome Profiling of Extracellular Vesicles and Their Roles in Cancer Biology. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef] [Green Version]
- García-Silva, S.; Benito-Martín, A.; Sánchez-Redondo, S.; Hernández-Barranco, A.; Ximénez-Embún, P.; Nogués, L.; Mazariegos, M.S.; Brinkmann, K.; Amor López, A.; Meyer, L.; et al. Use of Extracellular Vesicles from Lymphatic Drainage as Surrogate Markers of Melanoma Progression and BRAF V600E Mutation. J. Exp. Med. 2019, 216, 1061–1070. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.-W.; Kim, H.; Hur, W.; Jung, J.; Jeong, S.J.; Shin, H.; Seo, D.; Jeong, H.; Choi, B.; Hong, S.; et al. A Proteomic Approach to Understand the Clinical Significance of Acute Myeloid Leukemia–Derived Extracellular Vesicles Reflecting Essential Characteristics of Leukemia. Mol. Cell. Proteom. 2021, 20, 100017. [Google Scholar] [CrossRef]
- Hodge, K.; Have, S.T.; Hutton, L.; Lamond, A.I. Cleaning up the Masses: Exclusion Lists to Reduce Contamination with HPLC-MS/MS. J. Proteom. 2013, 88, 92–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, H.; Wong, D.T.W. Proteomic Analysis of Microvesicles in Human Saliva by Gel Electrophoresis with Liquid Chromatography-Mass Spectrometry. Anal. Chim. Acta 2012, 723, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Turay, D.; Khan, S.; Diaz Osterman, C.J.; Curtis, M.P.; Khaira, B.; Neidigh, J.W.; Mirshahidi, S.; Casiano, C.A.; Wall, N.R. Proteomic Profiling of Serum-Derived Exosomes from Ethnically Diverse Prostate Cancer Patients. Cancer Investig. 2016, 34, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Ciatto, S.; Bonardi, R.; Zappa, M.; Giorgi, D. Risk of Breast Cancer Subsequent to Histological or Clinical Diagnosis of Fibroadenoma—Retrospective Longitudinal Study of 3938 Cases. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 1997, 8, 297–300. [Google Scholar] [CrossRef]
- Dupont, W.D.; Page, D.L.; Parl, F.F.; Vnencak-Jones, C.L.; Plummer, W.D.; Rados, M.S.; Schuyler, P.A. Long-Term Risk of Breast Cancer in Women with Fibroadenoma. N. Engl. J. Med. 1994, 331, 10–15. [Google Scholar] [CrossRef]
- Thorat, M.A.; Balasubramanian, R. Breast Cancer Prevention in High-Risk Women. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 18–31. [Google Scholar] [CrossRef]
- El-Wakeel, H.; Umpleby, H.C. Systematic Review of Fibroadenoma as a Risk Factor for Breast Cancer. Breast Edinb. Scotl. 2003, 12, 302–307. [Google Scholar] [CrossRef]
- Kabat, G.C.; Jones, J.G.; Olson, N.; Negassa, A.; Duggan, C.; Ginsberg, M.; Kandel, R.A.; Glass, A.G.; Rohan, T.E. A Multi-Center Prospective Cohort Study of Benign Breast Disease and Risk of Subsequent Breast Cancer. Cancer Causes Control CCC 2010, 21, 821–828. [Google Scholar] [CrossRef] [Green Version]
- Nassar, A.; Visscher, D.W.; Degnim, A.C.; Frank, R.D.; Vierkant, R.A.; Frost, M.; Radisky, D.C.; Vachon, C.M.; Kraft, R.A.; Hartmann, L.C.; et al. Complex Fibroadenoma and Breast Cancer Risk: A Mayo Clinic Benign Breast Disease Cohort Study. Breast Cancer Res. Treat. 2015, 153, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Rosa-Fernandes, L.; Rocha, V.B.; Carregari, V.C.; Urbani, A.; Palmisano, G. A Perspective on Extracellular Vesicles Proteomics. Front. Chem. 2017, 5, 102. [Google Scholar] [CrossRef] [Green Version]
- Alves, G.; Wu, W.W.; Wang, G.; Shen, R.-F.; Yu, Y.-K. Enhancing Peptide Identification Confidence by Combining Search Methods. J. Proteome Res. 2008, 7, 3102–3113. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; Searle, B.C.; Clauser, K.R.; Tabb, D.L. A Face in the Crowd: Recognizing Peptides Through Database Search. Mol. Cell. Proteom. MCP 2011, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helbig, A.O.; Heck, A.J.R.; Slijper, M. Exploring the Membrane Proteome—Challenges and Analytical Strategies. J. Proteom. 2010, 73, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Dahms, T.E.S.; Babu, M. Chromatographic Separation Strategies for Precision Mass Spectrometry to Study Protein-Protein Interactions and Protein Phosphorylation. J. Chromatogr. B 2018, 1102, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Esposito, C.L.; Quintavalle, C.; Ingenito, F.; Rotoli, D.; Roscigno, G.; Nuzzo, S.; Thomas, R.; Catuogno, S.; de Franciscis, V.; Condorelli, G. Identification of a Novel RNA Aptamer That Selectively Targets Breast Cancer Exosomes. Mol. Ther. Nucleic Acids 2021, 23, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, I.; Cocca, L.; Puoti, I.; Palma, F.; Ingenito, F.; Quintavalle, C.; Affinito, A.; Roscigno, G.; Nuzzo, S.; Chianese, R.V.; et al. Exosomal microRNAs synergistically trigger stromal fibroblasts in breast cancer. Mol. Ther. Nucleic Acids 2022, 28, 17–31. [Google Scholar] [CrossRef]

| Sample | Mean Diameter (nm) ± S.D. | PDI | (D ± S.D.) * 10−12 m2 s−1 |
|---|---|---|---|
| #37 | 346 ± 70 | 0.356 | 1.15 ± 0.23 |
| #44 | 270 ± 74 8 ± 2 | 0.395 | 1.47 ± 0.40 49.6 ± 12.4 |
| #148 | 298 ± 64 | 0.365 | 1.33 ± 0.29 |
| MDA-MB-231 | 295 ± 58 | 0.310 | 1.39 ± 0.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pane, K.; Quintavalle, C.; Nuzzo, S.; Ingenito, F.; Roscigno, G.; Affinito, A.; Scognamiglio, I.; Pattanayak, B.; Gallo, E.; Accardo, A.; et al. Comparative Proteomic Profiling of Secreted Extracellular Vesicles from Breast Fibroadenoma and Malignant Lesions: A Pilot Study. Int. J. Mol. Sci. 2022, 23, 3989. https://doi.org/10.3390/ijms23073989
Pane K, Quintavalle C, Nuzzo S, Ingenito F, Roscigno G, Affinito A, Scognamiglio I, Pattanayak B, Gallo E, Accardo A, et al. Comparative Proteomic Profiling of Secreted Extracellular Vesicles from Breast Fibroadenoma and Malignant Lesions: A Pilot Study. International Journal of Molecular Sciences. 2022; 23(7):3989. https://doi.org/10.3390/ijms23073989
Chicago/Turabian StylePane, Katia, Cristina Quintavalle, Silvia Nuzzo, Francesco Ingenito, Giuseppina Roscigno, Alessandra Affinito, Iolanda Scognamiglio, Birlipta Pattanayak, Enrico Gallo, Antonella Accardo, and et al. 2022. "Comparative Proteomic Profiling of Secreted Extracellular Vesicles from Breast Fibroadenoma and Malignant Lesions: A Pilot Study" International Journal of Molecular Sciences 23, no. 7: 3989. https://doi.org/10.3390/ijms23073989
APA StylePane, K., Quintavalle, C., Nuzzo, S., Ingenito, F., Roscigno, G., Affinito, A., Scognamiglio, I., Pattanayak, B., Gallo, E., Accardo, A., Thomas, G., Minic, Z., Berezovski, M. V., Franzese, M., & Condorelli, G. (2022). Comparative Proteomic Profiling of Secreted Extracellular Vesicles from Breast Fibroadenoma and Malignant Lesions: A Pilot Study. International Journal of Molecular Sciences, 23(7), 3989. https://doi.org/10.3390/ijms23073989











