Multiple Pre-Treatment miRNAs Levels in Untreated Major Depressive Disorder Patients Predict Early Response to Antidepressants and Interact with Key Pathways
Abstract
1. Introduction
2. Results
2.1. Baseline Analysis
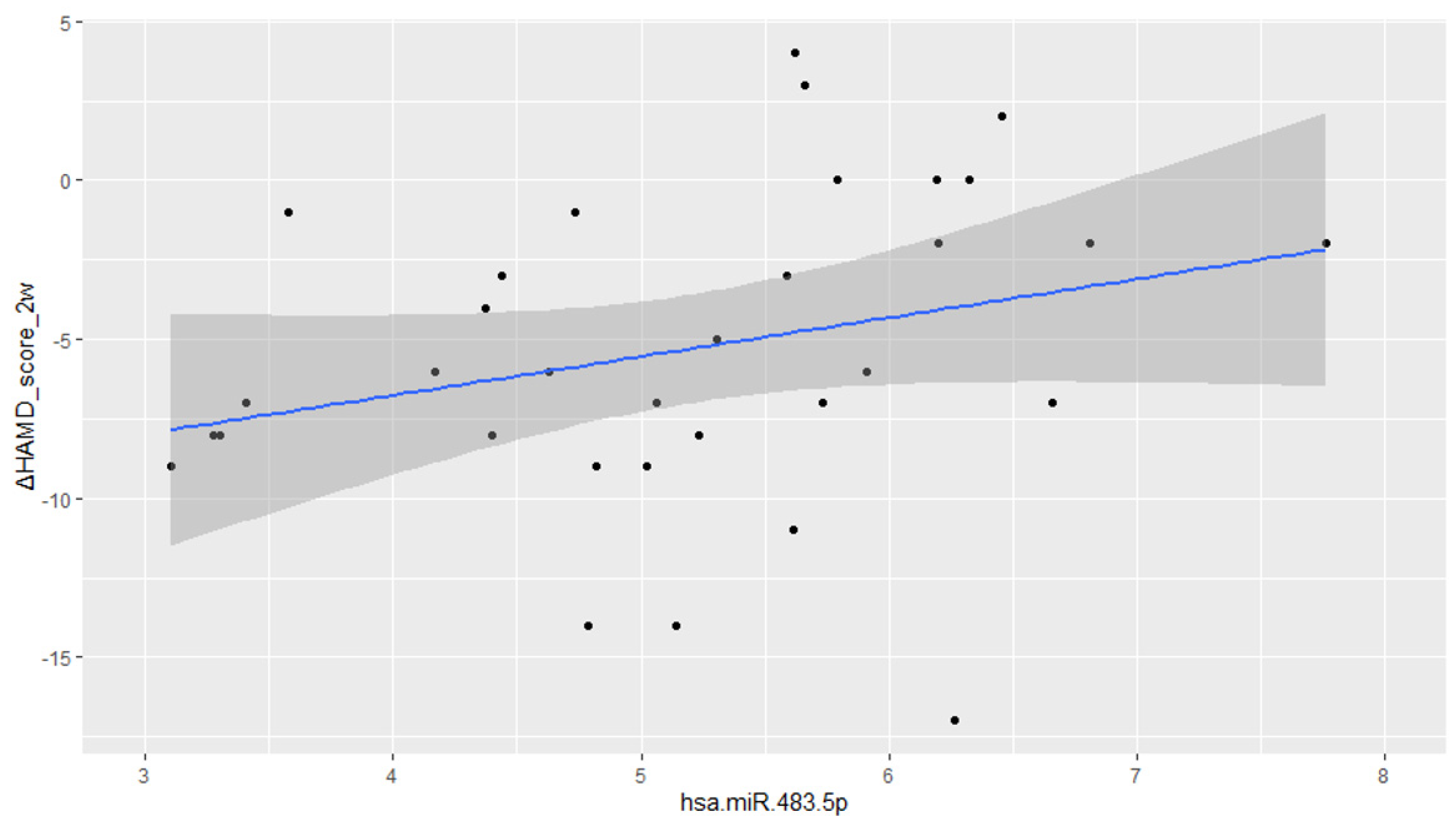
2.2. Associations between miRNA and Treatment Response
2.2.1. SSRIs
2.2.2. Mirtazapine
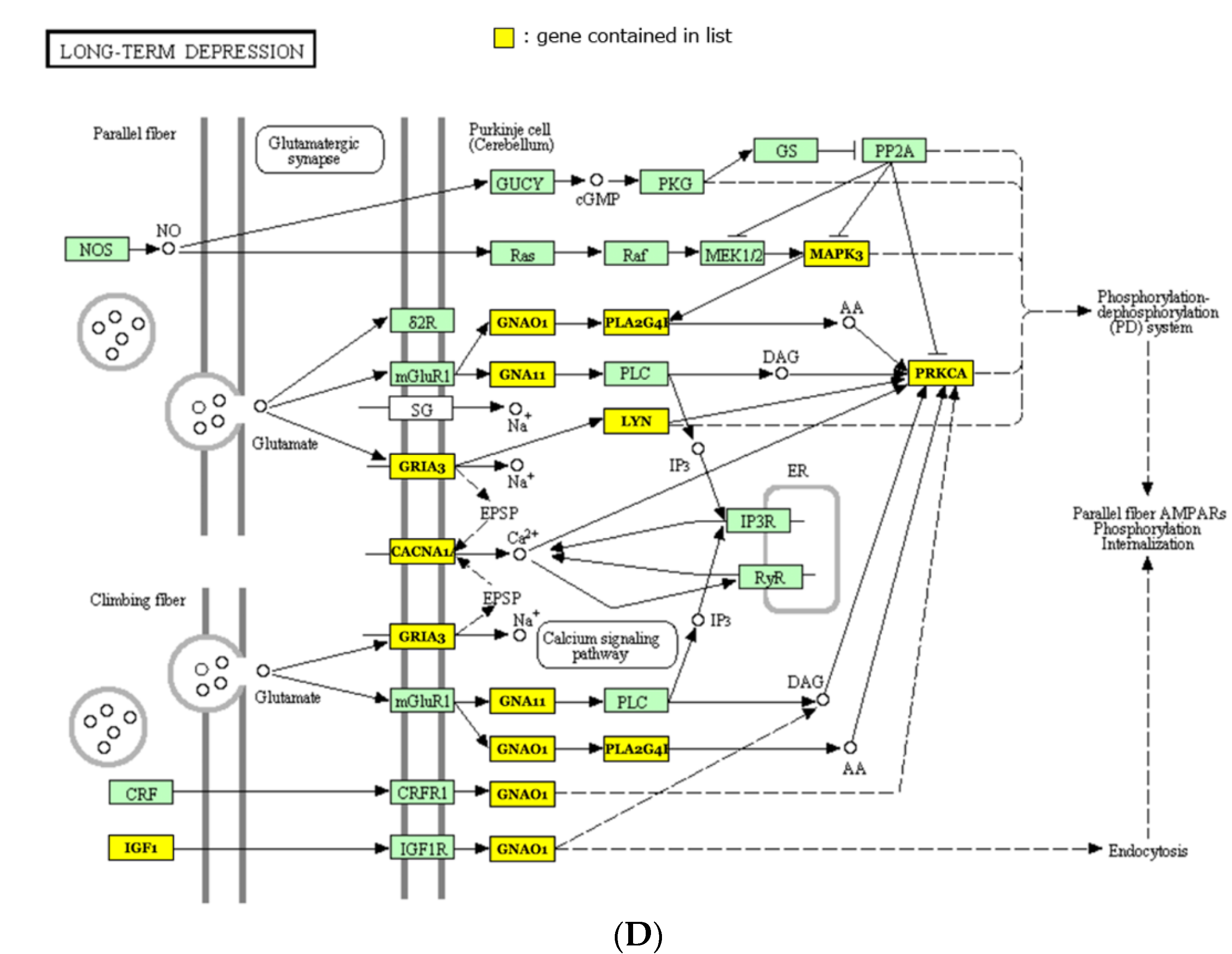
2.3. miRNA Target Prediction and Pathway Analysis
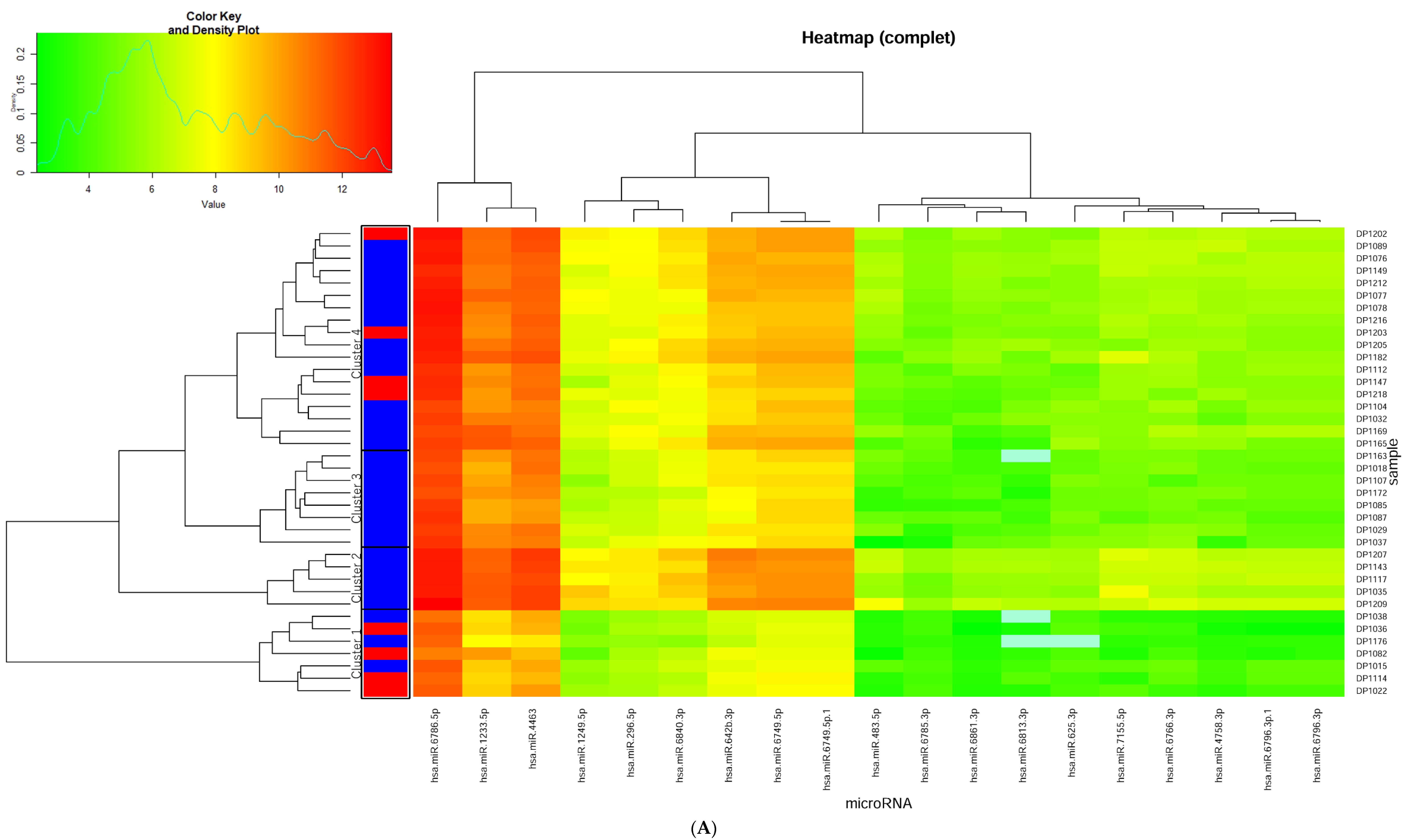
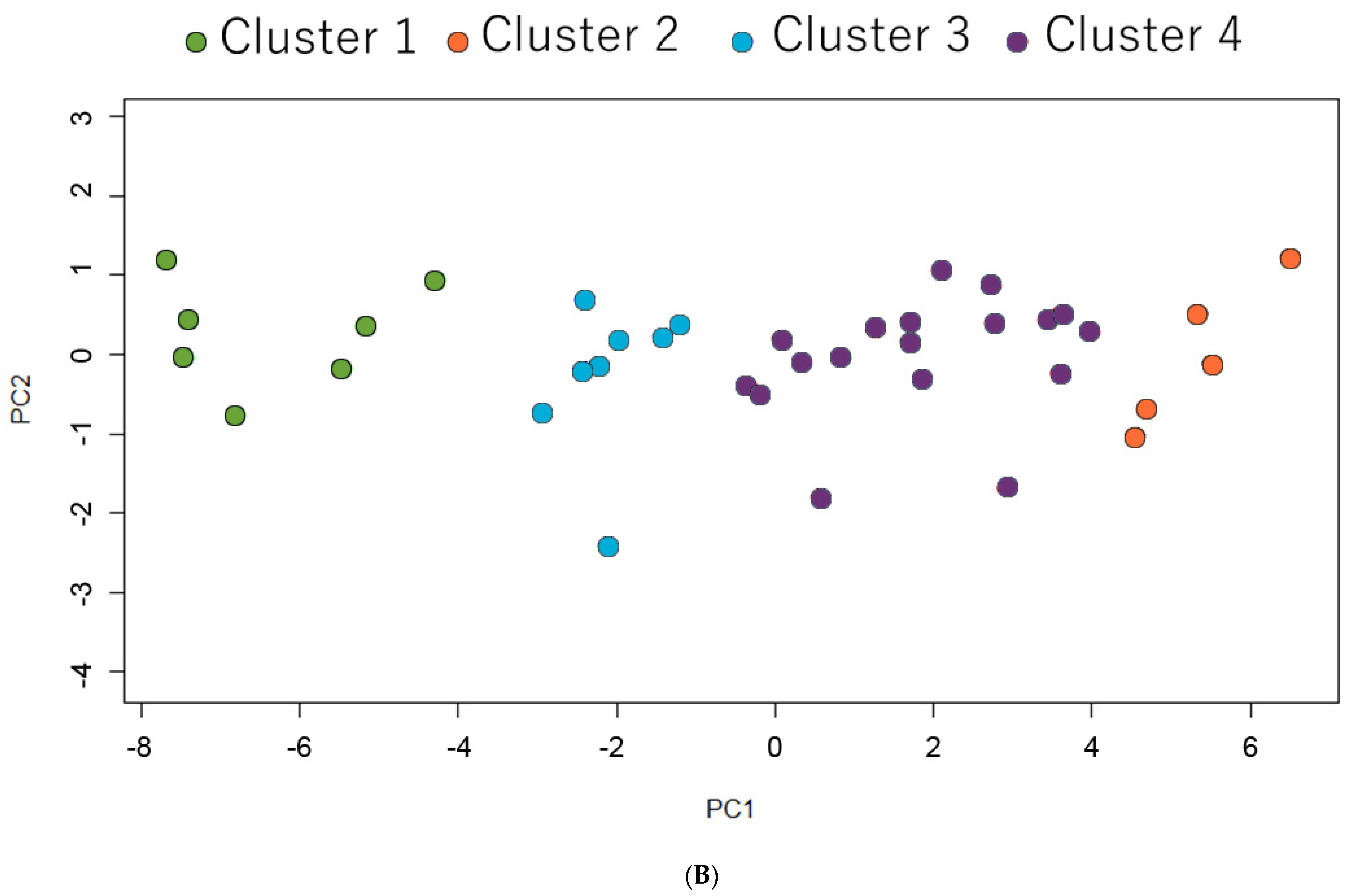
2.4. Heatmap Analysis
3. Discussion
4. Methods
4.1. Study Design and Participants
4.2. Microarray Analysis of miRNA Expression
4.3. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ferrari, A.J.; Somerville, A.J.; Baxter, A.J.; Norman, R.; Patten, S.B.; Vos, T.; Whiteford, H.A. Global variation in the prevalence and incidence of major depressive disorder: A systematic review of the epidemiological literature. Psychol. Med. 2013, 43, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Bromet, E.J.; Andrade, L.H.; Bruffaerts, R.; Williams, D.R. Major Depressive Disorder. In Mental Disorders Around the World: Facts and Figures from the WHO World Mental Health Surveys; Stein, D.J., Scott, K.M., de Jonge, P., Kessler, R.C., Eds.; Cambridge University Press: Cambridge, UK, 2018; pp. 41–56. [Google Scholar] [CrossRef]
- Moffitt, T.E.; Caspi, A.; Taylor, A.; Kokaua, J.; Milne, B.J.; Polanczyk, G.; Poulton, R. How common are common mental disorders? Evidence that lifetime prevalence rates are doubled by prospective versus retrospective ascertainment. Psychol. Med. 2010, 40, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Herrman, H.; Patel, V.; Kieling, C.; Berk, M.; Buchweitz, C.; Cuijpers, P.; Furukawa, T.A.; Kessler, R.C.; Kohrt, B.A.; Maj, M.; et al. Time for united action on depression: A Lancet-World Psychiatric Association Commission. Lancet 2022, 399, 957–1022. [Google Scholar] [CrossRef]
- Gold, S.M.; Kohler-Forsberg, O.; Moss-Morris, R.; Mehnert, A.; Miranda, J.J.; Bullinger, M.; Steptoe, A.; Whooley, M.A.; Otte, C. Comorbid depression in medical diseases. Nat. Rev. Dis. Primers 2020, 6, 69. [Google Scholar] [CrossRef]
- Walker, J.; Burke, K.; Wanat, M.; Fisher, R.; Fielding, J.; Mulick, A.; Puntis, S.; Sharpe, J.; Esposti, M.D.; Harriss, E.; et al. The prevalence of depression in general hospital inpatients: A systematic review and meta-analysis of interview-based studies. Psychol. Med. 2018, 48, 2285–2298. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.; Lai, W.; Long, E.; Zhang, X.; Li, W.; Zhu, Y.; Chen, C.; Zhong, X.; Liu, Z.; et al. Prevalence of depression and depressive symptoms among outpatients: A systematic review and meta-analysis. BMJ Open 2017, 7, e017173. [Google Scholar] [CrossRef]
- World Health Organization. The Mental Health Gap Action Program (mhGAP). Available online: https://www.who.int/publications/i/item/9789241596206: (accessed on 13 December 2018).
- Fava, M.; Detke, M.J.; Balestrieri, M.; Wang, F.; Raskin, J.; Perahia, D. Management of depression relapse: Re-initiation of duloxetine treatment or dose increase. J. Psychiatry Res. 2006, 40, 328–336. [Google Scholar] [CrossRef]
- Kato, M.; Takekita, Y.; Koshikawa, Y.; Sakai, S.; Bandou, H.; Nishida, K.; Sunada, N.; Onohara, A.; Hatashita, Y.; Serretti, A.; et al. Non response at week 4 as clinically useful indicator for antidepressant combination in major depressive disorder. A sequential RCT. J. Psychiatry Res. 2017, 89, 97–104. [Google Scholar] [CrossRef]
- Kato, M.; Hori, H.; Inoue, T.; Iga, J.; Iwata, M.; Inagaki, T.; Shinohara, K.; Imai, H.; Murata, A.; Mishima, K.; et al. Discontinuation of antidepressants after remission with antidepressant medication in major depressive disorder: A systematic review and meta-analysis. Mol. Psychiatry 2021, 26, 118–133. [Google Scholar] [CrossRef]
- Rush, A.J.; Trivedi, M.H.; Wisniewski, S.R.; Nierenberg, A.A.; Stewart, J.W.; Warden, D.; Niederehe, G.; Thase, M.E.; Lavori, P.W.; Lebowitz, B.D.; et al. Acute and Longer-Term Outcomes in Depressed Outpatients Requiring One or Several Treatment Steps: A STAR*D Report. Am. J. Psychiatry 2006, 163, 1905–1917. [Google Scholar] [CrossRef]
- Penninx, B.W.; Milaneschi, Y.; Lamers, F.; Vogelzangs, N. Understanding the somatic consequences of depression: Biological mechanisms and the role of depression symptom profile. BMC Med. 2013, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Rodriquez, M.S. Pathways linking affective disturbances and physical disorders. Health Psychol. 1995, 14, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Sáiz-Vázquez, O.; Gracia-García, P.; Ubillos-Landa, S.; Puente-Martínez, A.; Casado-Yusta, S.; Olaya, B.; Santabárbara, J. Depression as a Risk Factor for Alzheimer’s Disease: A Systematic Review of Longitudinal Meta-Analyses. J. Clin. Med. 2021, 10, 1809. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, M.H.; Morris, D.W.; Wisniewski, S.R.; Lesser, I.; Nierenberg, A.A.; Daly, E.; Kurian, B.T.; Gaynes, B.N.; Balasubramani, G.K.; Rush, A.J. Increase in work productivity of depressed individuals with improvement in depressive symptom severity. Am J. Psychiatry 2013, 170, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Habert, J.; Katzman, M.A.; Oluboka, O.J.; McIntyre, R.S.; McIntosh, D.; MacQueen, G.M.; Khullar, A.; Milev, R.V.; Kjernisted, K.D.; Chokka, P.R.; et al. Functional Recovery in Major Depressive Disorder: Focus on Early Optimized Treatment. Prim. Care Companion CNS Disord. 2016, 18, 295–305. [Google Scholar] [CrossRef]
- Kato, M.; Serretti, A.; Nonen, S.; Takekita, Y.; Wakeno, M.; Azuma, J.; Kinoshita, T. Genetic variants in combination with early partial improvement as a clinical utility predictor of treatment outcome in major depressive disorder: The result of two pooled RCTs. Transl. Psychiatry 2015, 5, e513. [Google Scholar] [CrossRef]
- Szegedi, A.; Jansen, W.T.; van Willigenburg, A.P.; van der Meulen, E.; Stassen, H.H.; Thase, M.E. Early improvement in the first 2 weeks as a predictor of treatment outcome in patients with major depressive disorder: A meta-analysis including 6562 patients. J. Clin. Psychiatry 2009, 70, 344–353. [Google Scholar] [CrossRef]
- Bauer, M.; Pfennig, A.; Severus, E.; Whybrow, P.C.; Angst, J.; Moller, H.J. World Federation of Societies of Biological Psychiatry (WFSBP) guidelines for biological treatment of unipolar depressive disorders, part 1: Update 2013 on the acute and continuation treatment of unipolar depressive disorders. World J. Biol. Psychiatry 2013, 14, 334–385. [Google Scholar] [CrossRef]
- Kennedy, S.H.; Lam, R.W.; McIntyre, R.S.; Tourjman, S.V.; Bhat, V.; Blier, P.; Hasnain, M.; Jollant, F.; Levitt, A.J.; MacQueen, G.M.; et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 Clinical Guidelines for the Management of Adults with Major Depressive Disorder: Section 3. Pharmacological Treatments. Can. J. Psychiatry 2016, 61, 540–560. [Google Scholar] [CrossRef]
- Watanabe, N.; Omori, I.M.; Nakagawa, A.; Cipriani, A.; Barbui, C.; McGuire, H.; Churchill, R.; Furukawa, T.A. Mirtazapine versus other antidepressants in the acute-phase treatment of adults with major depression: Systematic review and meta-analysis. J. Clin. Psychiatry 2008, 69, 1404–1415. [Google Scholar] [CrossRef]
- Anttila, S.A.; Leinonen, E.V. A review of the pharmacological and clinical profile of mirtazapine. CNS Drug Rev. 2001, 7, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Haddjeri, N.; Blier, P.; de Montigny, C. Effect of the alpha-2 adrenoceptor antagonist mirtazapine on the 5-hydroxytryptamine system in the rat brain. J. Pharmacol. Exp. Ther. 1996, 277, 861–871. [Google Scholar] [PubMed]
- Otte, C.; Gold, S.M.; Penninx, B.W.; Pariante, C.M.; Etkin, A.; Fava, M.; Mohr, D.C.; Schatzberg, A.F. Major depressive disorder. Nat. Rev. Dis. Primers 2016, 2, 16065. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Tóth, F.; Polyák, H.; Szabó, Á.; Mándi, Y.; Vécsei, L. Immune Influencers in Action: Metabolites and Enzymes of the Tryptophan-Kynurenine Metabolic Pathway. Biomedicines 2021, 9, 734. [Google Scholar] [CrossRef]
- Nemeroff, C.B. The State of Our Understanding of the Pathophysiology and Optimal Treatment of Depression: Glass Half Full or Half Empty? Am. J. Psychiatry 2020, 177, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, H.; Yonezawa, K.; Tani, H.; Mimura, M.; Bauer, M.; Uchida, H. Novel Antidepressants in the Pipeline (Phase II and III): A Systematic Review of the US Clinical Trials Registry. Pharmacopsychiatry 2022. [Google Scholar] [CrossRef]
- Hill, A.S.; Sahay, A.; Hen, R. Increasing Adult Hippocampal Neurogenesis is Sufficient to Reduce Anxiety and Depression-Like Behaviors. Neuropsychopharmacology 2015, 40, 2368–2378. [Google Scholar] [CrossRef]
- Atake, K.; Hori, H.; Kageyama, Y.; Koshikawa, Y.; Igata, R.; Tominaga, H.; Katsuki, A.; Bando, H.; Sakai, S.; Nishida, K.; et al. Pretreatment plasma cytokine levels as potential predictors of short-term remission of depression. World J. Biol. Psychiatry 2022, 40, 1–25. [Google Scholar] [CrossRef]
- Sharp, T. Molecular and cellular mechanisms of antidepressant action. Curr. Top Behav. Neurosci. 2013, 14, 309–325. [Google Scholar] [CrossRef]
- Pena-Vargas, C.; Armaiz-Pena, G.; Castro-Figueroa, E. A Biopsychosocial Approach to Grief, Depression, and the Role of Emotional Regulation. Behav. Sci. 2021, 11, 110. [Google Scholar] [CrossRef]
- Tanaka, M.; Vecsei, L. Editorial of Special Issue “Crosstalk between Depression, Anxiety, and Dementia: Comorbidity in Behavioral Neurology and Neuropsychiatry”. Biomedicines 2021, 9, 517. [Google Scholar] [CrossRef] [PubMed]
- Sanacora, G.; Zarate, C.A.; Krystal, J.H.; Manji, H.K. Targeting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nat. Rev. Drug Discov. 2008, 7, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K. Ketamine’s antidepressant action: Beyond NMDA receptor inhibition. Expert Opin. Ther. Targets 2016, 20, 1389–1392. [Google Scholar] [CrossRef] [PubMed]
- Noto, C.; Rizzo, L.B.; Mansur, R.B.; McIntyre, R.S.; Maes, M.; Brietzke, E. Targeting the inflammatory pathway as a therapeutic tool for major depression. Neuroimmunomodulation 2014, 21, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, I.A.; Mehler, M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012, 13, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Wehrspaun, C.C.; Ponting, C.P.; Marques, A.C. Brain-expressed 3′UTR extensions strengthen miRNA cross-talk between ion channel/transporter encoding mRNAs. Front. Genet. 2014, 5, 41. [Google Scholar] [CrossRef][Green Version]
- Belzeaux, R.; Lin, C.W.; Ding, Y.; Bergon, A.; Ibrahim, E.C.; Turecki, G.; Tseng, G.; Sibille, E. Predisposition to treatment response in major depressive episode: A peripheral blood gene coexpression network analysis. J. Psychiatry Res. 2016, 81, 119–126. [Google Scholar] [CrossRef]
- Tavakolizadeh, J.; Roshanaei, K.; Salmaninejad, A.; Yari, R.; Nahand, J.S.; Sarkarizi, H.K.; Mousavi, S.M.; Salarinia, R.; Rahmati, M.; Mousavi, S.F.; et al. MicroRNAs and exosomes in depression: Potential diagnostic biomarkers. J. Cell Biochem. 2018, 119, 3783–3797. [Google Scholar] [CrossRef]
- Smalheiser, N.R.; Lugli, G.; Rizavi, H.S.; Torvik, V.I.; Turecki, G.; Dwivedi, Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE 2012, 7, e33201. [Google Scholar] [CrossRef]
- Fan, H.M.; Sun, X.Y.; Guo, W.; Zhong, A.F.; Niu, W.; Zhao, L.; Dai, Y.H.; Guo, Z.M.; Zhang, L.Y.; Lu, J. Differential expression of microRNA in peripheral blood mononuclear cells as specific biomarker for major depressive disorder patients. J. Psychiatry Res. 2014, 59, 45–52. [Google Scholar] [CrossRef]
- Yoshino, Y.; Roy, B.; Dwivedi, Y. Differential and unique patterns of synaptic miRNA expression in dorsolateral prefrontal cortex of depressed subjects. Neuropsychopharmacology 2021, 46, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.D.; Li, L.; Chan, W.Y. MicroRNAs: Key Regulators in the Central Nervous System and Their Implication in Neurological Diseases. Int. J. Mol. Sci. 2016, 17, 842. [Google Scholar] [CrossRef] [PubMed]
- Belzeaux, R.; Fiori, L.M.; Lopez, J.P.; Boucekine, M.; Boyer, L.; Blier, P.; Farzan, F.; Frey, B.N.; Giacobbe, P.; Lam, R.W.; et al. Predicting Worsening Suicidal Ideation with Clinical Features and Peripheral Expression of Messenger RNA and MicroRNA during Antidepressant Treatment. J. Clin. Psychiatry 2019, 80, 556. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.P.; Lim, R.; Cruceanu, C.; Crapper, L.; Fasano, C.; Labonte, B.; Maussion, G.; Yang, J.P.; Yerko, V.; Vigneault, E.; et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat. Med. 2014, 20, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Belzeaux, R.; Bergon, A.; Jeanjean, V.; Loriod, B.; Formisano-Treziny, C.; Verrier, L.; Loundou, A.; Baumstarck-Barrau, K.; Boyer, L.; Gall, V.; et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl. Psychiatry 2012, 2, e185. [Google Scholar] [CrossRef]
- Issler, O.; Haramati, S.; Paul, E.D.; Maeno, H.; Navon, I.; Zwang, R.; Gil, S.; Mayberg, H.S.; Dunlop, B.W.; Menke, A.; et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 2014, 83, 344–360. [Google Scholar] [CrossRef]
- He, S.; Liu, X.; Jiang, K.; Peng, D.; Hong, W.; Fang, Y.; Qian, Y.; Yu, S.; Li, H. Alterations of microRNA-124 expression in peripheral blood mononuclear cells in pre- and post-treatment patients with major depressive disorder. J. Psychiatry Res. 2016, 78, 65–71. [Google Scholar] [CrossRef]
- Fiori, L.M.; Lopez, J.P.; Richard-Devantoy, S.; Berlim, M.; Chachamovich, E.; Jollant, F.; Foster, J.; Rotzinger, S.; Kennedy, S.H.; Turecki, G. Investigation of miR-1202, miR-135a, and miR-16 in Major Depressive Disorder and Antidepressant Response. Int. J. Neuropsychopharmacol. 2017, 20, 619–623. [Google Scholar] [CrossRef]
- Wagner, S.; Engel, A.; Engelmann, J.; Herzog, D.; Dreimuller, N.; Muller, M.B.; Tadic, A.; Lieb, K. Early improvement as a resilience signal predicting later remission to antidepressant treatment in patients with Major Depressive Disorder: Systematic review and meta-analysis. J. Psychiatry Res. 2017, 94, 96–106. [Google Scholar] [CrossRef]
- Lopez, J.P.; Pereira, F.; Richard-Devantoy, S.; Berlim, M.; Chachamovich, E.; Fiori, L.M.; Niola, P.; Turecki, G.; Jollant, F. Co-Variation of Peripheral Levels of miR-1202 and Brain Activity and Connectivity during Antidepressant Treatment. Neuropsychopharmacology 2017, 42, 2043–2051. [Google Scholar] [CrossRef]
- Dadkhah, T.; Rahimi-Aliabadi, S.; Jamshidi, J.; Ghaedi, H.; Taghavi, S.; Shokraeian, P.; Akhavan-Niaki, H.; Tafakhori, A.; Ohadi, M.; Darvish, H. A genetic variant in miRNA binding site of glutamate receptor 4, metabotropic (GRM4) is associated with increased risk of major depressive disorder. J. Affect. Disord. 2017, 208, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Katsura, A.; Yasuda, T.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol. 2015, 22, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Roth, A.; Yu, M.; Morris, R.; Bersani, F.; Rivera, M.N.; Lu, J.; Shioda, T.; Vasudevan, S.; Ramaswamy, S.; et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013, 27, 2543–2548. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; He, H.; Xie, Y.; Zhao, L.; Zhao, S.; Wan, X.; Yang, W.; Mo, Z. miR-125a-3p and miR-483-5p promote adipogenesis via suppressing the RhoA/ROCK1/ERK1/2 pathway in multiple symmetric lipomatosis. Sci. Rep. 2015, 5, 11909. [Google Scholar] [CrossRef] [PubMed]
- Pepe, F.; Visone, R.; Veronese, A. The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer. Cancers 2018, 10, 181. [Google Scholar] [CrossRef]
- Lopez, J.P.; Fiori, L.M.; Cruceanu, C.; Lin, R.; Labonte, B.; Cates, H.M.; Heller, E.A.; Vialou, V.; Ku, S.M.; Gerald, C.; et al. MicroRNAs 146a/b-5 and 425-3p and 24-3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nat. Commun. 2017, 8, 15497. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Association: Washington, DC, USA, 1994. [Google Scholar]
- Hamilton, M. Development of a rating scale for primary depressive illness. Br. J. Soc. Clin. Psychol. 1967, 6, 278–296. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Caraci, F.; Spampinato, S.F.; Morgese, M.G.; Tascedda, F.; Salluzzo, M.G.; Giambirtone, M.C.; Caruso, G.; Munafo, A.; Torrisi, S.A.; Leggio, G.M.; et al. Neurobiological links between depression and AD: The role of TGF-β1 signaling as a new pharmacological target. Pharmacol. Res. 2018, 130, 374–384. [Google Scholar] [CrossRef]
- Liput, D.J.; Puhl, H.L.; Dong, A.; He, K.; Li, Y.; Lovinger, D.M. 2-Arachidonoylglycerol mobilization following brief synaptic stimulation in the dorsal lateral striatum requires glutamatergic and cholinergic neurotransmission. Neuropharmacology 2022, 205, 108916. [Google Scholar] [CrossRef] [PubMed]
- Hiew, L.F.; Poon, C.H.; You, H.Z.; Lim, L.W. TGF-β/Smad Signalling in Neurogenesis: Implications for Neuropsychiatric Diseases. Cells 2021, 10, 1382. [Google Scholar] [CrossRef] [PubMed]
- Krieglstein, K.; Zheng, F.; Unsicker, K.; Alzheimer, C. More than being protective: Functional roles for TGF-β/activin signaling pathways at central synapses. Trends Neurosci. 2011, 34, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Behl, T.; Rana, T.; Alotaibi, G.H.; Shamsuzzaman, M.; Naqvi, M.; Sehgal, A.; Singh, S.; Sharma, N.; Almoshari, Y.; Abdellatif, A.A.H.; et al. Polyphenols inhibiting MAPK signalling pathway mediated oxidative stress and inflammation in depression. Biomed. Pharmacother. 2022, 146, 112545. [Google Scholar] [CrossRef]
- Wang, X.L.; Yuan, K.; Zhang, W.; Li, S.X.; Gao, G.F.; Lu, L. Regulation of Circadian Genes by the MAPK Pathway: Implications for Rapid Antidepressant Action. Neurosci. Bull. 2020, 36, 66–76. [Google Scholar] [CrossRef]
- Zhou, X.; Teng, T.; Zhang, Y.; Del Giovane, C.; Furukawa, T.A.; Weisz, J.R.; Li, X.; Cuijpers, P.; Coghill, D.; Xiang, Y.; et al. Comparative efficacy and acceptability of antidepressants, psychotherapies, and their combination for acute treatment of children and adolescents with depressive disorder: A systematic review and network meta-analysis. Lancet Psychiatry 2020, 7, 581–601. [Google Scholar] [CrossRef]
- Borgomaneri, S.; Battaglia, S.; Sciamanna, G.; Tortora, F.; Laricchiuta, D. Memories are not written in stone: Re-writing fear memories by means of non-invasive brain stimulation and optogenetic manipulations. Neurosci. Biobehav. Rev. 2021, 127, 334–352. [Google Scholar] [CrossRef]
- McGrath, C.L.; Kelley, M.E.; Holtzheimer, P.E.; Dunlop, B.W.; Craighead, W.E.; Franco, A.R.; Craddock, R.C.; Mayberg, H.S. Toward a neuroimaging treatment selection biomarker for major depressive disorder. JAMA Psychiatry 2013, 70, 821–829. [Google Scholar] [CrossRef]


| Total (n = 78) | Mirtazapine (n = 40) | SSRIs (n = 38) | p | ||||
|---|---|---|---|---|---|---|---|
| % | % | % | |||||
| Sex (female) | 48.7% | 50.0% | 47.3% | n.s. | |||
| First episode | 68.4% | 71.1% | 65.8% | n.s. | |||
| Family psychiatric history | 29.2% | 25.7% | 32.4% | n.s. | |||
| Physical comorbidity | 38.7% | 35.1% | 42.1% | n.s. | |||
| Smoking | 2.7% | 5.4% | 0.0% | n.s. | |||
| Drinking | 22.7% | 23.7% | 21.6% | n.s. | |||
| Occupational status: Employed | 79.5% | 80.0% | 78.9% | n.s. | |||
| Mean | SD | Mean | SD | Mean | SD | ||
| Age | 47.7 | 16.8 | 48.4 | 16.4 | 47 | 17.4 | n.s. |
| Duration of current MDD episode (months) | 8.6 | 18.1 | 6 | 10 | 11.4 | 23.7 | 0.015 |
| HAM-D 17 items total score | 21 | 4.7 | 21.5 | 5.1 | 20.5 | 4.4 | n.s. |
| Pathway | Database | p-Value | #Genes | #miRNAs |
|---|---|---|---|---|
| TGF-beta signaling pathway | microT-CDS/Tarbase | 0.006/0.036 | 21/6 | 8/1 |
| Proteoglycans in cancer | microT-CDS | <0.001 | 41 | 9 |
| Long-term depression | TargetScan | 0.002 | 13 | 5 |
| Glutamatergic synapse | microT-CDS | 0.006 | 26 | 8 |
| Thyroid hormone signaling pathway | microT-CDS | 0.006 | 26 | 9 |
| Amphetamine addiction | microT-CDS | 0.024 | 16 | 7 |
| Morphine addiction | microT-CDS | 0.025 | 22 | 8 |
| Endocrine and other factor-regulated calcium reabsorption | microT-CDS | 0.027 | 13 | 6 |
| Calcium signaling pathway | microT-CDS | 0.027 | 36 | 8 |
| Hippo signaling pathway | microT-CDS | 0.027 | 28 | 8 |
| Signaling pathways regulating pluripotency of stem cells | microT-CDS | 0.027 | 27 | 9 |
| Dilated cardiomyopathy | microT-CDS | 0.027 | 22 | 9 |
| MAPK signaling pathway | microT-CDS | 0.028 | 49 | 9 |
| Circadian entrainment | microT-CDS | 0.028 | 24 | 9 |
| Colorectal cancer | microT-CDS | 0.038 | 13 | 5 |
| ErbB signaling pathway | microT-CDS | 0.038 | 17 | 7 |
| ECM-receptor interaction | microT-CDS | 0.038 | 16 | 7 |
| Axon guidance | microT-CDS | 0.038 | 25 | 8 |
| Cytokine-cytokine receptor interaction | microT-CDS | 0.038 | 37 | 9 |
| Endocytosis | microT-CDS | 0.038 | 37 | 10 |
| Focal adhesion | microT-CDS | 0.049 | 40 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, M.; Ogata, H.; Tahara, H.; Shimamoto, A.; Takekita, Y.; Koshikawa, Y.; Nishida, K.; Nonen, S.; Higasa, K.; Kinoshita, T. Multiple Pre-Treatment miRNAs Levels in Untreated Major Depressive Disorder Patients Predict Early Response to Antidepressants and Interact with Key Pathways. Int. J. Mol. Sci. 2022, 23, 3873. https://doi.org/10.3390/ijms23073873
Kato M, Ogata H, Tahara H, Shimamoto A, Takekita Y, Koshikawa Y, Nishida K, Nonen S, Higasa K, Kinoshita T. Multiple Pre-Treatment miRNAs Levels in Untreated Major Depressive Disorder Patients Predict Early Response to Antidepressants and Interact with Key Pathways. International Journal of Molecular Sciences. 2022; 23(7):3873. https://doi.org/10.3390/ijms23073873
Chicago/Turabian StyleKato, Masaki, Haruhiko Ogata, Hidetoshi Tahara, Akira Shimamoto, Yoshiteru Takekita, Yosuke Koshikawa, Keiichiro Nishida, Shinpei Nonen, Koichiro Higasa, and Toshihiko Kinoshita. 2022. "Multiple Pre-Treatment miRNAs Levels in Untreated Major Depressive Disorder Patients Predict Early Response to Antidepressants and Interact with Key Pathways" International Journal of Molecular Sciences 23, no. 7: 3873. https://doi.org/10.3390/ijms23073873
APA StyleKato, M., Ogata, H., Tahara, H., Shimamoto, A., Takekita, Y., Koshikawa, Y., Nishida, K., Nonen, S., Higasa, K., & Kinoshita, T. (2022). Multiple Pre-Treatment miRNAs Levels in Untreated Major Depressive Disorder Patients Predict Early Response to Antidepressants and Interact with Key Pathways. International Journal of Molecular Sciences, 23(7), 3873. https://doi.org/10.3390/ijms23073873









