Identification and In Silico Characterization of a Novel COLGALT2 Gene Variant in a Child with Mucosal Rectal Prolapse
Abstract
:1. Introduction
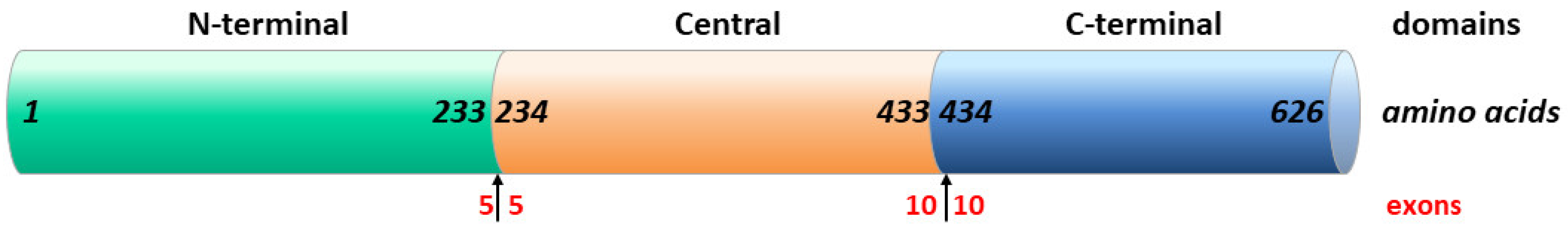
2. Results
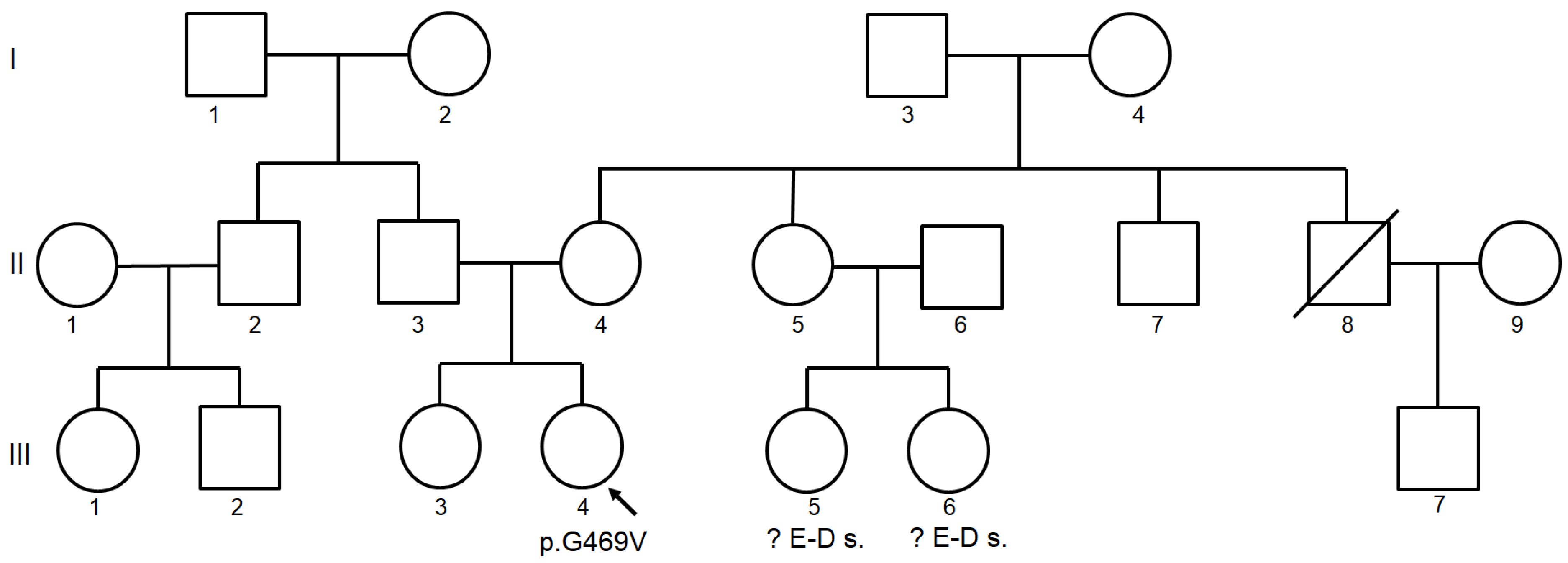
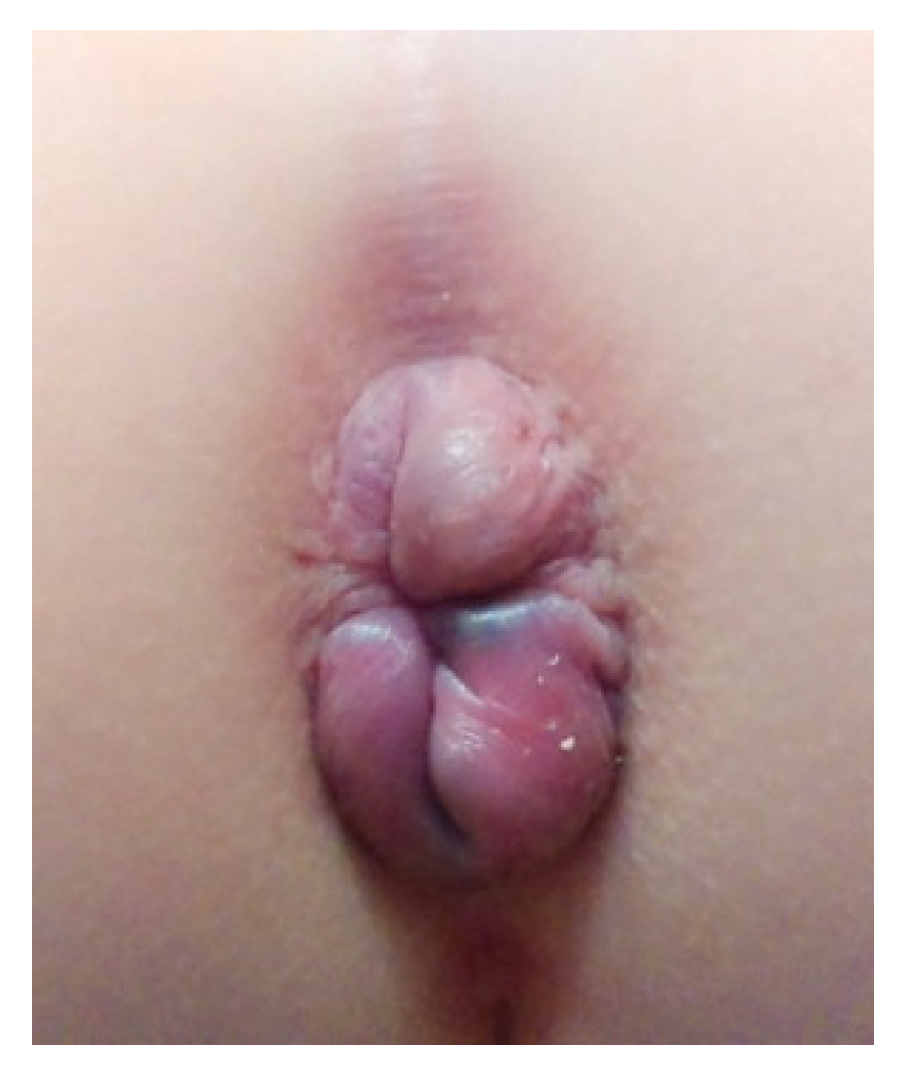
2.1. Clinical Findings
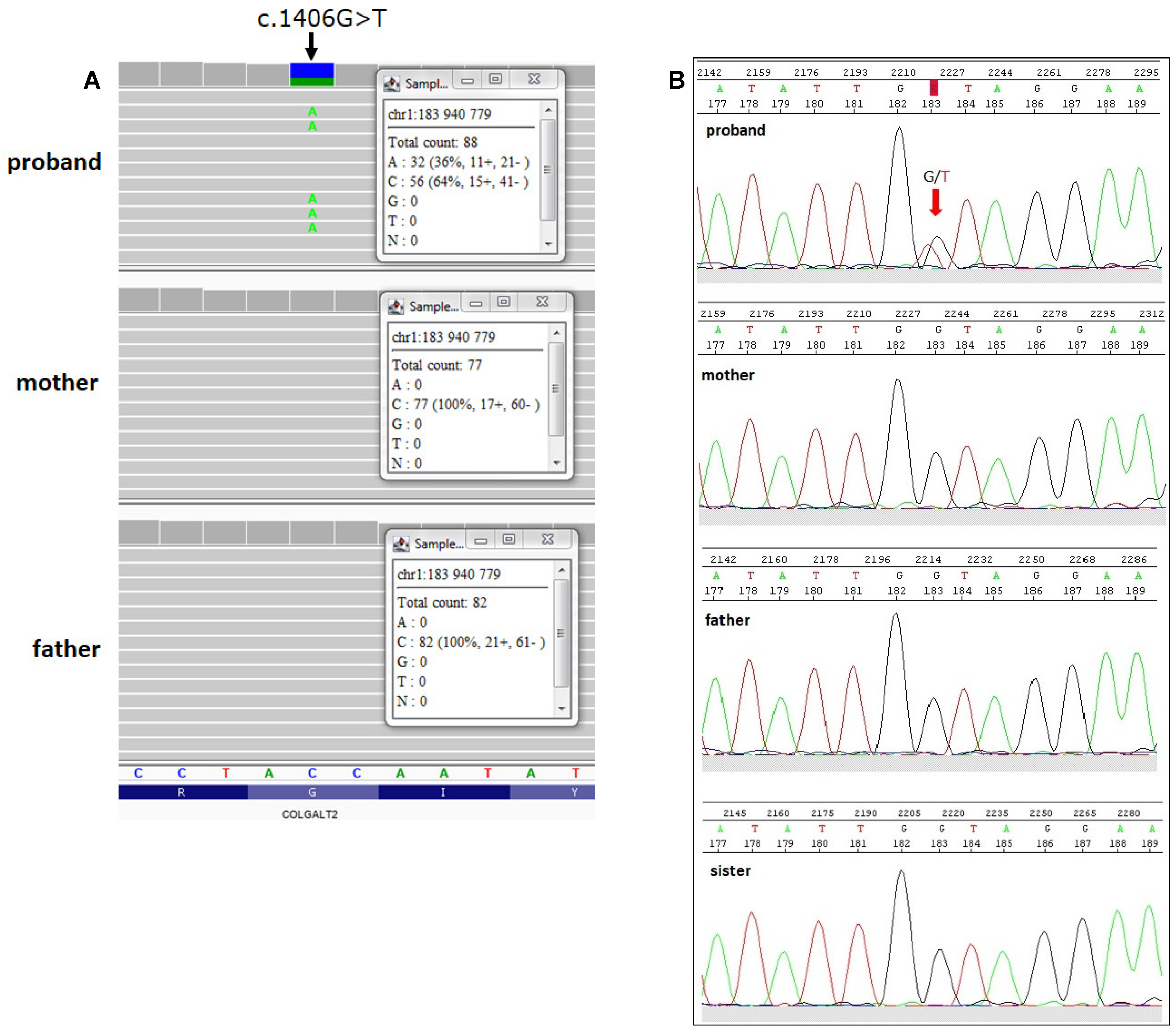
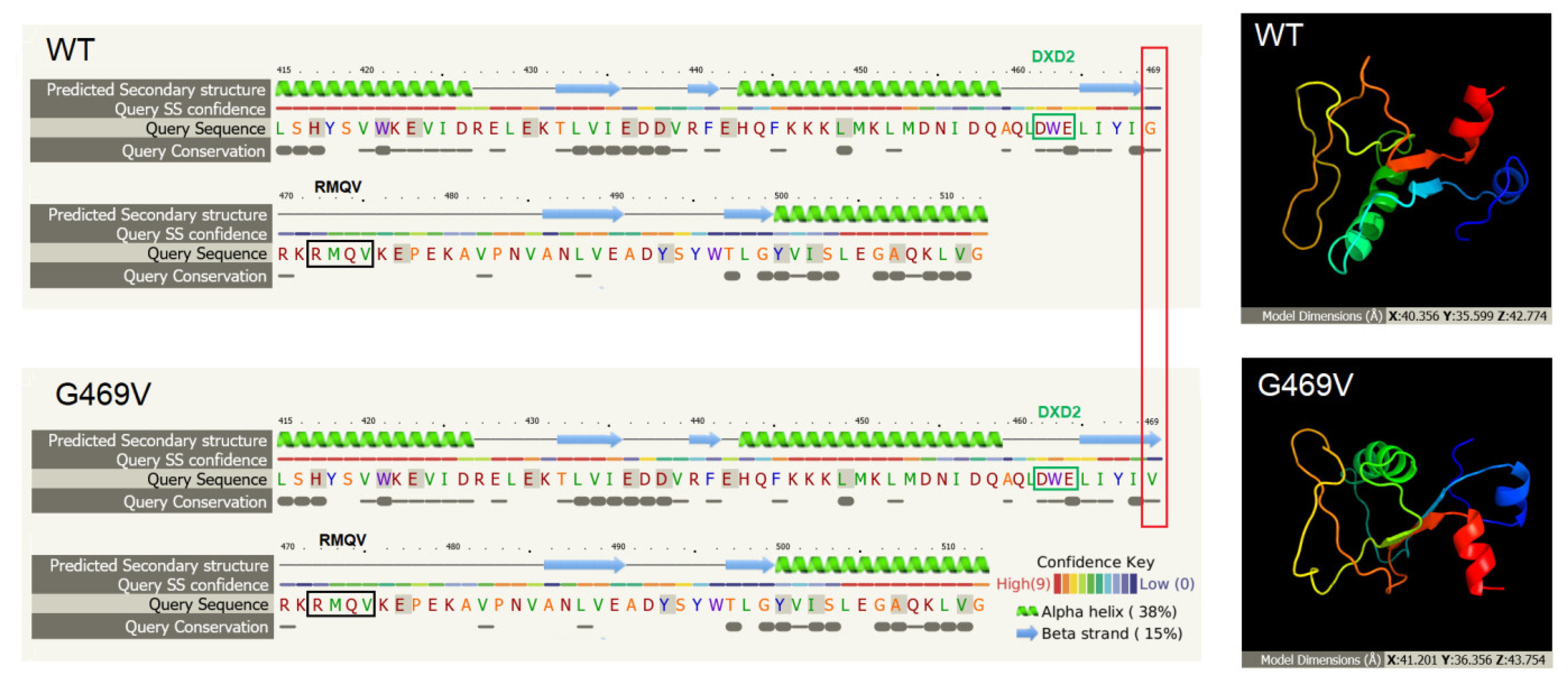
2.2. Identification of COLGALT2 Variant
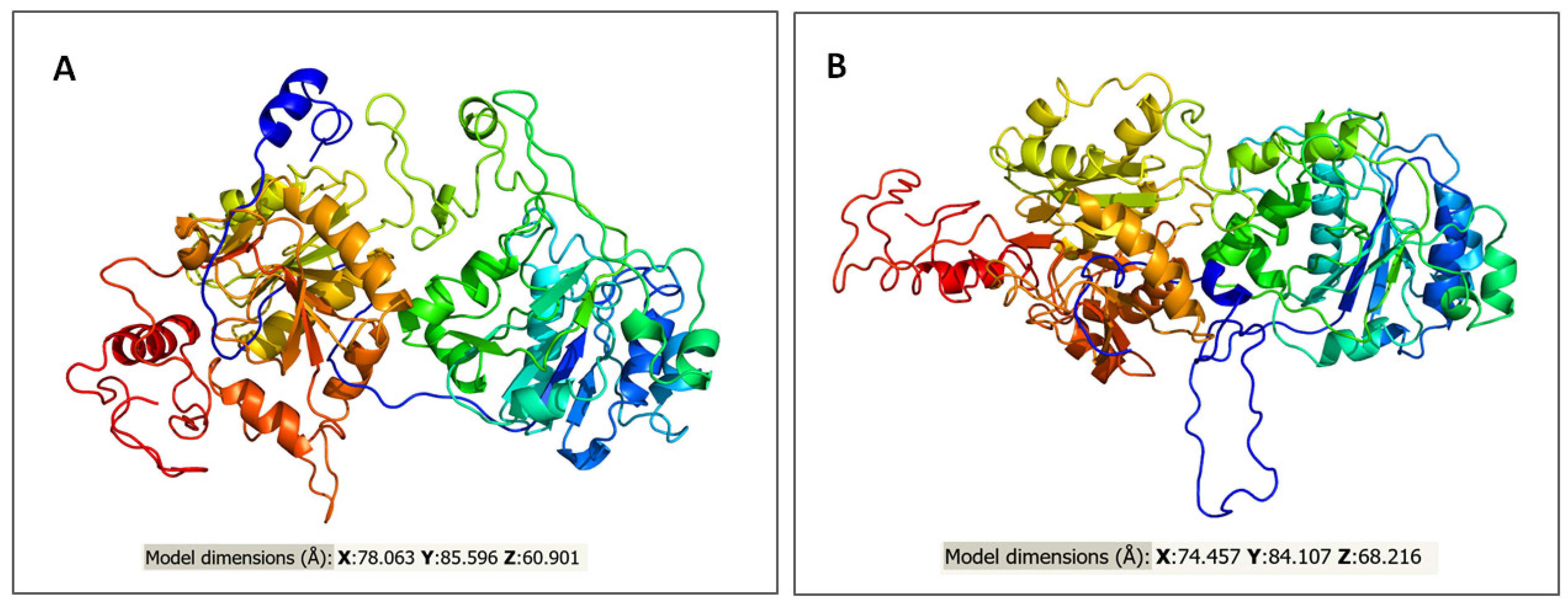
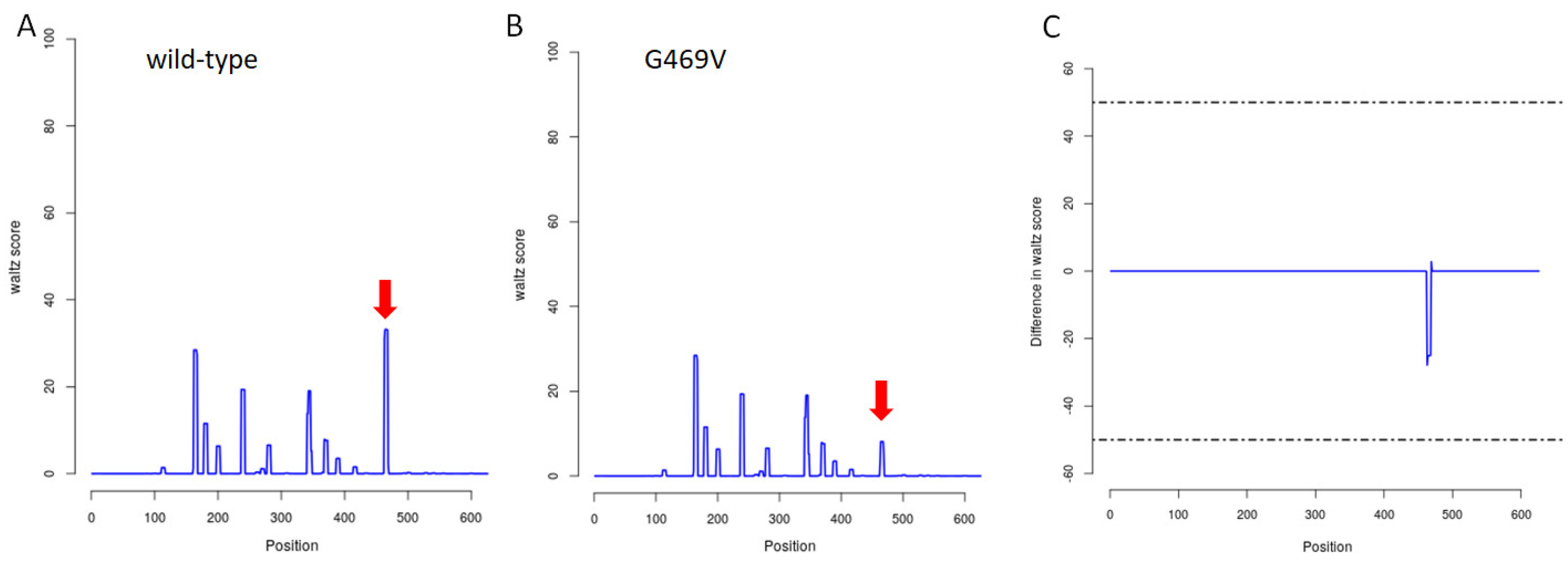
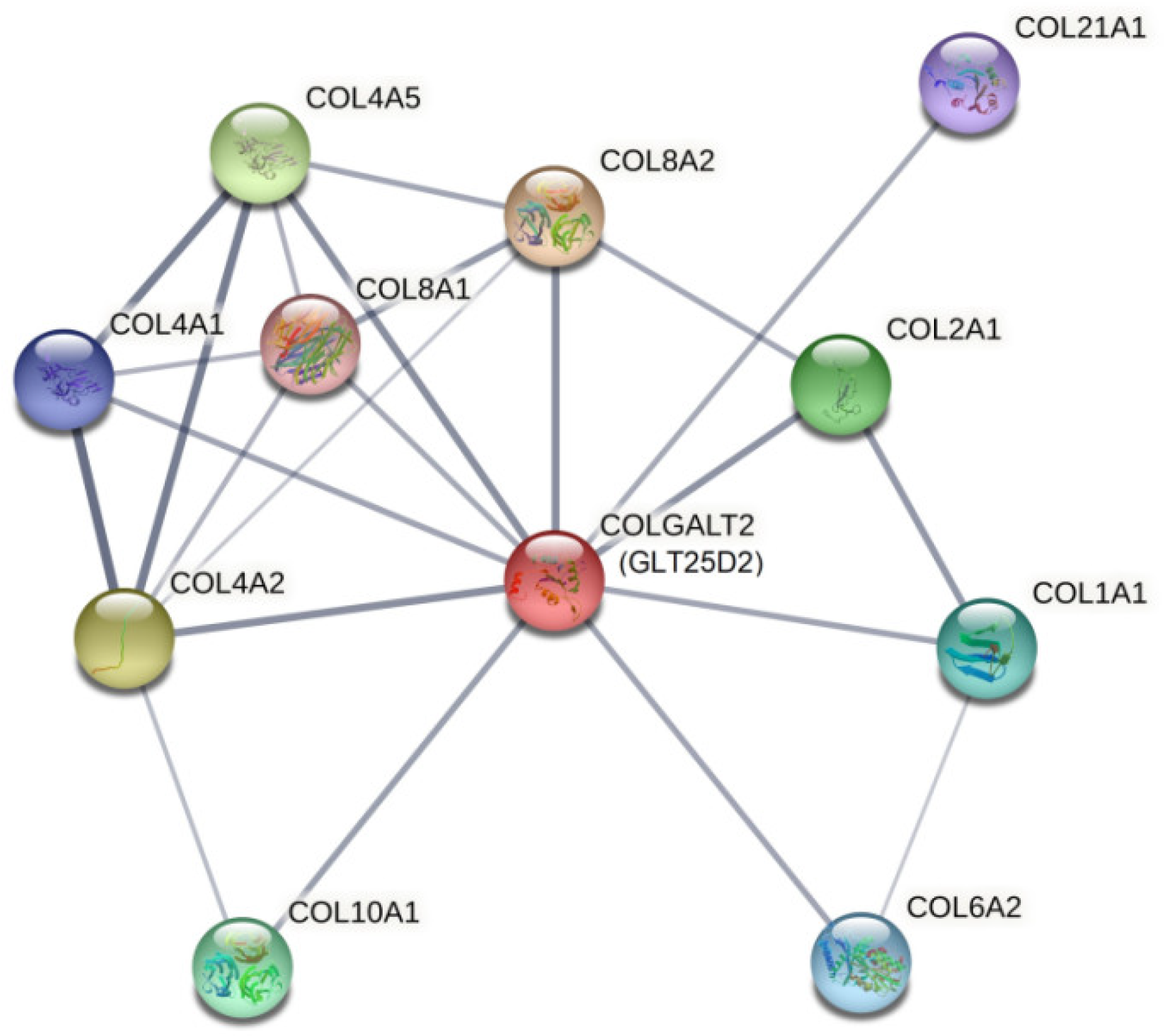
2.3. In Silico Assessment of Protein Structure, Stability and Interactions
3. Discussion
4. Materials and Methods
4.1. Patient
4.2. DNA Extraction
4.3. WES Sequencing
4.4. Validation by Sanger Sequencing
4.5. In Silico Tools
4.5.1. Prediction of Variant Impact on the Protein Structure, Function and Stability
4.5.2. Molecular Phenotypic Characterization of Protein Variants
4.5.3. Prediction of Protein Interactions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corman, M.L. Rectal prolapse in children. Dis. Colon Rectum 1985, 28, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Rentea, R.M.; Peter, S.D.S. Pediatric rectal prolapse. Clin. Colon Rectal Surg. 2018, 31, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Kerkhof, M.; Hendriks, L.; Brölmann, H. Changes in connective tissue in patients with pelvic organ prolapse-A review of the current literature. Int. Urogynecol. J. Pelvic Floor Dysfunct. 2009, 20, 461–474. [Google Scholar] [CrossRef] [Green Version]
- Hennet, T. Collagen glycosylation. Curr. Opin. Struct. Biol. 2019, 56, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Salo, A.; Myllyharju, J. Prolyl and lysyl hydroxylases in collagen synthesis. Exp. Dermatol. 2021, 30, 38–49. [Google Scholar] [CrossRef]
- Basak, T.; Vega-Montoto, L.; Zimmerman, L.; Tabb, D.; Hudson, B.; Vanacore, R. Comprehensive characterization of glycosylation and hydroxylation of basement membrane collagen iv by high-resolution mass spectrometry. J. Proteome Res. 2016, 15, 245–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, S.; Hennet, T. Collagen accumulation in osteosarcoma cells lacking glt25d1 collagen galactosyltransferase. J. Biol. Chem. 2016, 291, 18514–18524. [Google Scholar] [CrossRef] [Green Version]
- Perrin-Tricaud, C.; Rutschmann, C.; Hennet, T. Identification of domains and amino acids essential to the collagen galactosyltransferase activity of glt25d1. PLoS ONE 2011, 6, e29390. [Google Scholar] [CrossRef] [Green Version]
- Venselaar, H.; Te Beek, T.A.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef] [Green Version]
- Yan, B.; Sun, Y. Glycine residues provide flexibility for enzyme active sites. J. Biol. Chem. 2022, 272, 3190–3194. [Google Scholar] [CrossRef] [Green Version]
- Deller, M.; Kong, L.; Rupp, B. Protein stability: A crystallographer’s perspective. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2016, 72, 72–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schegg, B.; Hülsmeier, A.; Rutschmann, C.; Maag, C.; Hennet, T. Core glycosylation of collagen is initiated by two beta(1-o)galactosyltransferases. Mol. Cell. Biol. 2009, 29, 943–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, M.; Sricholpech, M. Lysine post-translational modifications of collagen. Essays Biochem. 2012, 52, 113–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhakar, A.; Kalluri, R. Molecular Mechanisms of Angiostasis; Academic Press: Cambridge, MA, USA, 2010; pp. 52–59. [Google Scholar]
- Abreu-Velez, A.M.; Howard, M.S. Collagen IV in normal skin and in pathological processes. N. Am. J. Med Sci. 2012, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Valdoz, J.C.; Johnson, B.C.; Jacobs, D.J.; Franks, N.A.; Dodson, E.L.; Sanders, C.; Cribbs, C.G.; Van Ry, P.M. The ECM: To Scaffold, or Not to Scaffold, That Is the Question. Int. J. Mol. Sci. 2021, 22, 12690. [Google Scholar] [CrossRef]
- Aumailley, M.; Timpl, R. Attachment of cells to basement membrane collagen type IV. J. Cell Biol. 1986, 103, 1569–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudko, S.P.; Danylevych, N.; Hudson, B.G.; Pedchenko, V.K. Basement membrane collagen IV: Isolation of functional domains. In Methods in Cell Biology; Academic Press: Cambridge, MA, USA, 2017; Volume 143, pp. 171–185. [Google Scholar] [CrossRef]
- Sipilä, L.; Ruotsalainen, H.; Sormunen, R.; Baker, N.L.; Lamande, S.; Vapola, M.; Wang, C.; Sado, Y.; Aszodi, A.; Myllylä, R. Secretion and Assembly of Type IV and VI Collagens Depend on Glycosylation of Hydroxylysines. J. Biol. Chem. 2007, 282, 33381–33388. [Google Scholar] [CrossRef] [Green Version]
- Jürgensen, H.J.; Madsen, D.H.; Ingvarsen, S.; Melander, M.C.; Gårdsvoll, H.; Patthy, L.; Engelholm, L.H.; Behrendt, N. A Novel Functional Role of Collagen Glycosylation. J. Biol. Chem. 2011, 286, 32736–32748. [Google Scholar] [CrossRef] [Green Version]
- Herzog, C.; Zhuang, L.; Gorgan, L.; Segal, Y.; Zhou, J. Tissue- and developmental stage-specific activation of α5 and α6(IV) collagen expression in the upper gastrointestinal tract of transgenic mice. Biochem. Biophys. Res. Commun. 2003, 311, 553–560. [Google Scholar] [CrossRef]
- Kendall, C.H.; Sanderson, P.R.; Cope, J.; Talbot, I.C. Follicular thyroid tumours: A study of laminin and type IV collagen in basement membrane and endothelium. J. Clin. Pathol. 1985, 38, 1100–1105. [Google Scholar] [CrossRef] [Green Version]
- Kehayova, Y.S.; Watson, E.; Wilkinson, J.M.; Loughlin, J.; Rice, S.J. Genetic and Epigenetic Interplay Within a COLGALT2 Enhancer Associated With Osteoarthritis. Arthritis Rheumatol. 2021, 73, 1856–1865. [Google Scholar] [CrossRef]
- Miyatake, S.; Schneeberger, S.; Koyama, N.; Yokochi, K.; Ohmura, K.; Shiina, M.; Mori, H.; Koshimizu, E.; Imagawa, E.; Uchiyama, Y.; et al. Biallelic COLGALT1 variants are associated with cerebral small vessel disease. Ann. Neurol. 2018, 84, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Śmigiel, R.; Biela, M.; Szmyd, K.; Błoch, M.; Szmida, E.; Skiba, P.; Walczak, A.; Gasperowicz, P.; Kosińska, J.; Rydzanicz, M.; et al. Rapid Whole-Exome Sequencing as a Diagnostic Tool in a Neonatal/Pediatric Intensive Care Unit. J. Clin. Med. 2020, 9, 2220. [Google Scholar] [CrossRef] [PubMed]
- De Baets, G.; Van Durme, J.; Reumers, J.; Maurer-Stroh, S.; Vanhee, P.; Dopazo, J.; Schymkowitz, J.; Rousseau, F. SNPeffect 4.0: On-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2011, 40, D935–D939. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadakierska-Chudy, A.; Szymanowski, P.; Lebioda, A.; Płoski, R. Identification and In Silico Characterization of a Novel COLGALT2 Gene Variant in a Child with Mucosal Rectal Prolapse. Int. J. Mol. Sci. 2022, 23, 3670. https://doi.org/10.3390/ijms23073670
Sadakierska-Chudy A, Szymanowski P, Lebioda A, Płoski R. Identification and In Silico Characterization of a Novel COLGALT2 Gene Variant in a Child with Mucosal Rectal Prolapse. International Journal of Molecular Sciences. 2022; 23(7):3670. https://doi.org/10.3390/ijms23073670
Chicago/Turabian StyleSadakierska-Chudy, Anna, Paweł Szymanowski, Arleta Lebioda, and Rafał Płoski. 2022. "Identification and In Silico Characterization of a Novel COLGALT2 Gene Variant in a Child with Mucosal Rectal Prolapse" International Journal of Molecular Sciences 23, no. 7: 3670. https://doi.org/10.3390/ijms23073670
APA StyleSadakierska-Chudy, A., Szymanowski, P., Lebioda, A., & Płoski, R. (2022). Identification and In Silico Characterization of a Novel COLGALT2 Gene Variant in a Child with Mucosal Rectal Prolapse. International Journal of Molecular Sciences, 23(7), 3670. https://doi.org/10.3390/ijms23073670





