Cross-Omics Analysis of Fenugreek Supplementation Reveals Beneficial Effects Are Caused by Gut Microbiome Changes Not Mammalian Host Physiology
Abstract
:1. Introduction
2. Results
2.1. Significant Differences between Small Intestine, Large Intestine, Liver, and Serum
2.2. Metabolomics Data Indicate Significant Differences as a Result of Fenugreek Supplementation
2.2.1. Small Intestine
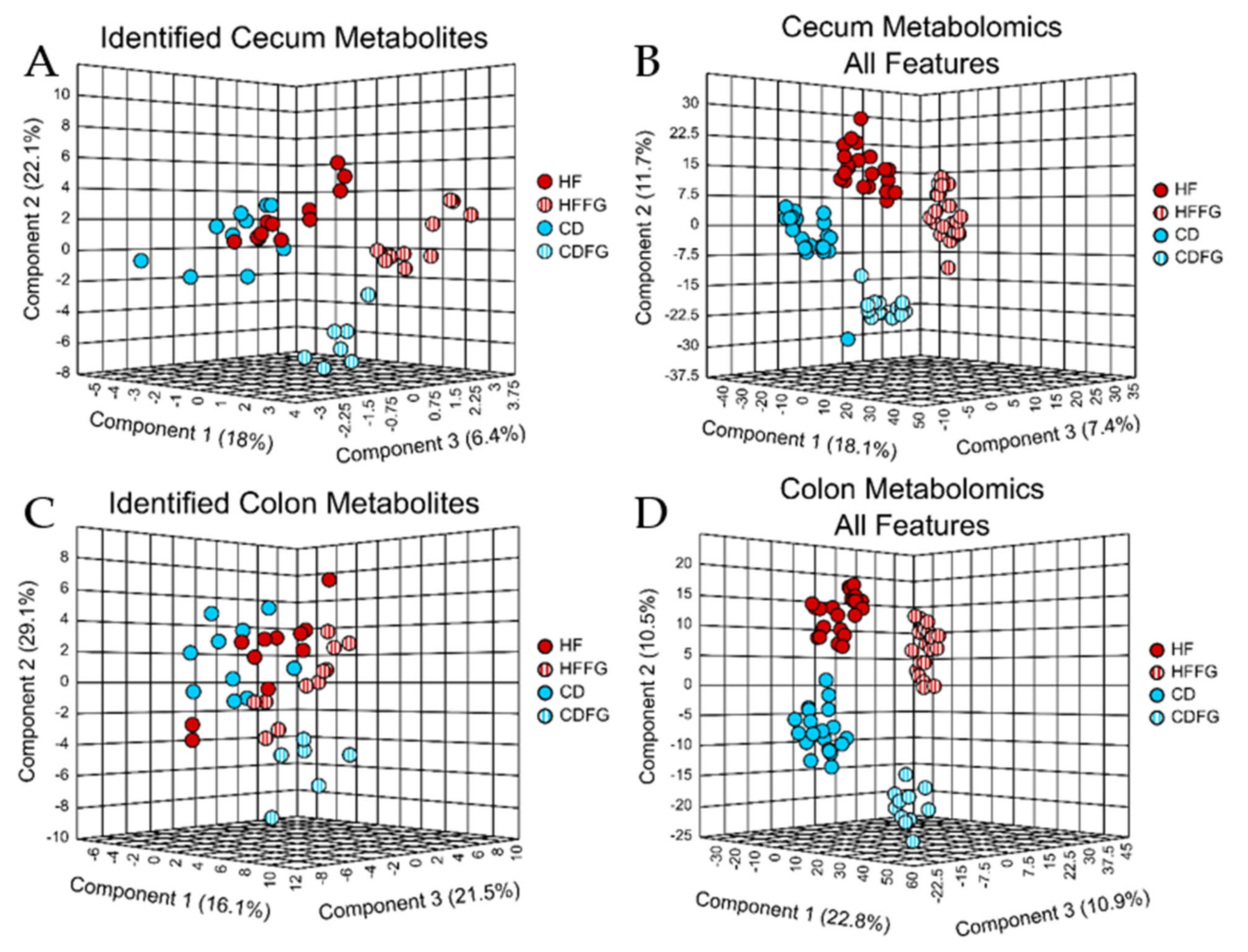
2.2.2. Large Intestine
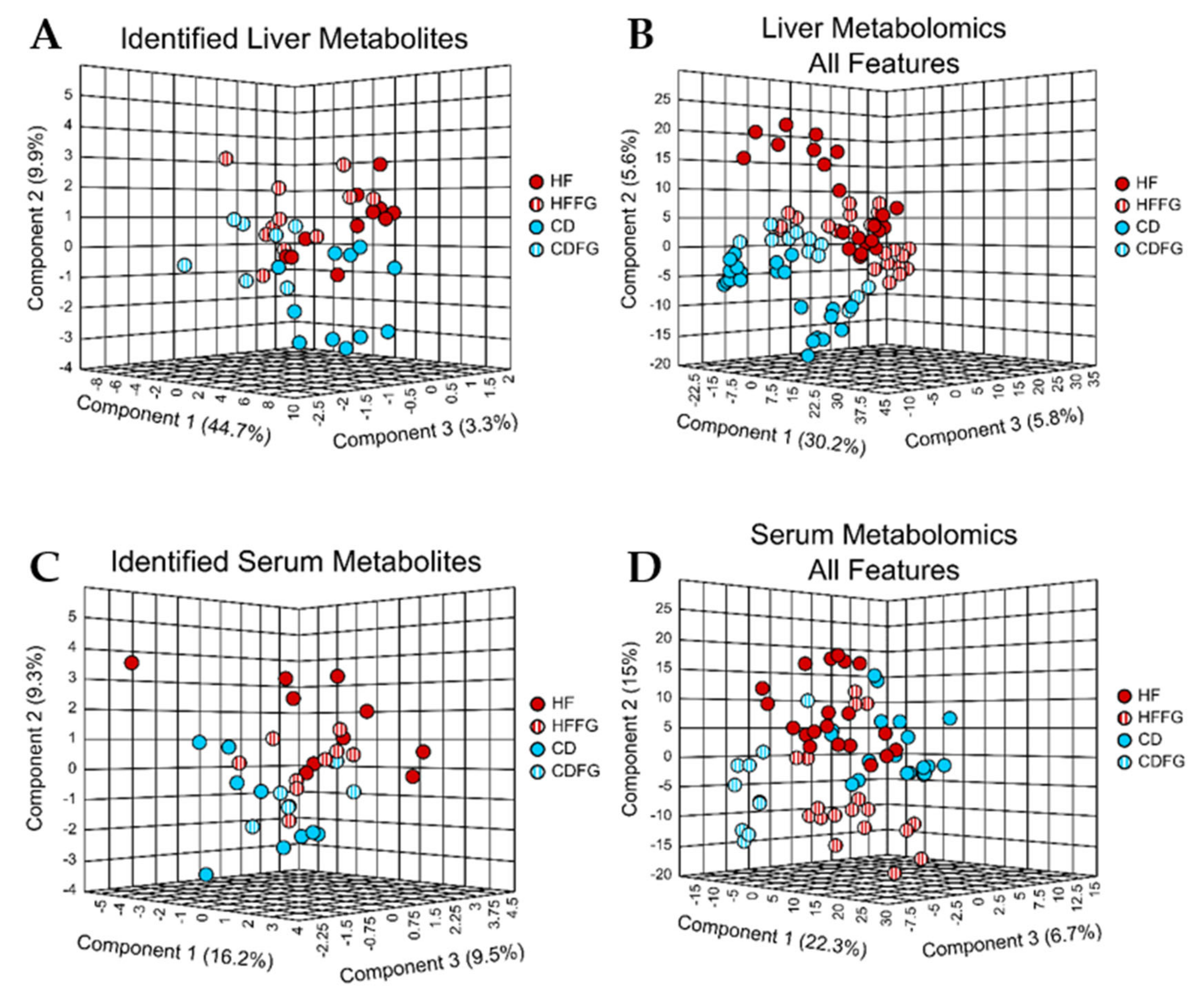
2.2.3. Liver
2.2.4. Serum
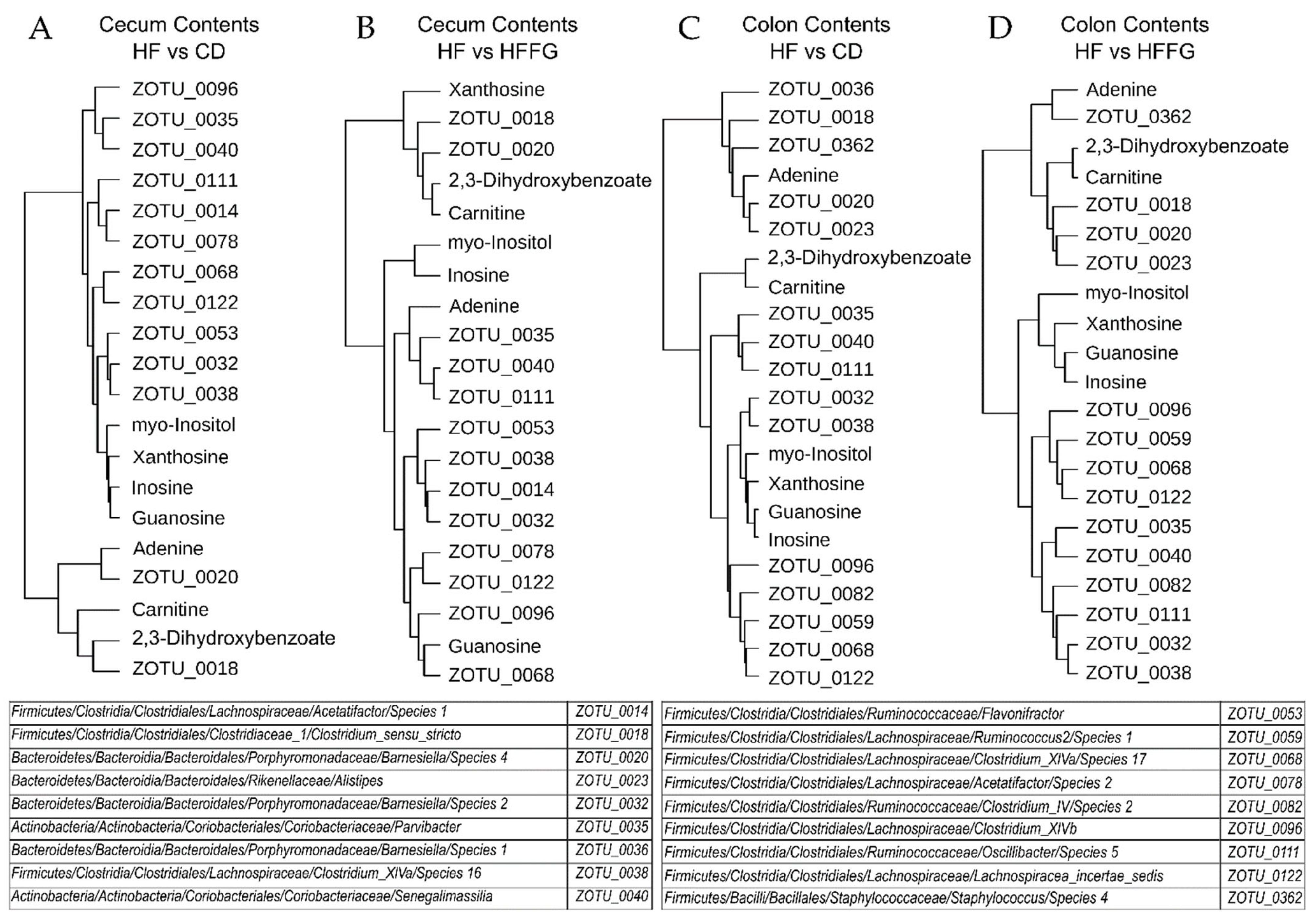
2.3. Metagenomic Analysis Reveal Notable Modifications in Gut Microbiome of FG Supplemented Mice
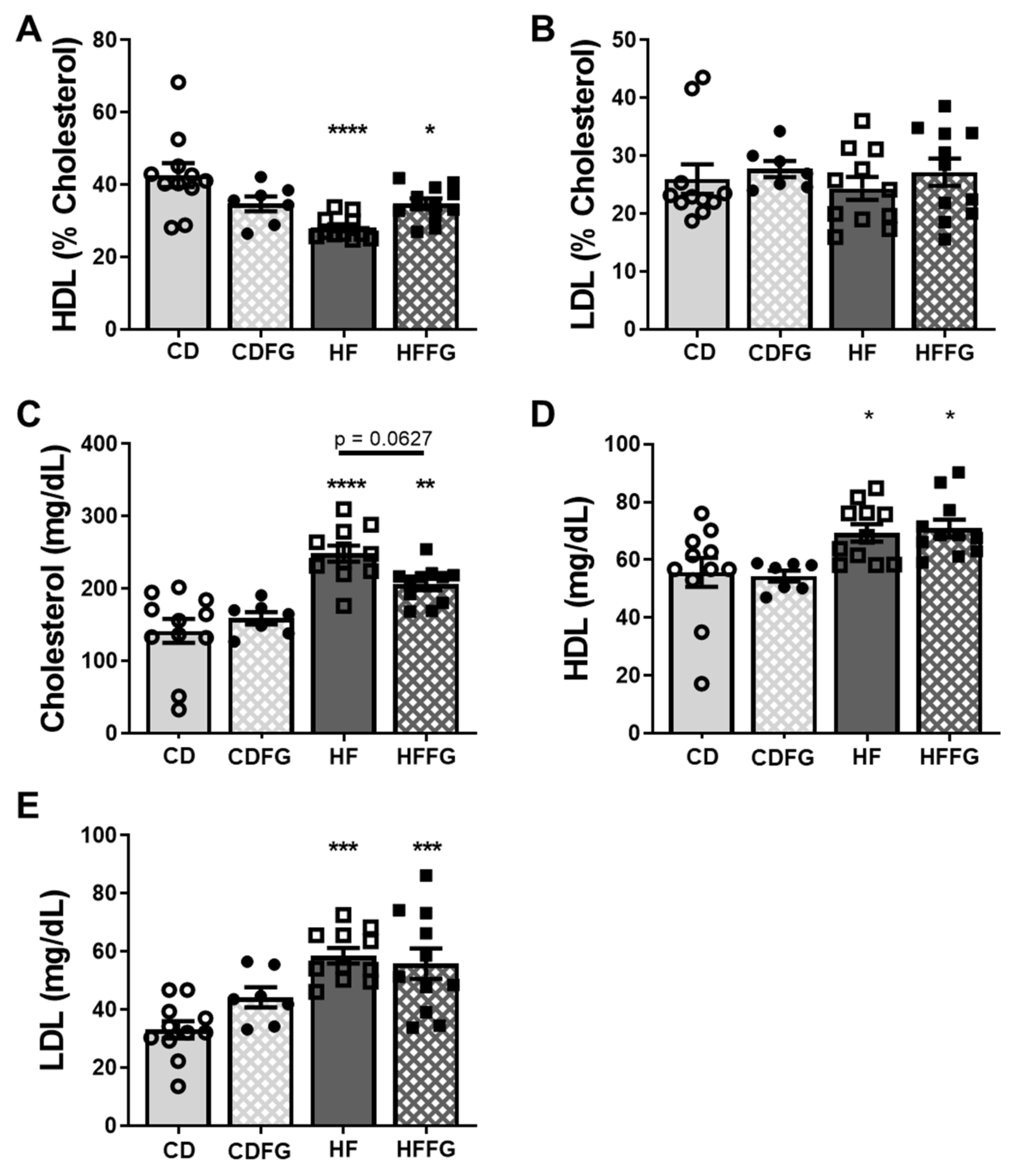
2.4. Fenugreek Supplementation Modulates HDL Balance and Total Cholesterol in HF Diet-Fed Mice
3. Discussion
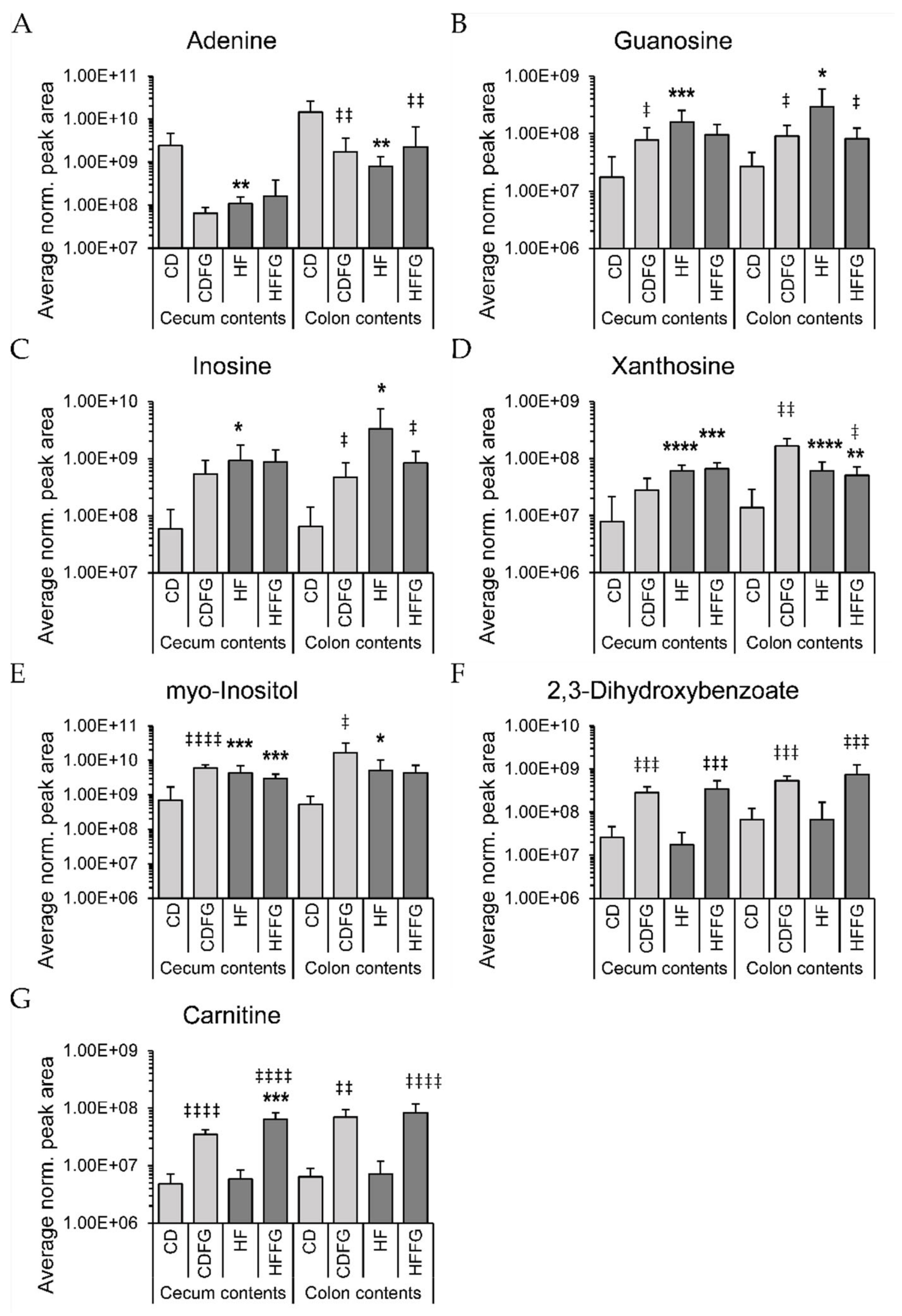
3.1. High Fat Diet Influences Purine Metabolite Abundances
3.2. Fenugreek Induces Considerable Changes in the Large Intestines
3.3. Liver Metabolome Is Affected by Fenugreek
3.4. Fenugreek Impacts Specific Pathways by Location
4. Methods
4.1. Animals and Diets
4.2. Metabolic Phenotyping
4.3. Metagenomic Sequencing
4.4. Metabolite Extractions
4.5. UHPLC-HRMS
4.6. Metabolomics Data Processing
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. NIH/NCCIH Product Integrity Form for Fenugreek Seed in Animal Studies
Appendix A.1. Identify the Source Plant for the Product Using the Scientific Taxonomic Nomenclature and Author Citation
Appendix A.2. Describe the Parts of the Plant from Which the Product Is Derived. If an Extract Is Used, Identify the Extraction Solvent
Appendix A.3. Name the Supplier of the Product. Include the Brand Name of the Product, If Applicable
Appendix A.4. Present Data on the Characterization (e.g., Chemical Profile or Fingerprint) of the Final Product as Thoroughly as the State of the Science Allows
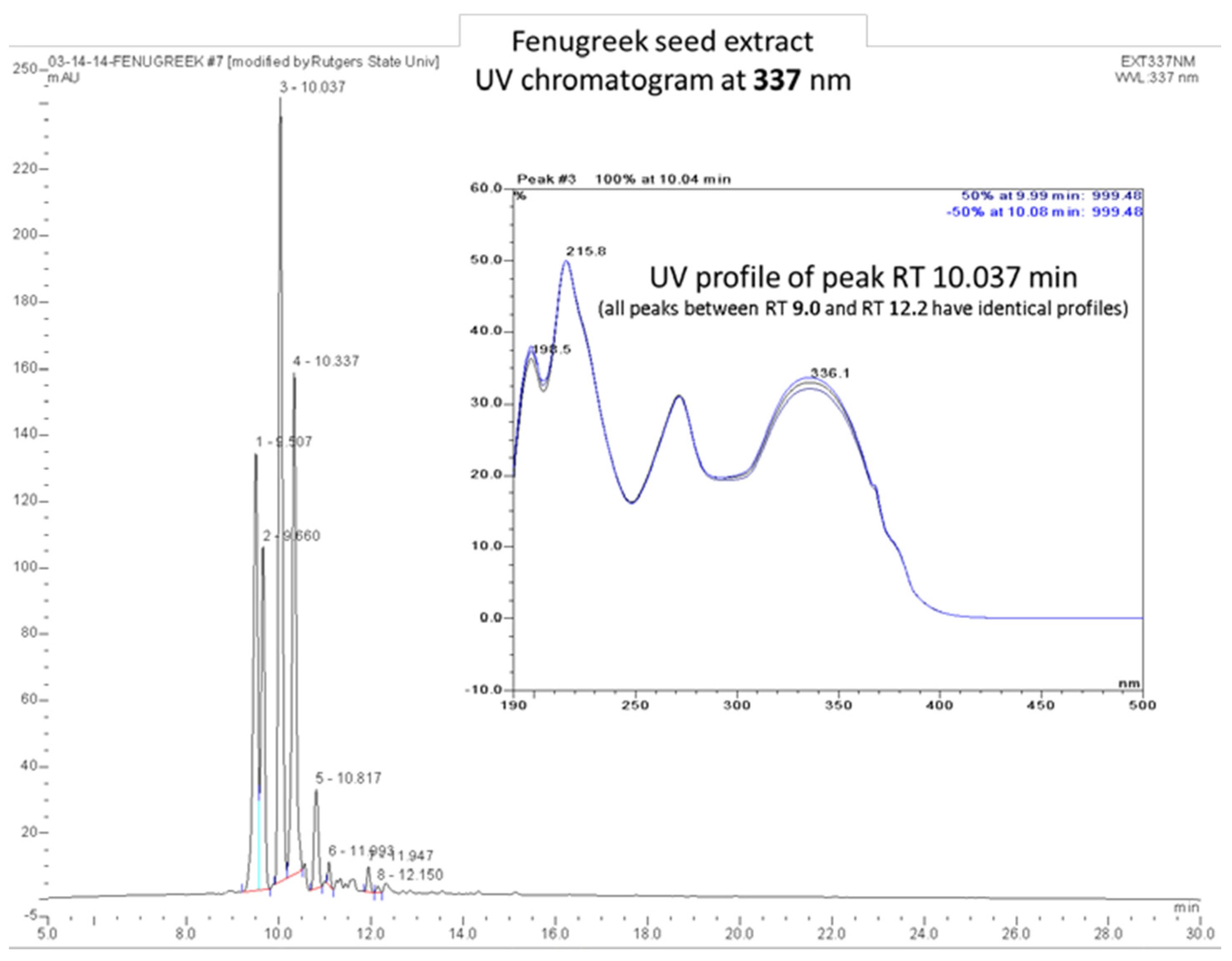
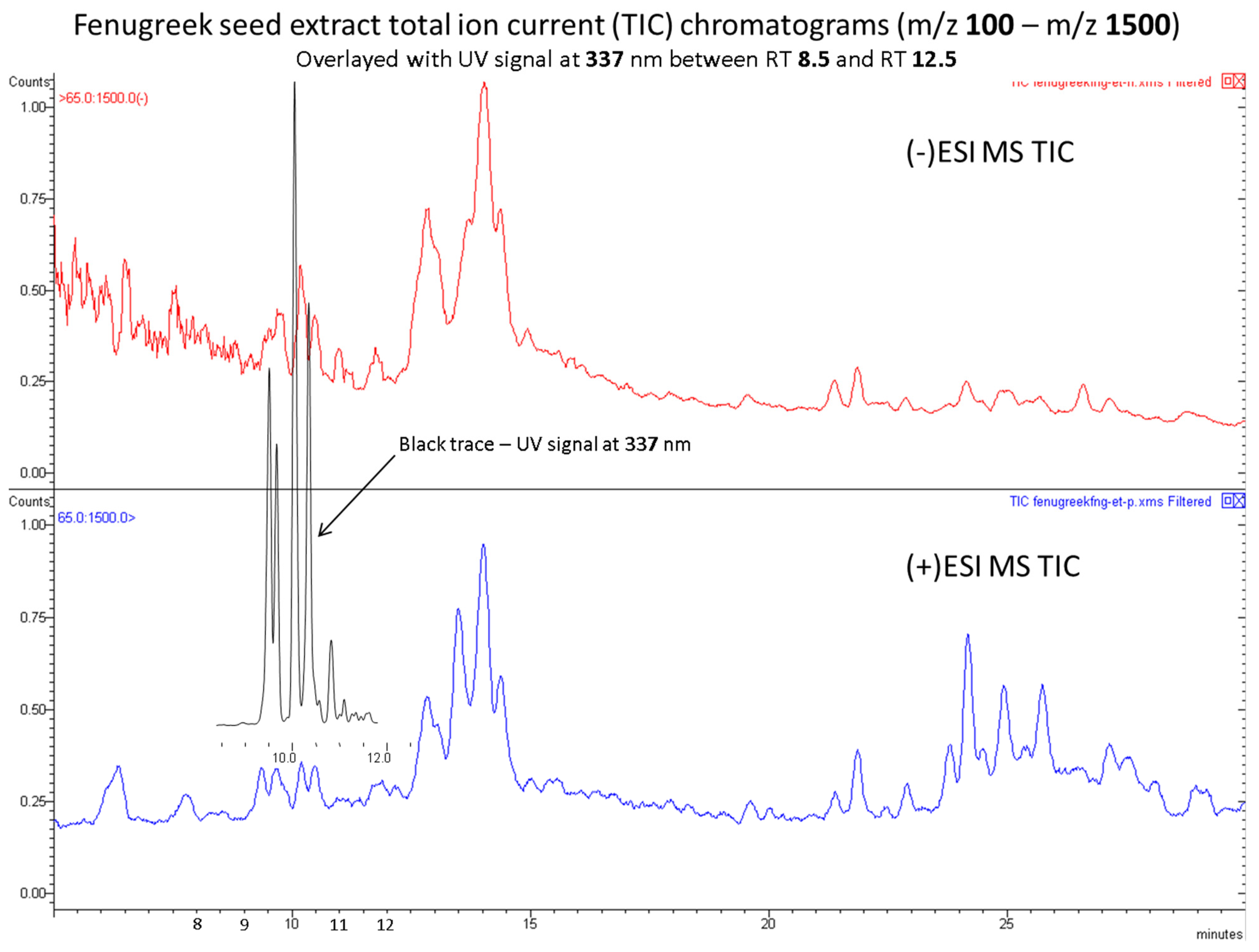
Appendix A.5. Provide a Certificate of Analysis from the Supplier to Show Compliance to Their Specifications for Content and Any Other Supporting Information Relating to the Batches to Be Used in the Study. This Should Include Data on the Analysis of the Product for Contaminants/Impurities Such as Pesticides, Heavy Metals, and Residual Solvents
Appendix A.6. Provide Data on Batch-To-Batch Reproducibility. These Data May Be Required Separately for the Botanical Alone, the Vehicle Alone, and the Final Product, as Appropriate

| Peak No. | Mass | Formula | Compound(s) | Reported from Trigonella (Reaxys.com) |
|---|---|---|---|---|
| 1 | Mix of 324, 444 | C21H32O10, C13H24O9 | Organic acid glycosides? | No; 32 structures |
| 2 | 594 | C27H30O15 | Flavone (most likely Quercetin) disaccharide | Yes; 2 structures |
| 3 and 4 | 564 | C26H28O14 | Flavone disaccharides? | No; >100 structures |
| 5 and 6 | 448 | C21H20O11 | Flavone (Quercitrin, Luteolin, Vitexin) glycosides | Yes; 4 structures |
| 7 and 8 | 534 | C25H26O13 | Possibly flavone disaccharides, Caffeoyl-syringoyl quinic acids, etc. | No; 40 structures |
| 9 | 432 | C21H20O10 | Vitexin and/or Kaempferol-rhamnoside | Yes; 2 structures |
| 10 | 548 | C26H28O13 | Wide variety of polyphenolic (di-)saccharides | No; 47 structures |
| 11 | 504 | C24H30O11 | Megastigmane glycoside(s)? | No; 24 structures |
| 12 | 904 | C44H72O19 | Furostane and/or spirostane trisaccharides | No; 5 structures |
| 13 and 14 | 906 | C44H74O19 | Furostane and/or spirostane trisaccharides (Trigoneosides Ia, Ib) | Yes; 3 structures |
| 15 | 920 | C45H76O19 | Furostane trisaccharides | Yes; 2 structures |
| 16 | 738 | C39H64O13 | Spirostane disaccharides, saponins, etc. | No; 51 structures |
| 17 | 888 | C44H72O18 | Furostane and/or spirostane trisaccharides, saponins, etc. | No; 23 structures |
| 890 | C44H74O18 | Furostane trisaccharides | Yes; 2 structures | |
| 1048 | C51H84O22 | Furostane tetrasaccharides (Trigoneosides, or Trigonellosides) | Yes; 2 structures | |
| 18 | 902 | C45H74O18 | Furostane trisaccharides; Trigofoenoside A | Yes; 4 structures |
| 19 | 1046 | C51H82O22 | Furo-/spirostane tetrasaccharides | No; 40 structures |
| 872 | C44H72O17 | Spirostane trisaccharides | No; 27 structures | |
| 20 | 888 | C45H76O17 | Cholestane trisaccharides | No; 9 structures |
| 21 | 644 | C34H44O12 | Cyclopentane perhydro-phenanthrene derivatives | No; 18 structures |
Appendix A.7. Present a Plan to Monitor the Stability of Product Samples from All Batches Used during the Course of the Study
Appendix A.8. Describe the Method of Authentication of the Raw Material and Where and How an Authenticated Voucher Specimen of the Plant Material Is Reserved
Appendix A.9. Describe the Manufacturing Process
Appendix A.10. If the Product Is Combined with a Diet, a Certificate of Analysis and Specifications for the Vehicle Will Also Be Necessary to Assure Purity, Consistency, Absence of Bioactive Components (e.g., Soy), and/or Reproducibility of the Vehicle. Furthermore, Analysis of the Formulated Diet May Be Required to Assess Consistency, Stability, etc. of the Product in this Matrix
| D12450J | D16020408 | D12492 | D16020408 | |
|---|---|---|---|---|
| Product # | 10 kcal% Fat | 10 kcal% Fat | 60 kcal% Fat | 60 kcal% Fat |
| Fenugreek | Fenugreek | |||
| Ingredient | Seeds | Seeds | ||
| Casein | 200 | 193.4 | 200 | 193.4 |
| L-Cystine | 3 | 3 | 3 | 3 |
| Corn Starch | 506.2 | 495.4 | ||
| Maltodextrin 10 | 125 | 125 | 125 | 114.2 |
| Sucrose | 68.8 | 68.8 | 68.8 | 68.8 |
| Cellulose | 50 | 48.6 | 50 | 48.6 |
| Lard | 20 | 18.7 | 245 | 243.7 |
| Soybean Oil | 25 | 25 | 25 | 25 |
| Mineral Mix S10026 | 10 | 10 | 10 | 10 |
| Dicalcium Phosphate | 13 | 13 | 13 | 13 |
| Calcium Carbonate | 5.5 | 5.5 | 5.5 | 5.5 |
| Potassium Citrate, 1 H2O | 16.5 | 16.5 | 16.5 | 16.5 |
| Vitamin Mix V10001 | 10 | 10 | 10 | 10 |
| Choline Bitartrate | 2 | 2 | 2 | 2 |
| Fenugreek Seeds | 0 | 21.1 | 0 | 21.1 |
| Red Dye #40, FD&C | 0 | 0.025 | 0 | 0.05 |
| Blue Dye #1, FD&C | 0.01 | 0.025 | 0.05 | 0 |
| Yellow Dye #5, FD&C | 0.04 | 0 | 0 | 0 |
| Total | 1055.05 | 1056.05 | 773.85 | 774.85 |
| D12492 | D16020408 | D12492 | D16020408 | |
| gm | ||||
| Protein | 179 | 179 | 179 | 179 |
| Carbohydrate | 710 | 710 | 203.8 | 203.8 |
| Fat | 47.4 | 47.4 | 272.4 | 272.4 |
| Fiber | 50 | 50 | 50 | 50 |
| gm% | ||||
| Protein | 17 | 16.9 | 23.1 | 23.1 |
| Carbohydrate | 0 | 0 | 26.3 | 26.3 |
| Fat | 4.5 | 4.5 | 35.2 | 35.2 |
| Fiber | 4.7 | 4.7 | 6.5 | 6.5 |
| Fenugreek seeds | 0 | 2 | 0 | 2.72 |
| kcals | ||||
| Protein | 716 | 716 | 716 | 716 |
| Carbohydrate | 2840 | 2840 | 815 | 815 |
| Fat | 427 | 427 | 2452 | 2452 |
| Total | 3983 | 3983 | 3983 | 3983 |
| kcal% | ||||
| Protein | 18 | 18 | 18 | 18 |
| Carbohydrate | 71 | 71 | 20 | 20 |
| Fat | 11 | 11 | 62 | 62 |
| Total | 100 | 100 | 100 | 100 |
| kcal/gm | 3.8 | 3.8 | 5.1 | 5.1 |
References
- Caballero, B. Humans against Obesity: Who Will Win? Adv. Nutr. 2019, 10, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.V.; Donahue, M.; Pi-Sunyer, F.X.; Fuster, V. Obesity as a risk factor in coronary artery disease. Am. Heart J. 2001, 142, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Khaodhiar, L.; McCowen, K.C.; Blackburn, G.L. Obesity and its comorbid conditions. Clin. Cornerstone 1999, 2, 17–31. [Google Scholar] [CrossRef]
- Gregor, M.F.; Hotamisligil, G.S. Inflammatory Mechanisms in Obesity. Annu. Rev. Immunol. 2011, 29, 415–445. [Google Scholar] [CrossRef] [Green Version]
- Simon, G.E.; Von Korff, M.; Saunders, K.; Miglioretti, D.L.; Crane, P.K.; van Belle, G.; Kessler, R.C. Association Between Obesity and Psychiatric Disorders in the US Adult Population. Arch. Gen. Psychiatry 2006, 63, 824–830. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Xiao, L. Obesity and Depression in US Women: Results From the 2005–2006 National Health and Nutritional Examination Survey. Obesity 2010, 18, 347–353. [Google Scholar] [CrossRef]
- Nissen, S.E.; Wolski, K. Effect of Rosiglitazone on the Risk of Myocardial Infarction and Death from Cardiovascular Causes. N. Engl. J. Med. 2007, 356, 2457–2471. [Google Scholar] [CrossRef] [Green Version]
- Filippatos, T.D.; Panagiotopoulou, T.V.; Elisaf, M.S. Adverse Effects of GLP-1 Receptor Agonists. Rev. Diabet. Stud. 2014, 11, 202–230. [Google Scholar] [CrossRef] [Green Version]
- Piette, J.D.; Heisler, M.; Wagner, T.H. Problems Paying Out-of-Pocket Medication Costs Among Older Adults with Diabetes. Diabetes Care 2004, 27, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Payab, M.; Hasani-Ranjbar, S.; Shahbal, N.; Qorbani, M.; Aletaha, A.; Haghi-Aminjan, H.; Soltani, A.; Khatami, F.; Nikfar, S.; Hassani, S.; et al. Effect of the herbal medicines in obesity and metabolic syndrome: A systematic review and meta-analysis of clinical trials. Phytother. Res. 2020, 34, 526–545. [Google Scholar] [CrossRef]
- Fuller, S.; Stephens, J.M. Diosgenin, 4-Hydroxyisoleucine, and Fiber from Fenugreek: Mechanisms of Actions and Potential Effects on Metabolic Syndrome. Adv. Nutr. 2015, 6, 189–197. [Google Scholar] [CrossRef] [Green Version]
- Garg, R.C. Chapter 44—Fenugreek: Multiple Health Benefits. In Nutraceuticals; Gupta, R.C., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 599–617. [Google Scholar]
- Nagulapalli Venkata, K.C.; Swaroop, A.; Bagchi, D.; Bishayee, A. A small plant with big benefits: Fenugreek (Trigonella foenum-graecum Linn.) for disease prevention and health promotion. Mol. Nutr. Food Res. 2017, 61, 1600950. [Google Scholar] [CrossRef]
- Knott, E.J.; Richard, A.J.; Mynatt, R.L.; Ribnicky, D.; Stephens, J.M.; Bruce-Keller, A. Fenugreek supplementation during high-fat feeding improves specific markers of metabolic health. Sci. Rep. 2017, 7, 12770. [Google Scholar] [CrossRef] [Green Version]
- Bruce-Keller, A.J.; Richard, A.J.; Fernandez-Kim, S.-O.; Ribnicky, D.M.; Salbaum, J.M.; Newman, S.; Carmouche, R.; Stephens, J.M. Fenugreek Counters the Effects of High Fat Diet on Gut Microbiota in Mice: Links to Metabolic Benefit. Sci. Rep. 2020, 10, 1245. [Google Scholar] [CrossRef] [Green Version]
- Zentek, J.; Gärtner, S.; Tedin, L.; Männer, K.; Mader, A.; Vahjen, W. Fenugreek seed affects intestinal microbiota and immunological variables in piglets after weaning. Br. J. Nutr. 2013, 109, 859–866. [Google Scholar] [CrossRef] [Green Version]
- Shtriker, M.G.; Hahn, M.; Taieb, E.; Nyska, A.; Moallem, U.; Tirosh, O.; Madar, Z. Fenugreek galactomannan and citrus pectin improve several parameters associated with glucose metabolism and modulate gut microbiota in mice. Nutrition 2018, 46, 134–142.e3. [Google Scholar] [CrossRef]
- Antharam, V.C.; McEwen, D.C.; Garrett, T.J.; Dossey, A.T.; Li, E.C.; Kozlov, A.N.; Mesbah, Z.; Wang, G.P. An Integrated Metabolomic and Microbiome Analysis Identified Specific Gut Microbiota Associated with Fecal Cholesterol and Coprostanol in Clostridium difficile Infection. PLoS ONE 2016, 11, e0148824. [Google Scholar] [CrossRef] [Green Version]
- Cornejo-Pareja, I.; Muñoz-Garach, A.; Clemente-Postigo, M.; Tinahones, F.J. Importance of gut microbiota in obesity. Eur. J. Clin. Nutr. 2019, 72, 26–37. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [Green Version]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Gaskins, H.R.; McIntosh, M.K. Influence of dietary fat on intestinal microbes, inflammation, barrier function and metabolic outcomes. J. Nutr. Biochem. 2014, 25, 270–280. [Google Scholar] [CrossRef]
- Wells, A.; Barrington, W.T.; Dearth, S.; May, A.; Threadgill, D.W.; Campagna, S.R.; Voy, B.H. Tissue Level Diet and Sex-by-Diet Interactions Reveal Unique Metabolite and Clustering Profiles Using Untargeted Liquid Chromatography–Mass Spectrometry on Adipose, Skeletal Muscle, and Liver Tissue in C57BL6/J Mice. J. Proteome Res. 2018, 17, 1077–1090. [Google Scholar] [CrossRef]
- Pendyala, S.; Walker, J.M.; Holt, P.R. A High-Fat Diet Is Associated with Endotoxemia That Originates from the Gut. Gastroenterology 2012, 142, 1100–1101. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Douglas-Escobar, M.; Elliott, E.; Neu, J. Effect of Intestinal Microbial Ecology on the Developing Brain. JAMA Pediatr. 2013, 167, 374–379. [Google Scholar] [CrossRef]
- Dinan, T.G.; Quigley, E.M. Probiotics in the Treatment of Depression: Science or Science Fiction? Aust. N. Z. J. Psychiatry 2011, 45, 1023–1025. [Google Scholar] [CrossRef]
- Bruce-Keller, A.J.; Salbaum, J.M.; Berthoud, H.-R. Harnessing Gut Microbes for Mental Health: Getting from Here to There. Biol. Psychiatry 2018, 83, 214–223. [Google Scholar] [CrossRef]
- Tillisch, K. The effects of gut microbiota on CNS function in humans. Gut Microbes 2014, 5, 404–410. [Google Scholar] [CrossRef]
- Neufeld, K.M.; Kang, N.; Bienenstock, J.; Foster, J.A. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 2011, 23, 255-e119. [Google Scholar] [CrossRef]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannan, J.M.A.; Rokeya, B.; Faruque, O.; Nahar, N.; Mosihuzzaman, M.; Azad Khan, A.K.; Ali, L. Effect of soluble dietary fibre fraction of Trigonella foenum graecum on glycemic, insulinemic, lipidemic and platelet aggregation status of Type 2 diabetic model rats. J. Ethnopharmacol. 2003, 88, 73–77. [Google Scholar] [CrossRef]
- Narender, T.; Puri, A.; Shweta; Khaliq, T.; Saxena, R.; Bhatia, G.; Chandra, R. 4-Hydroxyisoleucine an unusual amino acid as antidyslipidemic and antihyperglycemic agent. Bioorg. Med. Chem. Lett. 2006, 16, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Hamden, K.; Jaouadi, B.; Carreau, S.; Bejar, S.; Elfeki, A. Inhibitory effect of fenugreek galactomannan on digestive enzymes related to diabetes, hyperlipidemia, and liver-kidney dysfunctions. Biotechnol. Bioprocess Eng. 2010, 15, 407–413. [Google Scholar] [CrossRef]
- Jiang, W.; Gao, L.; Li, P.; Kan, H.; Qu, J.; Men, L.; Liu, Z.; Liu, Z. Metabonomics study of the therapeutic mechanism of fenugreek galactomannan on diabetic hyperglycemia in rats, by ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. J. Chromatogr. B 2017, 1044–1045, 8–16. [Google Scholar] [CrossRef]
- Pedley, A.M.; Benkovic, S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Leal, J.L.; Torralbo, F.; Antonio Quiles, F.; Pineda, M.; Alamillo, J.M. Molecular and functional characterization of allantoate amidohydrolase from Phaseolus vulgaris. Physiol. Plant. 2014, 152, 43–58. [Google Scholar] [CrossRef]
- Witte, C.-P. Urea metabolism in plants. Plant Sci. 2011, 180, 431–438. [Google Scholar] [CrossRef]
- Finkelstein, J.D.; Martin, J.J.; Harris, B.J. Methionine metabolism in mammals. The methionine-sparing effect of cystine. J. Biol. Chem. 1988, 263, 11750–11754. [Google Scholar] [CrossRef]
- Hoppel, C. The role of carnitine in normal and altered fatty acid metabolism. Am. J. Kidney Dis. 2003, 41, S4–S12. [Google Scholar] [CrossRef]
- Bene, J.; Hadzsiev, K.; Melegh, B. Role of carnitine and its derivatives in the development and management of type 2 diabetes. Nutr. Diabetes 2018, 8, 8. [Google Scholar] [CrossRef]
- Flanagan, J.L.; Simmons, P.A.; Vehige, J.; Willcox, M.D.P.; Garrett, Q. Role of carnitine in disease. Nutr. Metab. 2010, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Ouwerkerk, J.P.; Aalvink, S.; Belzer, C.; de Vos, W.M. Akkermansia glycaniphila sp. nov. an anaerobic mucin-degrading bacterium isolated from reticulated python faeces. Int. J. Syst. Evol. Microbiol. 2016, 66, 4614–4620. [Google Scholar] [CrossRef]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov. sp. nov. a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, J.; Lin, J.; Isnard, S.; Fombuena, B.; Peng, X.; Marette, A.; Routy, B.; Messaoudene, M.; Chen, Y.; Routy, J.-P. The Bacterium Akkermansia muciniphila: A Sentinel for Gut Permeability and Its Relevance to HIV-Related Inflammation. Front. Immunol. 2020, 11, 645. [Google Scholar] [CrossRef]
- Zhou, K. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J. Funct. Foods 2017, 33, 194–201. [Google Scholar] [CrossRef]
- Tu, P.; Bian, X.; Chi, L.; Gao, B.; Ru, H.; Knobloch, T.J.; Weghorst, C.M.; Lu, K. Characterization of the Functional Changes in Mouse Gut Microbiome Associated with Increased Akkermansia muciniphila Population Modulated by Dietary Black Raspberries. ACS Omega 2018, 3, 10927–10937. [Google Scholar] [CrossRef]
- Iglesias-Carres, L.; Hughes, M.D.; Steele, C.N.; Ponder, M.A.; Davy, K.P.; Neilson, A.P. Use of dietary phytochemicals for inhibition of trimethylamine N-oxide formation. J. Nutr. Biochem. 2021, 91, 108600. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Marc Rhoads, J.; Carey Satterfield, M.; Smith, S.B.; Spencer, T.E.; Yin, Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allerton, T.D.; Proctor, D.N.; Stephens, J.M.; Dugas, T.R.; Spielmann, G.; Irving, B.A. l-Citrulline Supplementation: Impact on Cardiometabolic Health. Nutrients 2018, 10, 921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; Kimball, E. Acidic Acetonitrile for Cellular Metabolome Extraction from Escherichia coli. Anal. Chem. 2007, 79, 6167–6173. [Google Scholar] [CrossRef]
- Bazurto, J.V.; Dearth, S.P.; Tague, E.D.; Campagna, S.R.; Downs, D.M. Untargeted metabolomics confirms and extends the understanding of the impact of aminoimidazole carboxamide ribotide (AICAR) in the metabolic network of Salmonella enterica. Microb. Cell 2017, 5, 74–87. [Google Scholar] [CrossRef] [Green Version]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Clasquin, M.F.; Melamud, E.; Rabinowitz, J.D. LC-MS Data Processing with MAVEN: A Metabolomic Analysis and Visualization Engine. Curr. Protoc. Bioinform. 2012, 37, 11–14. [Google Scholar]
- Melamud, E.; Vastag, L.; Rabinowitz, J.D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 2010, 82, 9818–9826. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; 3.5.1; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [Green Version]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An Integrated Strategy for Compound Spectra Extraction and Annotation of Liquid Chromatography/Mass Spectrometry Data Sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [Green Version]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2019, 48, D440–D444. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; 4.0.3; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [Green Version]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Worley, B.; Powers, R. PCA as a practical indicator of OPLS-DA model reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams; 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 12 December 2021).
- Kraus, D. Consolidated data analysis and presentation using an open-source add-in for the Microsoft Excel® spreadsheet software. Med. Writ. 2014, 23, 25–28. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007, 8, 105. [Google Scholar] [CrossRef] [Green Version]



| High Fat Diet Metabolomics | |||||||
|---|---|---|---|---|---|---|---|
| Jejunum | Ileum | Cecum | Colon | Liver | Serum | ||
| Total identified metabolites | 145 | 140 | 141 | 137 | 130 | 120 | |
| HF-altered | 11 | 2 | 41 | 30 | 2 | 7 | |
| HF vs. CD | increased w/HF | 5 | 0 | 35 | 9 | 0 | 6 |
| decreased w/HF | 6 | 2 | 6 | 21 | 2 | 1 | |
| HF-altered & FG-altered | 6 | 4 | 1 | 6 | 3 | 0 | |
| CD vs. CDFG | increased w/FG | 2 | 4 | 0 | 0 | 1 | 0 |
| decreased w/FG | 4 | 0 | 0 | 0 | 2 | 0 | |
| HF vs. HFFG | increased w/FG | 0 | 0 | 0 | 5 | 0 | 0 |
| decreased w/FG | 0 | 0 | 1 | 1 | 0 | 0 | |
| Total unique spectral features | 6837 | 6823 | 11157 | 8974 | 3323 | 1772 | |
| HF-altered | 594 | 718 | 4294 | 2809 | 339 | 105 | |
| HF vs. CD | increased w/HF | 114 | 508 | 3671 | 1977 | 59 | 94 |
| decreased w/HF | 480 | 210 | 623 | 832 | 280 | 11 | |
| HF-altered & FG-altered | 104 | 473 | 2917 | 1913 | 111 | 37 | |
| CD vs. CDFG | increased w/FG | 18 | 0 | 1685 | 1243 | 0 | 16 |
| decreased w/FG | 72 | 255 | 678 | 282 | 23 | 2 | |
| HF vs. HFFG | increased w/FG | 12 | 11 | 217 | 238 | 51 | 8 |
| decreased w/FG | 6 | 310 | 992 | 515 | 37 | 13 | |
| Weighted | Unweighted | ||||
|---|---|---|---|---|---|
| Intestinal Region | Comparison | Score | p-Value | Score | p-Value |
| Jejunum | CD vs. CDFG | 0.632386 | 0.158 | 0.794898 | 0.261 |
| HF vs. CD | 0.65695 | 0.121 | 0.849862 | 0.158 | |
| HF vs. HFFG | 0.70675 | 0.136 | 0.787471 | 0.134 | |
| Ileum | CD vs. CDFG | 0.620738 | 0.454 | 0.742053 | 0.78 |
| HF vs. CD | 0.654047 | 0.254 | 0.758081 | 0.574 | |
| HF vs. HFFG | 0.692661 | 0.584 | 0.83724 | 0.789 | |
| Cecum | CD vs. CDFG | 0.654342 | 0.069 * | 0.724363 | 0.222 |
| HF vs. CD | 0.534961 | 0.004 *** | 0.809126 | 0.031 ** | |
| HF vs. HFFG | 0.627892 | 0.008 *** | 0.893359 | 0.003 *** | |
| Colon | CD vs. CDFG | 0.376439 | 0.54 | 0.673875 | 0.284 |
| HF vs. CD | 0.589599 | <0.0010 *** | 0.908955 | <0.0010 *** | |
| HF vs. HFFG | 0.607401 | <0.0010*** | 0.908432 | 0.005*** | |
| Jejunum | Ileum | Cecum | Colon | |
|---|---|---|---|---|
| Core OTUs | 63 | 69 | 113 | 120 |
| HF-altered | 10 | 8 | 57 | 53 |
| Significantly increased by HF | 2 | 7 | 50 | 40 |
| Significantly decreased by HF | 8 | 1 | 7 | 13 |
| HF-altered and FG corrected | 0 | 0 | 13 | 15 |
| Significantly increased by HF | 0 | 0 | 11 | 10 |
| Significantly decreased by HF | 0 | 0 | 2 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, K.A.; Richard, A.J.; Salbaum, J.M.; Newman, S.; Carmouche, R.; Webb, S.; Bruce-Keller, A.J.; Stephens, J.M.; Campagna, S.R. Cross-Omics Analysis of Fenugreek Supplementation Reveals Beneficial Effects Are Caused by Gut Microbiome Changes Not Mammalian Host Physiology. Int. J. Mol. Sci. 2022, 23, 3654. https://doi.org/10.3390/ijms23073654
Jones KA, Richard AJ, Salbaum JM, Newman S, Carmouche R, Webb S, Bruce-Keller AJ, Stephens JM, Campagna SR. Cross-Omics Analysis of Fenugreek Supplementation Reveals Beneficial Effects Are Caused by Gut Microbiome Changes Not Mammalian Host Physiology. International Journal of Molecular Sciences. 2022; 23(7):3654. https://doi.org/10.3390/ijms23073654
Chicago/Turabian StyleJones, Katarina A., Allison J. Richard, J. Michael Salbaum, Susan Newman, Richard Carmouche, Sara Webb, Annadora J. Bruce-Keller, Jacqueline M. Stephens, and Shawn R. Campagna. 2022. "Cross-Omics Analysis of Fenugreek Supplementation Reveals Beneficial Effects Are Caused by Gut Microbiome Changes Not Mammalian Host Physiology" International Journal of Molecular Sciences 23, no. 7: 3654. https://doi.org/10.3390/ijms23073654
APA StyleJones, K. A., Richard, A. J., Salbaum, J. M., Newman, S., Carmouche, R., Webb, S., Bruce-Keller, A. J., Stephens, J. M., & Campagna, S. R. (2022). Cross-Omics Analysis of Fenugreek Supplementation Reveals Beneficial Effects Are Caused by Gut Microbiome Changes Not Mammalian Host Physiology. International Journal of Molecular Sciences, 23(7), 3654. https://doi.org/10.3390/ijms23073654






