Novel Insights on Nitric Oxide Synthase and NO Signaling in Ascidian Metamorphosis
Abstract
1. Introduction
2. Results
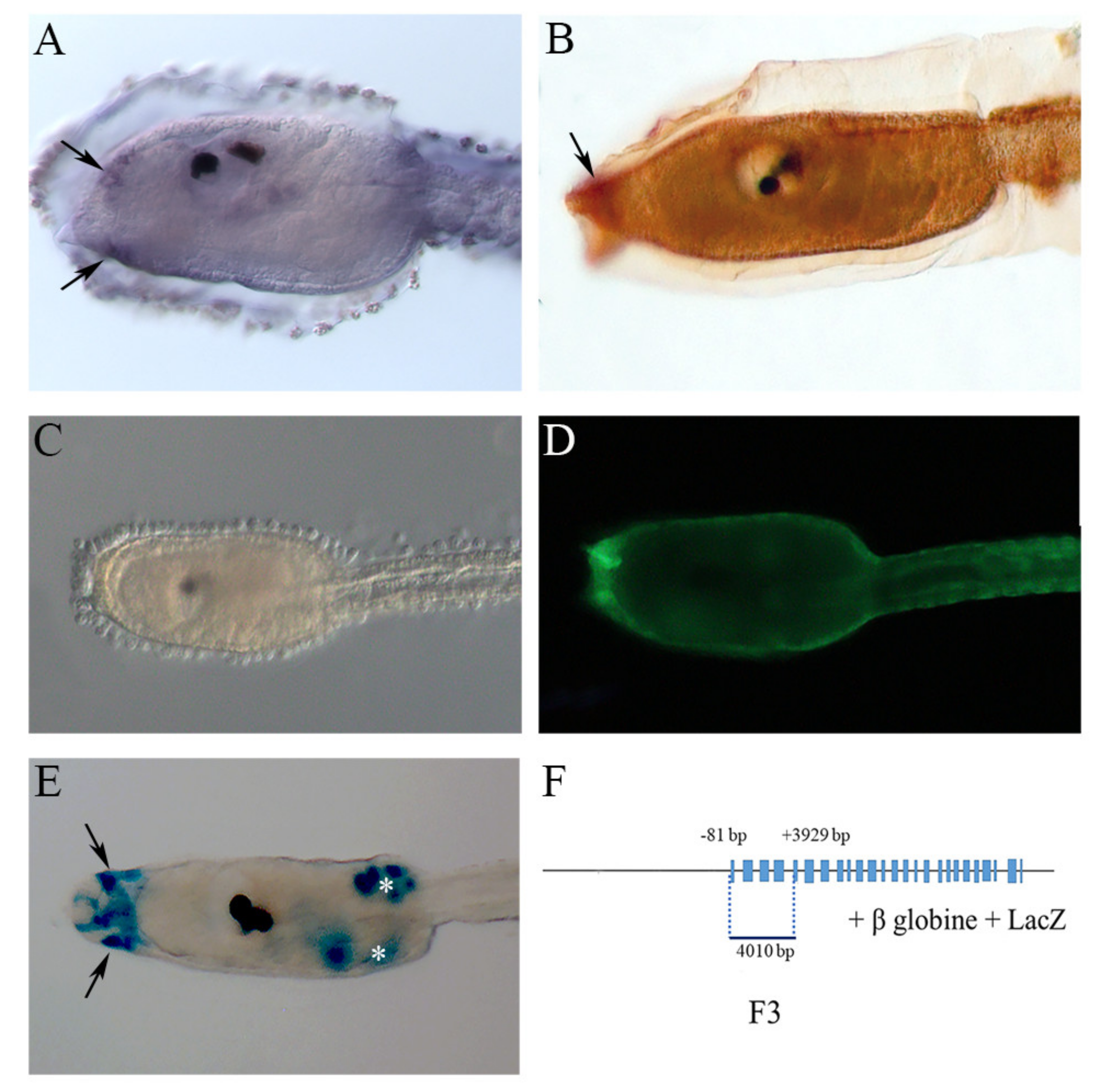
2.1. NOS and NO in the Larval Palps
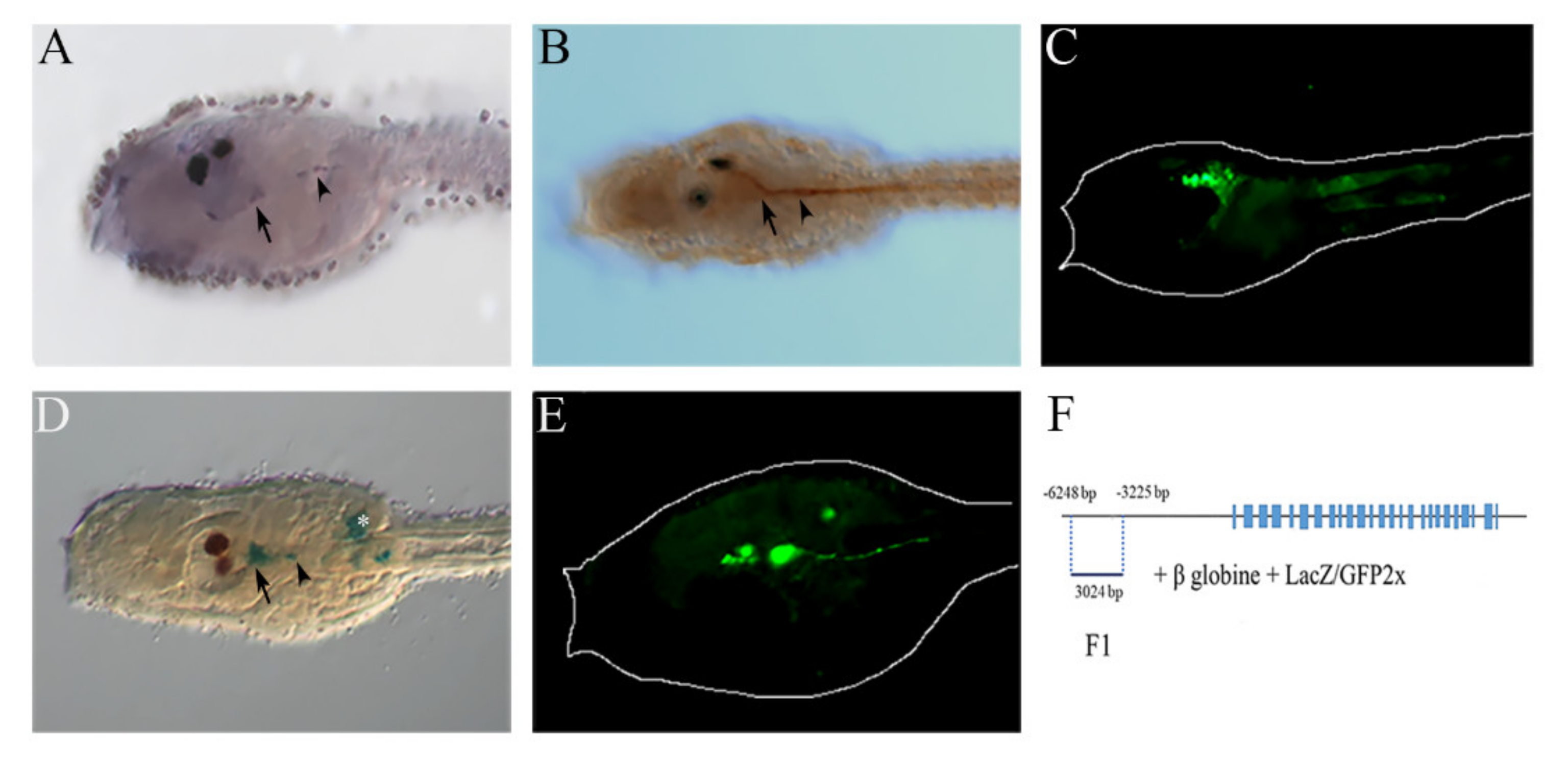
2.2. NOS and NO in the Anterior Nervous System
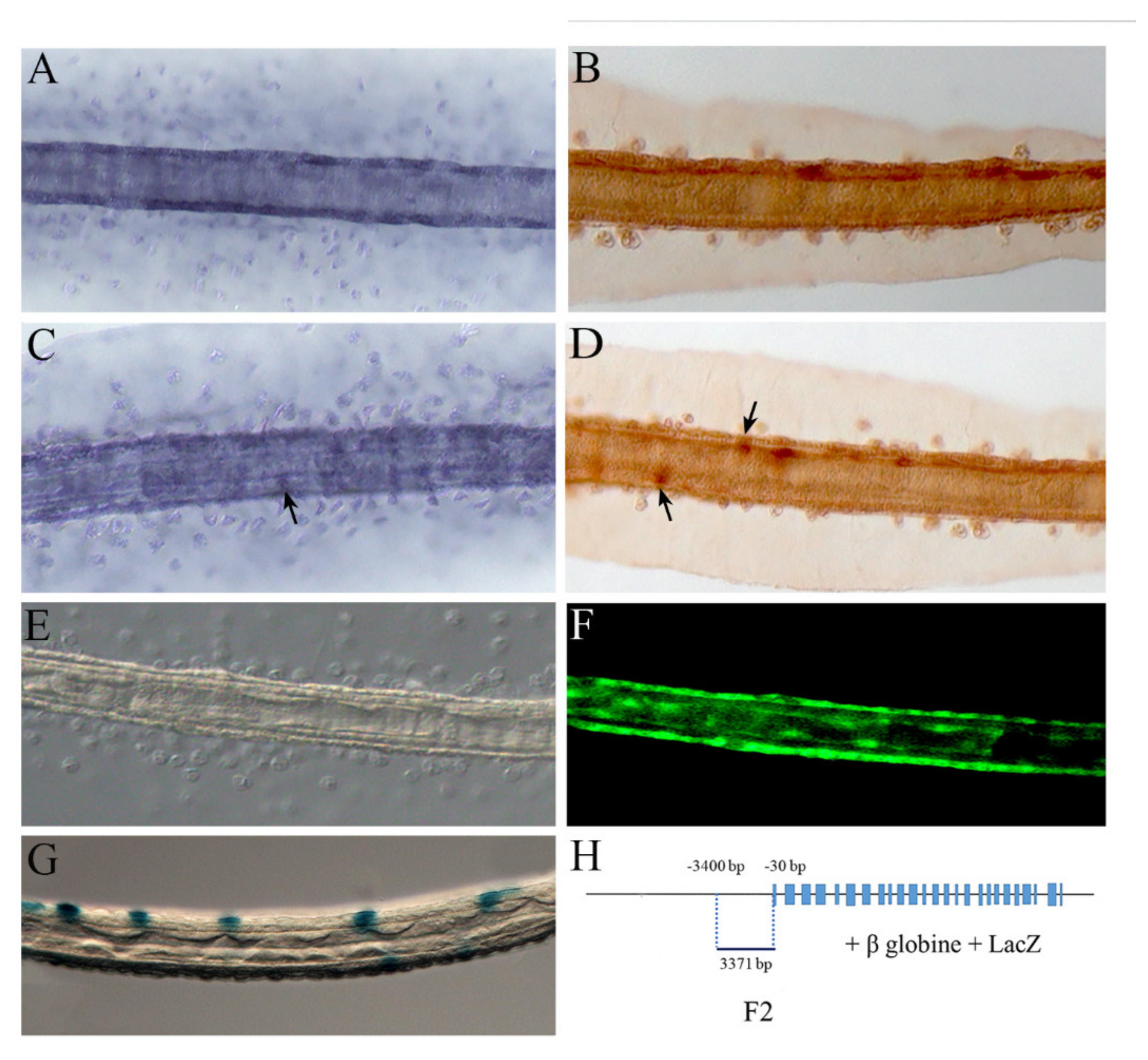
2.3. NOS and NO in the Tail
3. Discussion
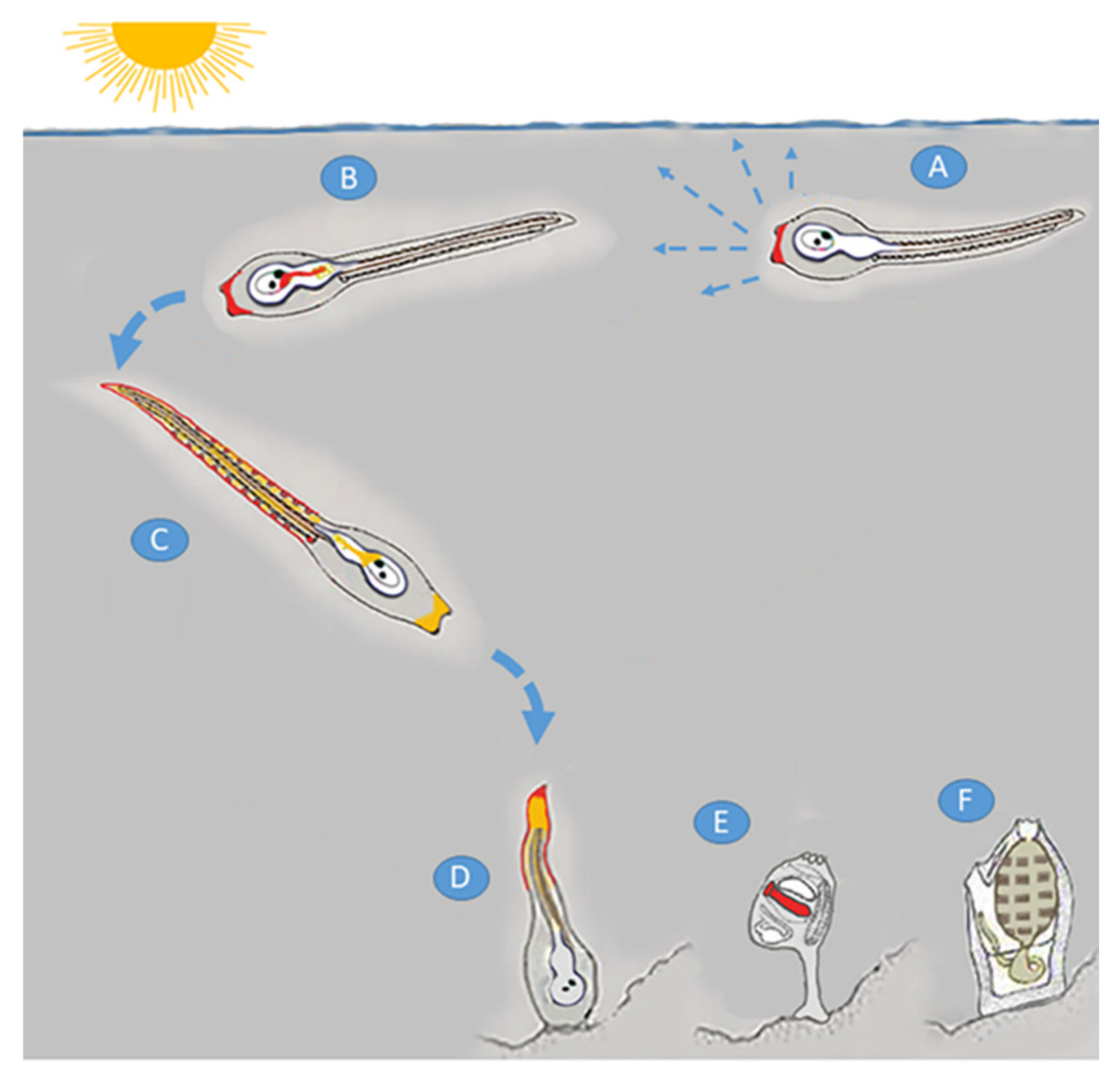
3.1. NO Signaling in the Different Phases of Metamorphosis
3.2. Conservation of NOS/NO Signaling during Evolution
4. Conclusions
5. Materials and Methods
5.1. Animal Care and Ethical Statement
5.2. Genomic Fragments Amplification and Preparation of Enhancer DNA Constructs
5.3. In Situ Hybridization
5.4. NO Detection
5.5. Immunochemistry
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andreakis, N.; D’Aniello, S.; Albalat, R.; Patti, F.P.; Garcia-Fernandez, J.; Procaccini, G.; Sordino, P.; Palumbo, A. Evolution of the nitric oxide synthase family in metazoans. Mol. Biol. Evol. 2011, 28, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Moroz, L.L.; Romanova, D.Y.; Nikitin, M.A.; Sohn, D.; Kohn, A.B.; Neveu, E.; Varoqueaux, F.; Fasshauer, D. The diversification and lineage-specific expansion of nitric oxide signaling in Placozoa: Insights in the evolution of gaseous transmission. Sci. Rep. 2020, 10, 13020. [Google Scholar] [CrossRef] [PubMed]
- Griffith, O.W.; Stuehr, D.J. Nitric oxide synthases: Properties and catalytic mechanism. Annu. Rev. Physiol. 1995, 57, 707–736. [Google Scholar] [CrossRef] [PubMed]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef]
- Giovine, M.; Pozzolini, M.; Favre, A.; Bavestrello, G.; Cerrano, C.; Ottaviani, F.; Chiarantini, L.; Cerasi, A.; Cangiotti, M.; Zocchi, E.; et al. Heat stress-activated, calcium-dependent nitric oxide synthase in sponges. Nitric Oxide 2001, 5, 427–431. [Google Scholar] [CrossRef]
- Romano, G.; Costantini, M.; Buttino, I.; Ianora, A.; Palumbo, A. Nitric oxide mediates the stress response induced by diatom aldehydes in the sea urchin Paracentrotus lividus. PLoS ONE 2011, 6, e25980. [Google Scholar] [CrossRef]
- Migliaccio, O.; Castellano, I.; Romano, G.; Palumbo, A. Stress response to cadmium and manganese in Paracentrotus lividus developing embryos is mediated by nitric oxide. Aquat. Toxicol. 2014, 156, 125–134. [Google Scholar] [CrossRef]
- Migliaccio, O.; Castellano, I.; Di Cioccio, D.; Tedeschi, G.; Negri, A.; Cirino, P.; Romano., G.; Zingone, A.; Palumbo, A. Subtle reproductive impairment through nitric oxide-mediated mechanisms in sea urchins from an area affected by harmful algal blooms. Sci. Rep. 2016, 6, 26086. [Google Scholar] [CrossRef]
- Castellano, I.; Ercolesi, E.; Romano, G.; Ianora, A.; Palumbo, A. The diatom-derived aldehyde decadienal affects life cycle transition in the ascidian Ciona intestinalis through nitric oxide/ERK signalling. Open Biol. 2015, 5, 140182. [Google Scholar] [CrossRef][Green Version]
- Castellano, I.; Migliaccio, O.; Ferraro, G.; Maffioli, E.; Marasco, D.; Merlino, A.; Zingone, A.; Tedeschi, G.; Palumbo, A. Biotic and environmental stress induces nitration and changes in structure and function of the sea urchin major yolk protein toposome. Sci. Rep. 2018, 8, 4610. [Google Scholar] [CrossRef]
- Salleo, A.; Musci, G.; Barra, P.; Calabrese, L. The discharge mechanism of acontial nematocytes involves the release of nitric oxide. J. Exp. Biol. 1996, 199, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Di Cosmo, A.; Gesualdo, I.; d’Ischia, M. A calcium-dependent nitric oxide synthase and NMDA R1 glutamate receptor in the ink gland of Sepia officinalis: A hint to a regulatory role of nitric oxide in melanogenesis? Biochem. Biophys. Res. Commun. 1997, 235, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Di Cosmo, A.; Poli, A.; Di Cristo, C.; d’Ischia, M. A calcium/calmodulin-dependent nitric oxide synthase, NMDAR2/3 receptor subunits, and glutamate in the CNS of the cuttlefish Sepia officinalis: Localization in specific neural pathways controlling the inking system. J. Neurochem. 1999, 73, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Poli, A.; Di Cosmo, A.; d’Ischia, M. N-Methyl-D-aspartate receptor stimulation activates tyrosinase and promotes melanin synthesis in the ink gland of the cuttlefish Sepia officinalis through the nitric oxide/cGMP signal transduction pathway. A novel possible role for glutamate as physiologic activator of melanogenesis. J. Biol. Chem. 2000, 275, 16885–16890. [Google Scholar]
- Stefano, G.B.; Ottaviani, E. The biochemical substrate of nitric oxide signaling is present in primitive non-cognitive organisms. Brain Res. 2002, 924, 82–89. [Google Scholar] [CrossRef]
- Tafalla, C.; Gomez-Leon, J.; Novoa, B.; Figueras, A. Nitric oxide production by carpet shell clam (Ruditapes decussatus) hemocytes. Dev. Comp. Immunol. 2003, 27, 197–205. [Google Scholar] [CrossRef]
- Fiore, G.; Poli, A.; Di Cosmo, A.; d’Ischia, M.; Palumbo, A. Dopamine in the ink defence system of Sepia officinalis: Biosynthesis, vesicular compartmentation in mature ink gland cells, nitric oxide (NO)/cGMP-induced depletion and fate in secreted ink. Biochem. J. 2004, 378, 785–791. [Google Scholar] [CrossRef]
- Palumbo, A. Nitric oxide in marine invertebrates: A comparative perspective. Comp. Biochem. Physiol. A 2005, 142, 241–248. [Google Scholar] [CrossRef]
- Palumbo, A.; D’Ischia, M. Nitric oxide biogenesis, signalling and roles in molluscs: The Sepia officinalis paradigm. Adv. Exp. Biol. 2007, 1, 45–64. [Google Scholar]
- Fiore, G.; Mattiello, T.; Tedeschi, G.; Nonnis, S.; d’Ischia, M.; Palumbo, A. Protein nitration is specifically associated with melanin production and reveals redox imbalance as a new correlate of cell maturation in the ink gland of Sepia officinalis. Pigment Cell Melanoma Res. 2009, 22, 857–859. [Google Scholar] [CrossRef]
- Mattiello, T.; Fiore, G.; Brown, E.R.; d’Ischia, M.; Palumbo, A. Nitric oxide mediates the glutamate-dependent pathway for neurotransmission in Sepia officinalis chromatophore organs. J. Biol. Chem. 2010, 285, 24154–24163. [Google Scholar] [CrossRef] [PubMed]
- Mattiello, T.; Costantini, M.; Di Matteo, B.; Livigni, S.; Andouche, A.; Bonnaud, L.; Palumbo, A. The dynamic nitric oxide pattern in developing cuttlefish Sepia officinalis. Dev. Dyn. 2012, 241, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Mattiello, T.; d’Ischia, M.; Palumbo, A. Nitric oxide in chromatic body patterning elements of Sepia officinalis. J. Exp. Mar. Biol. Ecol. 2013, 447, 128–131. [Google Scholar] [CrossRef]
- Di Cosmo, A.; Di Cristo, C.; Palumbo, A.; d’Ischia, M.; Messenger, J.B. Nitric oxide synthase (NOS) in the brain of the cephalopod Sepia officinalis. J. Comp. Neurol. 2000, 428, 411–427. [Google Scholar] [CrossRef]
- Moroz, L.L.; Meech, R.W.; Sweedler, J.V.; Mackie, G.O. Nitric oxide regulates swimming in the jellyfish Aglantha digitale. J. Comp. Neurol. 2004, 471, 26–36. [Google Scholar] [CrossRef]
- Moroz, L.L. Giant identified NO-releasing neurons and comparative histochemistry of putative nitrergic systems in gastropod molluscs. Microsc. Res. Tech. 2000, 49, 557–569. [Google Scholar] [CrossRef]
- Moroz, L.L.; Norekian, T.P.; Pirtle, T.J.; Robertson, K.J.; Satterlie, R.A. Distribution of NADPH-diaphorase reactivity and effects of nitric oxide on feeding and locomotory circuitry in the pteropod mollusc, Clione limacina. J. Comp. Neurol. 2000, 427, 274–284. [Google Scholar] [CrossRef]
- Davidson, S.K.; Koropatnick, T.A.; Kossmehl, R.; Sycuro, L.; McFall-Ngai, M.J. NO means “yes” in the squid-vibrio symbiosis: Nitric oxide (NO) during the initial stages of a beneficial association. Cell. Microbiol. 2004, 6, 1139–1151. [Google Scholar] [CrossRef]
- Grumetto, L.; Wilding, M.; De Simone, M.L.; Tosti, E.; Galione, A.; Dale, B. Nitric oxide gates fertilization channels in ascidian oocytes through nicotinamide nucleotide metabolism. Biochem. Biophys. Res. Commun. 1997, 239, 723–728. [Google Scholar] [CrossRef]
- Leckie, C.; Empson, R.; Becchetti, A.; Thomas, J.; Galione, A.; Whitaker, M. The NO pathway acts late during the fertilization response in sea urchin eggs. J. Biol. Chem. 2003, 278, 12247–12254. [Google Scholar] [CrossRef]
- Annona, G.; Caccavale, F.; Pascual-Anaya, J.; Kuratani, S.; de Luca, P.; Palumbo, A.; D’Aniello, S. Nitric oxide regulates mouth development in amphioxus. Sci. Rep. 2017, 7, 8432. [Google Scholar] [CrossRef] [PubMed]
- Caccavale, F.; Coppola, U.; Vassalli, Q.A.; La Vecchia, C.; Palumbo, A.; D’Aniello, E.; Locascio, A.; Ristoratore, F.; D’Aniello, S. Transphyletic conservation of nitric oxide synthase regulation in cephalochordates and tunicates. Dev. Genes Evol. 2020, 230, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Froggett, S.J.; Leise, E.M. Metamorphosis in the marine snail Ilyanassa obsoleta, Yes or NO? Biol. Bull. 1999, 196, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.D.; Bates, W.R.; Brandhorst, B.P. Regulation of metamorphosis in ascidians involves NO/cGMP signaling and HSP90. Exp. Zool. 2001, 289, 374–384. [Google Scholar] [CrossRef]
- Bishop, C.D.; Brandhorst, B.P. Development of nitric oxide synthase-defined neurons in the sea urchin larval ciliary band and evidence for a chemosensory function during metamorphosis. Dev. Dyn. 2007, 236, 1535–1546. [Google Scholar] [CrossRef]
- Comes, S.; Locascio, A.; Silvestre, F.; d’Ischia, M.; Russo, G.L.; Tosti, E.; Branno, M.; Palumbo, A. Regulatory roles of nitric oxide during larval development and metamorphosis in Ciona intestinalis. Dev. Biol. 2007, 306, 772–784. [Google Scholar] [CrossRef]
- Pechenik, J.A.; Cochrane, D.E.; Li, W.; West, E.T.; Pires, A.; Leppo, M. Nitric oxide inhibits metamorphosis in larvae of Crepidula fornicata, the slippershell snail. Biol. Bull. 2007, 213, 160–171. [Google Scholar] [CrossRef]
- Bishop, C.D.; Pires, A.; Norby, S.-W.; Boudko, D.; Moroz, L.L. Analysis of nitric oxide-cyclic guanosine monophosphate signaling during metamorphosis of the nudibranch Phestilla sibogae Bergh (Gastropoda: Opisthobranchia). Evol. Dev. 2008, 10, 288–299. [Google Scholar] [CrossRef]
- Biggers, W.J.; Pires, A.; Pechenik, J.A.; Johns, E.; Patel, P.; Polson, T.; Polson., J. Inhibitors of nitric oxide synthase induce larval settlement and metamorphosis of the polychaete annelid Capitella teleta. Invertebr. Reprod. Dev. 2011, 56, 1–13. [Google Scholar] [CrossRef]
- Ercolesi, E.; Tedeschi, G.; Fiore, G.; Negri, A.; Maffioli, E.; d’Ischia, M.; Palumbo, A. Protein nitration as footprint of oxidative stress-related nitric oxide signaling pathways in developing Ciona intestinalis. Nitric Oxide 2012, 27, 18–24. [Google Scholar] [CrossRef]
- Zhang, Y.; He, L.-S.; Zhang, G.; Xu, Y.; Lee, O.-O.; Matsumura, K.; Qian, P.-Y. The regulatory role of the NO/cGMP signal transduction cascade during larval attachment and metamorphosis of the barnacle Balanus (=Amphibalanus) amphitrite. J. Exp. Biol. 2012, 215, 3813–3822. [Google Scholar] [PubMed]
- Zhang, G.; Wong, Y.H.; Zhang, Y.; He, L.S.; Xu, Y.; Qian, P.Y. Nitric oxide inhibits larval settlement in Amphibalanus amphitrite cyprids by repressing muscle locomotion and molting. Proteomics 2015, 15, 3854–3864. [Google Scholar] [CrossRef] [PubMed]
- Ueda, N.; Degnan, S.M. Nitric oxide acts as a positive regulator to induce metamorphosis of the ascidian Herdmania momus. PLoS ONE 2013, 8, e72797. [Google Scholar]
- Romero, M.R.; Phuong, M.A.; Bishop, C.; Krug, P.J. Nitric oxide signaling differentially affects habitat choice by two larval morphs of the sea slug Alderia willowi: Mechanistic insight into evolutionary transitions in dispersal strategies. J. Exp. Biol. 2013, 216, 1114–1125. [Google Scholar] [PubMed]
- Castellano, I.; Ercolesi, E.; Palumbo, A. Nitric oxide affects ERK signaling through down-regulation of MAP kinase phosphatase levels during larval development of the ascidian Ciona intestinalis. PLoS ONE 2014, 9, e1029079. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ueda, N.; Richards, G.S.; Degnan, B.M.; Kranz, A.; Adamska, M.; Croll, R.P.; Degnan, S.M. An ancient role for nitric oxide in regulating the animal pelagobenthic life cycle: Evidence from a marine sponge. Sci. Rep. 2016, 6, 37546. [Google Scholar] [CrossRef]
- Yang, X.-X.; Zhang, Y.; Wong, Y.-H.; Qian, P.-Y. HSP90 regulates larval settlement of the bryozoan Bugula neritina through the nitric oxide pathway. J. Exp. Biol. 2018, 221, jeb167478. [Google Scholar] [CrossRef]
- Zhu, Y.-T.; Zhang, Y.; Liu, Y.-Z.; Li, Y.-F.; Yoshida, A.; Osatomi, K.; Yang, J.-L.; Liang, X. Nitric oxide negatively regulates larval metamorphosis in hard-shelled mussel (Mytilus coruscus). Front. Mar. Sci. 2020, 7, 356. [Google Scholar] [CrossRef]
- Silvagno, F.; Xia, H.; Bredt, D.S. Neuronal nitric-oxide synthase-μ, an alternatively spliced isoform expressed in differentiated skeletal muscle. J. Biol. Chem. 1996, 271, 11204–11208. [Google Scholar] [CrossRef]
- Wang, Y.; Newton, D.C.; Robb, G.B.; Kau, C.-L.; Miller, T.L.; Cheung, A.H.; Hall, A.V.; VanDamme, S.; Wilcox, J.N.; Marsden, P.A. RNA diversity has profound effects on the translation of neuronal nitric oxide synthase. Proc. Natl. Acad. Sci. USA 1999, 96, 12150–12155. [Google Scholar] [CrossRef]
- Saur, D.; Seidler, B.; Paehge, H.; Schusdziarra, V.; Allescher, H.D. Complex regulation of human neuronal nitric-oxide synthase exon 1c gene transcription. Essential role of Sp and ZNF family members of transcription factors. J. Biol. Chem. 2002, 277, 25798–25814. [Google Scholar] [CrossRef] [PubMed]
- Luckhart, S.; Li, K. Transcriptional complexity of the Anopheles stephensi nitric oxide synthase gene. Insect Biochem. Mol. Biol. 2001, 31, 249–256. [Google Scholar] [CrossRef]
- Korneev, S.; O’Shea, M. Evolution of Nitric Oxide Synthase regulatory genes by DNA inversion. Mol. Biol. Evol. 2002, 19, 1228–1233. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Korneev, S.A.; Straub, V.; Kemenes, I.; Korneeva, E.I.; Ott, S.R.; Benjamin, P.R.; O’Shea, M. Timed and targeted differential regulation of Nitric Oxide Synthase (NOS) and anti-NOS genes by reward conditioning leading to Long-Term Memory formation. J. Neurosci. 2005, 25, 1188–1192. [Google Scholar] [CrossRef] [PubMed]
- Stasiv, Y.; Kuzin, B.; Regulski, M.; Tully, T.; Enikolopov, G. Regulation of multimers via truncated isoforms: A novel mechanism to control nitric-oxide signaling. Genes Dev. 2004, 18, 1812–1823. [Google Scholar] [CrossRef][Green Version]
- Karaiskou, A.; Swalla, B.J.; Sasakura, Y.; Chambon, J.P. Metamorphosis in Solitary Ascidians. Genesis 2015, 53, 34–47. [Google Scholar] [CrossRef]
- Matsunobu, S.; Sasakura, Y. Time course for tail regression during metamorphosis of the ascidian Ciona intestinalis. Dev. Biol. 2015, 405, 71–81. [Google Scholar] [CrossRef]
- Hotta, K.; Dauga, D.; Manni, L. The ontology of the anatomy and development of the solitary ascidian Ciona: The swimming larva and its metamorphosis. Sci. Rep. 2020, 10, 17916. [Google Scholar] [CrossRef]
- Olivo, P.; Palladino, A.; Ristoratore, F.; Spagnuolo, A. Brain sensory organs of the Ascidian Ciona robusta: Structure, function and developmental mechanisms. Front. Cell Dev. Biol. 2021, 9, 701779. [Google Scholar] [CrossRef]
- Horie, T.; Sakurai, D.; Ohtsuki, H.; Terakita, A.; Shichida, Y.; Usukura, J.; Kusakabe, T.; Tsuda, M. Pigmented and nonpigmented ocelli in the brain vesicle of the ascidian larva. J. Comp. Neurol. 2008, 509, 88–102. [Google Scholar] [CrossRef]
- Ryan, K.; Lu, Z.; Meinertzhagen, I.A. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. eLife 2016, 5, e16962. [Google Scholar] [CrossRef] [PubMed]
- Imai, J.H.; Meinertzhagen, I.A. Neurons of the ascidian larval nervous system in Ciona intestinalis: II. Peripheral nervous system. J. Comp. Neurol. 2007, 501, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Nakagawa, M.; Sasakura, Y.; Kusakabe, T.G.; Tsuda, M. Simple motor system of the ascidian larva: Neuronal complex comprising putative cholinergic and GABAergic/glycinergic neurons. Zool. Sci. 2010, 272, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Meinertzhagen, I.A.; Okamura, Y. The larval ascidian nervous system: The chordate brain from its small beginnings. Trends Neurosci. 2001, 24, 401–410. [Google Scholar] [CrossRef]
- Meinertzhagen, I.A.; Lemaire, P.; Okamura, Y. The neurobiology of the ascidian tadpole larva: Recent developments in an ancient chordate. Annu. Rev. Neurosci. 2004, 27, 453–485. [Google Scholar] [CrossRef]
- Svane, I.B.; Young, C.M. The ecology and behaviour of ascidian larvae. Oceanogr. Mar. Biol. 1989, 27, 45–90. [Google Scholar]
- Brown, E.R.; Nishino, A.; Bone, Q.; Meinertzhagen, I.A.; Okamura, Y. GABAergic synaptic transmission modulates swimming in the ascidian larva. Eur. J. Neurosci. 2005, 22, 2541–2548. [Google Scholar] [CrossRef]
- Yamaji, S.; Hozumi, A.; Matsunobu, S.; Sasakura, Y. Orchestration of the distinct morphogenetic movements in different tissues drives tail regression during ascidian metamorphosis. Dev. Biol. 2020, 465, 66–78. [Google Scholar] [CrossRef]
- Krasovec, G.; Robine, K.; Quéinnec, E.; Karaiskou, A.; Chambon, J.P. Ci-hox12 tail gradient precedes and participates in the control of the apoptotic-dependent tail regression during Ciona larva metamorphosis. Dev. Biol. 2019, 448, 237–246. [Google Scholar] [CrossRef]
- Tsuda, M.; Kawakami, I.; Shiraishi, S. Sensitization and habituation of the swimming behavior in Ascidian larvae to light. Zool. Sci. 2003, 20, 13–22. [Google Scholar] [CrossRef]
- Bishop, C.D.; Brandhorst, B.P. NO/cGMP signaling and HSP90 activity represses metamorphosis in the sea urchin Lytechinus pictus. Biol. Bull. 2001, 201, 394–404. [Google Scholar] [CrossRef] [PubMed]
- García-Cardeña, G.; Fan, R.; Shah, V.; Sorrentino, R.; Cirino, G.; Papapetropoulos, A.; Sessa, W.C. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature 1998, 392, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Billecke, S.S.; Bender, A.T.; Kanelakis, K.C.; Murphy, P.J.M.; Lowe, E.R.; Kamada, Y.; Pratt, W.B.; Osawa, Y. Hsp90 is required for heme binding and activation of apo-neuronal nitric-oxide synthase. J. Biol. Chem. 2002, 277, 20504–20509. [Google Scholar] [CrossRef] [PubMed]
- Billecke, S.S.; Draganov, D.I.; Morishima, Y.; Murphy, P.J.M.; Dunbar, A.Y.; Pratt, W.B.; Osawa, Y. The role of hsp90 in heme-dependent activation of apo-neuronal nitric-oxide synthase. J. Biol. Chem. 2004, 279, 30252–30258. [Google Scholar] [CrossRef] [PubMed]
- Chambon, J.P.; Soule, J.; Pomies, P.; Fort, P.; Sahuquet, A.; Alexandre, D.; Mangeat, P.H.; Baghdiguian, S. Tail regression in Ciona intestinalis (Protochordate) involved a caspase-dependent apoptosis event associated with ERK activation. Development 2002, 129, 3105–3114. [Google Scholar] [CrossRef]
- Chambon, J.P.; Nakayama, A.; Takamura, K.; McDougall, A.; Satoh, N. ERK- and JNK-signalling regulate gene networks that stimulate metamorphosis and apoptosis in tail tissue of ascidian tadpoles. Development 2007, 134, 1203–1219. [Google Scholar] [CrossRef]
- Yang, X.-X.; Wong, Y.-H.; Zhang, Y.; Zhang, G.; Qian, P.-Y. Exploring the regulatory role of nitric oxide (NO) and the NO-p38MAPK/cGMP pathway in larval settlement of the bryozoan Bugula neritina. Biofouling 2018, 34, 545–556. [Google Scholar] [CrossRef]
- Kaji, T.; Shimizu, K.; Artinger, K.B.; Yasui, K. Dynamic modification of oral innervation during metamorphosis in Branchiostoma belcheri, the oriental lancelet. Biol. Bull. 2009, 217, 151–160. [Google Scholar] [CrossRef]
- Ueda, N.; Degnan, S.M. Nitric oxide is not a negative regulator of metamorphic induction in the abalone Haliotis asinina. Front. Mar. Sci. 2014, 1, 21. [Google Scholar] [CrossRef]
- Lin, M.-F.; Leise, E.M. NADPH-diaphorase activity changes during gangliogenesis and metamorphosis in the gastropod mollusc Ilyanassa obsoleta. J. Comp. Neurol. 1996, 374, 194–203. [Google Scholar] [CrossRef][Green Version]
- Hadfield, M.G.; Meleshkevitch, E.A.; Boudko, D.Y. The apical sensory organ of a gastropod veliger is a receptor for settlement cues. Biol. Bull. 2000, 198, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Jacox, L.; Sindelka, R.; Chen, J.; Rothman, A.; Dickinson, A.; Sive, H. The extreme anterior domain is an essential craniofacial organizer acting through Kinin-Kallikrein signaling. Cell Rep. 2014, 8, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Zega, G.; Thorndyke, M.C.; Brown, E.R. Development of swimming behaviour in the larva of the ascidian Ciona intestinalis. J. Exp. Biol. 2006, 209, 3405–3412. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, J.M.; Watson, W.H., III. Modulation of swimming in the gastropod Melibe leonina by nitric oxide. J. Exp. Biol. 2002, 205, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Kyriakatos, A.; Molinari, M.; Mahmood, R.; Grillner, S.; Sillar, K.T.; El Manira, A. Nitric Oxide potentiation of locomotor activity in the spinal cord of the lamprey. J. Neurosci. 2009, 29, 13283–13291. [Google Scholar] [CrossRef]
- McLean, D.L.; Sillar, K.T. Nitric Oxide selectively tunes inhibitory synapses to modulate vertebrate locomotion. J. Neurosci. 2002, 22, 4175–4184. [Google Scholar] [CrossRef]
- McLean, D.L.; Sillar, K.T. Metamodulation of a spinal locomotor network by Nitric Oxide. J. Neurosci. 2004, 24, 9561–9571. [Google Scholar] [CrossRef]
- Ramanathan, S.; Combes, D.; Molinari, M.; Simmers, J.; Sillar, K.T. Developmental and regional expression of NADPH-diaphorase/nitric oxide synthase in spinal cord neurons correlates with the emergence of limb motor networks in metamorphosing Xenopus laevis. Eur. J. Neurosci. 2006, 24, 1907–1922. [Google Scholar] [CrossRef]
- Newland, P.L.; Yates, P. Nitrergic modulation of an oviposition digging rhythm in locusts. J. Exp. Biol. 2007, 210, 4448–4456. [Google Scholar] [CrossRef]
- Severi, K.E.; Portugues, R.; Marques, J.C.; O’Malley, D.M.; Orger, M.B.; Engert, F. Neural control and modulation of swimming speed in the larval zebrafish. Neuron 2014, 83, 692–707. [Google Scholar] [CrossRef]
- Yoshida, M.; Nagayama, T.; Newland, P. Nitric oxide-mediated intersegmental modulation of cycle frequency in the crayfish swimmeret system. Biol. Open 2018, 7, bio032789. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.R.; Burrows, M. Nitric oxide synthase in the thoracic ganglia of the locust: Distribution in the neuropiles and morphology of neurones. J. Comp. Neurol. 1998, 395, 217–230. [Google Scholar] [CrossRef]
- Ott, S.R.; Jones, I.W.; Burrows, M.; Elphick, M.R. Sensory afferents and motor neurons as targets for nitric oxide in the locust. J. Comp. Neurol. 2000, 422, 521–532. [Google Scholar] [CrossRef]
- Poon., K.L.; Richardson, M.; Lam, C.S.; Khoo, H.E.; Korzh, V. Expression pattern of neuronal nitric oxide synthase in embryonic zebrafish. Gene Expr. Patterns 2003, 3, 463–466. [Google Scholar] [CrossRef]
- Chong, P.S.; Poon, C.H.; Fung, M.L.; Guan, L.; Steinbusch, H.W.M.; Chan, Y.S.; Lim, W.L.; Lim, L.W. Distribution of neuronal nitric oxide synthase immunoreactivity in adult male Sprague-Dawley rat brain. Acta Histochem. 2019, 121, 151437. [Google Scholar] [CrossRef]
- Mahmoud, M.A.; Fahmy, G.H.; Moftah, M.Z.; Sabry, I. Distribution of nitric oxide-producing cells along spinal cord in urodeles. Front. Cell. Neurosci. 2014, 8, 299. [Google Scholar] [CrossRef]
- Currie, S.P.; Combes, D.; Scott, N.W.; Simmers, J.; Sillar, K.T. A behaviorally related developmental switch in nitrergic modulation of locomotor rhythmogenesis in larval Xenopus tadpoles. J. Neurophysiol. 2016, 115, 1446–1457. [Google Scholar] [CrossRef]
- Currie, S.P.; Sillar, K.T. Developmental changes in spinal neuronal properties, motor network configuration, and neuromodulation at free-swimming stages of Xenopus tadpoles. J. Neurophysiol. 2018, 119, 786–795. [Google Scholar] [CrossRef]
- Brunelli, E.; Perrotta, I.; Talarico, E.; Tripepi, S. Localization of two nitric oxide synthase isoforms, eNOS and iNOS, in the skin of Triturus italicus (Amphibia, Urodela) during development. Comp. Biochem. Physiol. A 2005, 142, 249–255. [Google Scholar] [CrossRef]
- Wildling, S.; Kerschbaum, H.H. Nitric oxide decreases ammonium release in tadpoles of the clawed frog, Xenopus laevis, Daudin. J. Comp. Physiol. B 2007, 177, 401–411. [Google Scholar] [CrossRef]
- Tomankova, S.; Abaffy, P.; Sindelka, R. The role of nitric oxide during embryonic epidermis development of Xenopus laevis. Biol. Open 2017, 6, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Dang, E.; Man, G.; Zhang, J.; Lee, D.; Mauro, T.M.; Elias, P.M.; Man, M.-Q. Inducible nitric oxide synthase is required for epidermal permeability barrier homeostasis in mice. Exp. Dermatol. 2020, 29, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Cals-Grierson, M.M.; Ormerod, A.D. Nitric oxide function in the skin. Nitric Oxide 2004, 10, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.T.; Donizetti, A.; Locascio, A.; D’Aniello, S.; Amoroso, A.; Aniello, F.; Fucci, L.; Branno, M. Regulatory elements controlling Ci-msxb tissue-specific expression during Ciona intestinalis embryonic development. Dev. Biol. 2004, 267, 517–528. [Google Scholar] [CrossRef][Green Version]
- D’Aniello, S.; D’Aniello, E.; Locascio, A.; Memoli, A.; Corrado, M.; Russo, M.T.; Aniello, F.; Fucci, L.; Brown, E.R.; Branno, M. The ascidian homolog of the vertebrate homeobox gene Rx is essential for ocellus development and function. Differentiation 2006, 74, 222–234. [Google Scholar] [CrossRef]
- Hotta, K.; Mitsuhara, K.; Takahashi, H.; Inaba, K.; Oka, K.; Gojobori, T.; Ikeo, K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 2007, 236, 1790–1805. [Google Scholar] [CrossRef]
- Vassalli, Q.A.; Anishchenko, E.; Caputi, L.; Sordino, P.; D’Aniello, S.; Locascio, A. Regulatory elements retained during chordate evolution: Coming across tunicates. Genesis 2015, 53, 66–81. [Google Scholar] [CrossRef]
- Zeller, R.W.; Weldon, D.S.; Pellatiro, M.A.; Cone, A.C. Optimized green fluorescent protein variants provide improved single cell resolution of transgene expression in ascidian embryos. Dev. Dyn. 2006, 235, 456–467. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Locascio, A.; Vassalli, Q.A.; Castellano, I.; Palumbo, A. Novel Insights on Nitric Oxide Synthase and NO Signaling in Ascidian Metamorphosis. Int. J. Mol. Sci. 2022, 23, 3505. https://doi.org/10.3390/ijms23073505
Locascio A, Vassalli QA, Castellano I, Palumbo A. Novel Insights on Nitric Oxide Synthase and NO Signaling in Ascidian Metamorphosis. International Journal of Molecular Sciences. 2022; 23(7):3505. https://doi.org/10.3390/ijms23073505
Chicago/Turabian StyleLocascio, Annamaria, Quirino Attilio Vassalli, Immacolata Castellano, and Anna Palumbo. 2022. "Novel Insights on Nitric Oxide Synthase and NO Signaling in Ascidian Metamorphosis" International Journal of Molecular Sciences 23, no. 7: 3505. https://doi.org/10.3390/ijms23073505
APA StyleLocascio, A., Vassalli, Q. A., Castellano, I., & Palumbo, A. (2022). Novel Insights on Nitric Oxide Synthase and NO Signaling in Ascidian Metamorphosis. International Journal of Molecular Sciences, 23(7), 3505. https://doi.org/10.3390/ijms23073505






