Personalized Risk Schemes and Machine Learning to Empower Genomic Prognostication Models in Myelodysplastic Syndromes
Abstract
1. Introduction
2. MDS Prognostic Scoring Systems
2.1. The International Prognostic Scoring System (IPSS) and the Revised IPSS (IPSS-R)
2.2. Beyond IPSS-R
2.3. World Health Organization (WHO) Prognostic Scoring System (WPSS)
3. Recent Applications of Machine Learning Tools in Myelodysplastic Syndrome
3.1. Diagnostics
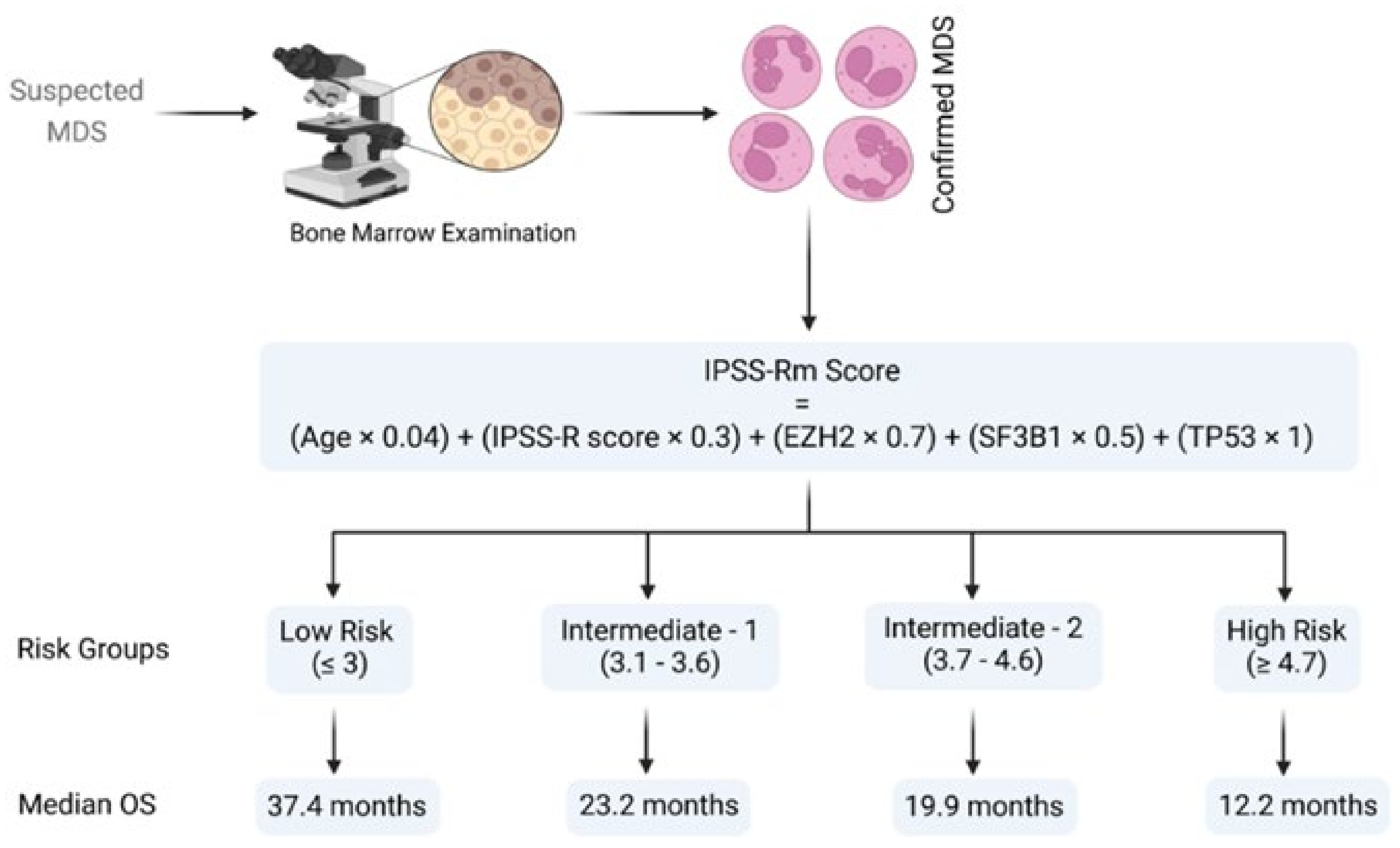
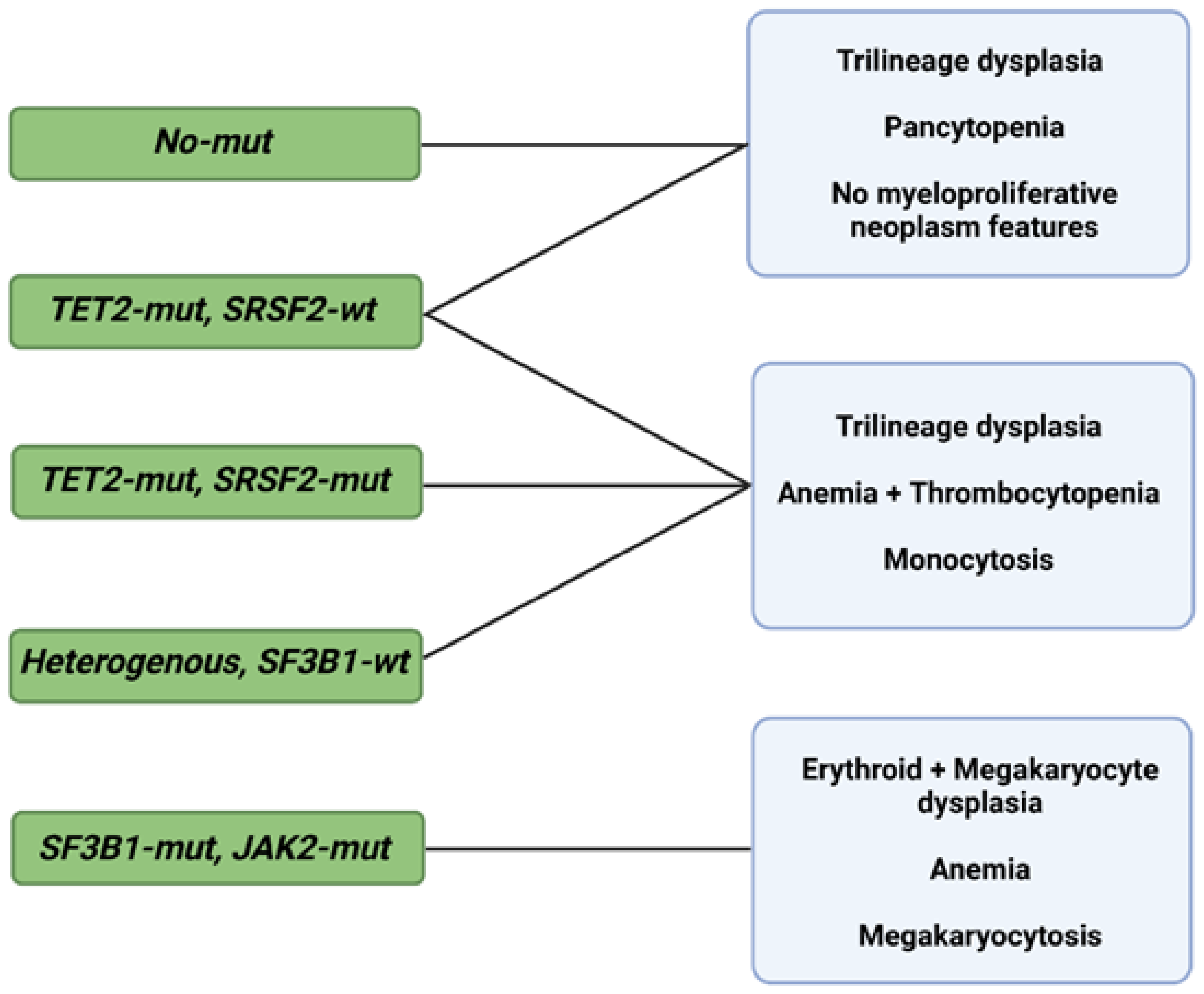
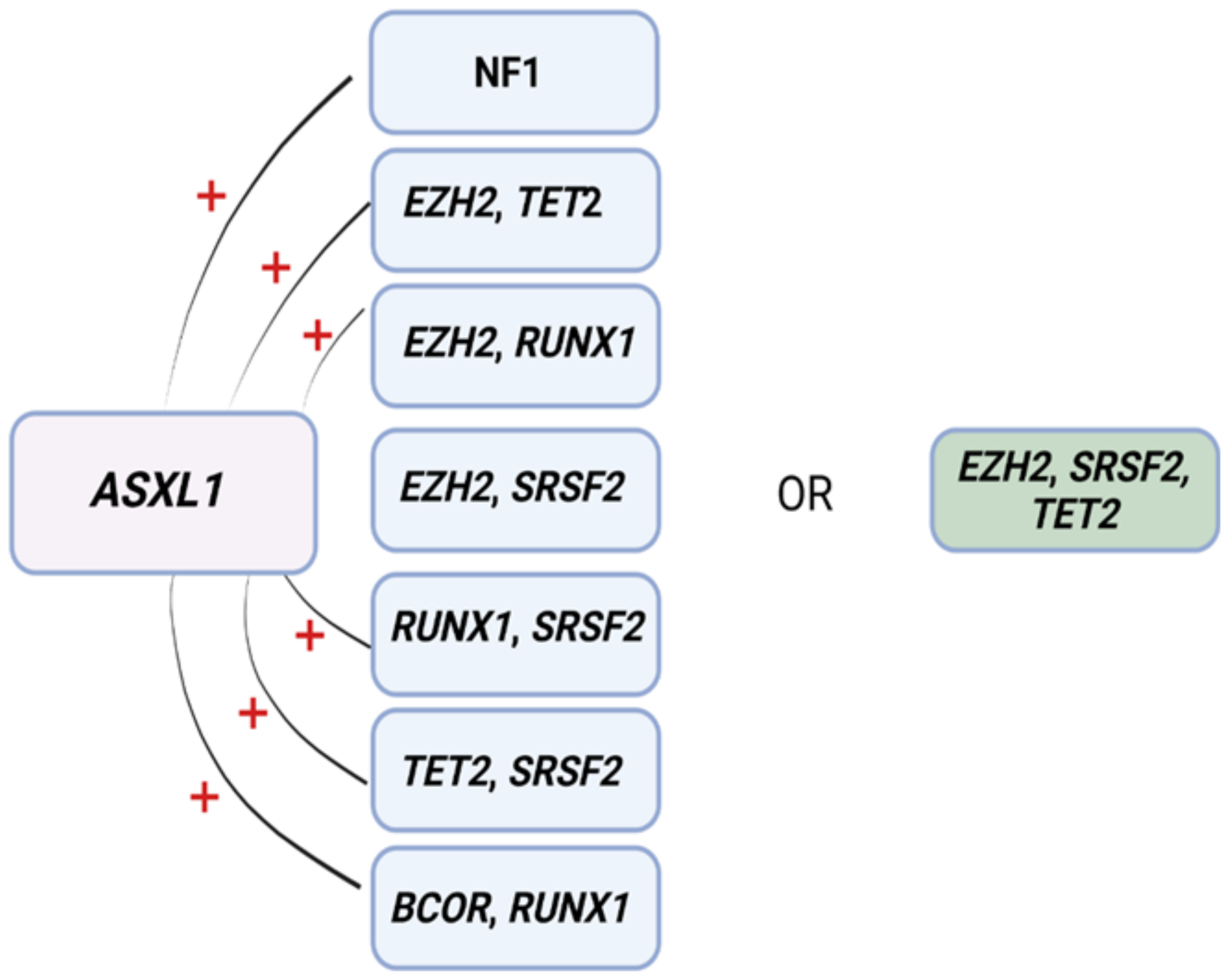
3.2. Risk Assessments and Prognostics
4. Future Endeavors and Perspective
- Hemoglobin, platelets, and BM blast percentage;
- IPSS-R cytogenetic categorization;
- 22 binary features derived from the presence of 21 predictive mutations;
- The number of mutations among 17 additional genes.
5. Limitations and Caveats
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Makishima, H.; Visconte, V.; Sakaguchi, H.; Jankowska, A.M.; Abu Kar, S.; Jerez, A.; Przychodzen, B.; Bupathi, M.; Guinta, K.; Afable, M.G.; et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 2012, 119, 3203–3210. [Google Scholar] [CrossRef] [PubMed]
- Damm, F.; Kosmider, O.; Gelsi-Boyer, V.; Renneville, A.; Carbuccia, N.; Hidalgo-Curtis, C.; Della Valle, V.; Couronné, L.; Scourzic, L.; Chesnais, V.; et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 2012, 119, 3211–3218. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Stevenson, K.E.; Caughey, B.; Lindsley, R.C.; Mar, B.G.; Stojanov, P.; Getz, G.; Steensma, D.P.; Ritz, J.; Soiffer, R.; et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J. Clin. Oncol. 2014, 32, 2691–2698. [Google Scholar] [CrossRef]
- Voso, M.T.; Gurnari, C. Have we reached a molecular era in myelodysplastic syndromes? Hematol. Am. Soc. Hematol. Educ. Program 2021, 2021, 418–427. [Google Scholar] [CrossRef]
- Gurnari, C.; Pagliuca, S.; Visconte, V. Alternative Splicing in Myeloid Malignancies. Biomedicines 2021, 9, 1844. [Google Scholar] [CrossRef]
- West, A.H.; Godley, L.A.; Churpek, J.E. Familial myelodysplastic syndrome/acute leukemia syndromes: A review and utility for translational investigations. Ann. N. Y. Acad. Sci. 2014, 1310, 111–118. [Google Scholar] [CrossRef]
- Gurnari, C.; Wahida, A.; Pagliuca, S.; Kewan, T.; Bahaj, W.; Mori, M.; Terkawi, L.; Pandit, I.; Zawit, M.; Carraway, H.E.; et al. TERT Rare Variants in Myeloid Neoplasia: Lack of Clinical Impact or Role as Risk Alleles. Blood 2021, 138, 1537. [Google Scholar] [CrossRef]
- Adès, L.; Itzykson, R.; Fenaux, P. Myelodysplastic syndromes. Lancet 2014, 383, 2239–2252. [Google Scholar] [CrossRef]
- Steensma, D.P. Myelodysplastic syndromes current treatment algorithm 2018. Blood Cancer J. 2018, 8, 47. [Google Scholar] [CrossRef]
- Meeus, P.; Michaux, L.; Bijnens, L.; Boogaerts, M.; Hagemeijer, A.; De Wolf-Peeters, C.; Verhoef, G. Application of the International Prognostic Scoring System for myelodysplastic syndromes. Ann. Oncol. 1999, 10, 825–829. [Google Scholar]
- Moreno Berggren, D.; Folkvaljon, Y.; Engvall, M.; Sundberg, J.; Lambe, M.; Antunovic, P.; Garelius, H.; Lorenz, F.; Nilsson, L.; Rasmussen, B.; et al. Prognostic scoring systems for myelodysplastic syndromes (MDS) in a population-based setting: A report from the Swedish MDS register. Br. J. Haematol. 2018, 181, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Corrales-Yepez, M.; Ali, N.A.; Kharfan-Dabaja, M.; Padron, E.; Zhang, L.; Epling-Burnette, P.K.; Pinilla-Ibarz, J.; Lancet, J.E.; List, A.F.; et al. Validation of the revised International Prognostic Scoring System in treated patients with myelodysplastic syndromes. Am. J. Hematol. 2013, 88, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Hasserjian, R.P.; Fox, P.S.; Stingo, F.; Zuo, Z.; Young, K.H.; Patel, K.; Medeiros, L.J.; Garcia-Manero, G.; Wang, S.A. Application of the international prognostic scoring system-revised in therapy-related myelodysplastic syndromes and oligoblastic acute myeloid leukemia. Leukemia 2014, 28, 185–189. [Google Scholar] [CrossRef]
- Voso, M.T.; Fenu, S.; Latagliata, R.; Buccisano, F.; Piciocchi, A.; Aloe-Spiriti, M.A.; Breccia, M.; Criscuolo, M.; Andriani, A.; Mancini, S.; et al. Revised International Prognostic Scoring System (IPSS) predicts survival and leukemic evolution of myelodysplastic syndromes significantly better than IPSS and WHO Prognostic Scoring System: Validation by the Gruppo Romano Mielodisplasie Italian Regional Database. J. Clin. Oncol. 2013, 31, 2671–2677. [Google Scholar]
- Della Porta, M.G.; Alessandrino, E.P.; Bacigalupo, A.; van Lint, M.T.; Malcovati, L.; Pascutto, C.; Falda, M.; Bernardi, M.; Onida, F.; Guidi, S.; et al. Predictive factors for the outcome of allogeneic transplantation in patients with MDS stratified according to the revised IPSS-R. Blood 2014, 123, 2333–2342. [Google Scholar] [CrossRef]
- Gangat, N.; Patnaik, M.M.; Begna, K.; Kourelis, T.; Al-Kali, A.; Elliott, M.A.; Hogan, W.J.; Letendre, L.; Litzow, M.R.; Knudson, R.A.; et al. Primary Myelodysplastic Syndromes: The Mayo Clinic Experience With 1000 Patients. Mayo Clin. Proc. 2015, 90, 1623–1638. [Google Scholar] [CrossRef]
- Greenberg, P.; Cox, C.; LeBeau, M.M.; Fenaux, P.; Morel, P.; Sanz, G.; Sanz, M.; Vallespi, T.; Hamblin, T.; Oscier, D.; et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997, 89, 2079–2088. [Google Scholar] [CrossRef]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Germing, U.; Kuendgen, A.; Della Porta, M.G.; Pascutto, C.; Invernizzi, R.; Giagounidis, A.; Hildebrandt, B.; Bernasconi, P.; Knipp, S.; et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J. Clin. Oncol. 2007, 25, 3503–3510. [Google Scholar] [CrossRef] [PubMed]
- Sekeres, M.A.; Swern, A.S.; Fenaux, P.; Greenberg, P.L.; Sanz, G.F.; Bennett, J.M.; Dreyfus, F.; List, A.F.; Li, J.S.; Sugrue, M.M. Validation of the IPSS-R in lenalidomide-treated, lower-risk myelodysplastic syndrome patients with del(5q). Blood Cancer J. 2014, 4, e242. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Breccia, M.; Salaroli, A.; Loglisci, G.; Alimena, G. Revised IPSS (IPSS-R) stratification and outcome of MDS patients treated with azacitidine. Ann. Hematol. 2013, 92, 411–412. [Google Scholar] [CrossRef] [PubMed]
- Lamarque, M.; Raynaud, S.; Itzykson, R.; Thepot, S.; Quesnel, B.; Dreyfus, F.; Rauzy, O.B.; Turlure, P.; Vey, N.; Recher, C.; et al. The revised IPSS is a powerful tool to evaluate the outcome of MDS patients treated with azacitidine: The GFM experience. Blood 2012, 120, 5084–5085. [Google Scholar] [CrossRef][Green Version]
- Neukirchen, J.; Lauseker, M.; Blum, S.; Giagounidis, A.; Lübbert, M.; Martino, S.; Siragusa, S.; Schlenk, R.F.; Platzbecker, U.; Hofmann, W.K.; et al. Validation of the revised international prognostic scoring system (IPSS-R) in patients with myelodysplastic syndrome: A multicenter study. Leuk. Res. 2014, 38, 57–64. [Google Scholar] [CrossRef]
- Itzykson, R.; Thépot, S.; Quesnel, B.; Dreyfus, F.; Beyne-Rauzy, O.; Turlure, P.; Vey, N.; Recher, C.; Dartigeas, C.; Legros, L.; et al. Prognostic factors for response and overall survival in 282 patients with higher-risk myelodysplastic syndromes treated with azacitidine. Blood 2011, 117, 403–411. [Google Scholar] [CrossRef]
- Nazha, A.; Komrokji, R.S.; Garcia-Manero, G.; Barnard, J.; Roboz, G.J.; Steensma, D.P.; DeZern, A.E.; Zell, K.; Zimmerman, C.; Ali, N.A.; et al. The efficacy of current prognostic models in predicting outcome of patients with myelodysplastic syndromes at the time of hypomethylating agent failure. Haematologica 2016, 101, e224–e227. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Sekeres, M.A.; Garcia-Manero, G.; Steensma, D.P.; Zell, K.; Barnard, J.; Ali, N.A.; Zimmerman, C.; Roboz, G.; DeZern, A.; et al. Comparison of risk stratification tools in predicting outcomes of patients with higher-risk myelodysplastic syndromes treated with azanucleosides. Leukemia 2016, 30, 649–657. [Google Scholar] [CrossRef]
- Nazha, A.; Narkhede, M.; Radivoyevitch, T.; Seastone, D.J.; Patel, B.J.; Gerds, A.T.; Mukherjee, S.; Kalaycio, M.; Advani, A.; Przychodzen, B.; et al. Incorporation of molecular data into the Revised International Prognostic Scoring System in treated patients with myelodysplastic syndromes. Leukemia 2016, 30, 2214–2220. [Google Scholar] [CrossRef]
- Hou, H.A.; Tsai, C.H.; Lin, C.C.; Chou, W.C.; Kuo, Y.Y.; Liu, C.Y.; Tseng, M.H.; Peng, Y.L.; Liu, M.C.; Liu, C.W.; et al. Incorporation of mutations in five genes in the revised International Prognostic Scoring System can improve risk stratification in the patients with myelodysplastic syndrome. Blood Cancer J. 2018, 8, 39. [Google Scholar] [CrossRef]
- Gu, S.; Xia, J.; Tian, Y.; Zi, J.; Ge, Z. A novel scoring system integrating molecular abnormalities with IPSS-R can improve the risk stratification in patients with MDS. BMC Cancer 2021, 21, 134. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Guo, J.; Wu, D.; Wu, L.Y.; Song, L.X.; Zhang, Z.; Zhao, Y.S.; Chang, C.K. Integration Analysis of JAK2 or RUNX1 Mutation With Bone Marrow Blast Can Improve Risk Stratification in the Patients With Lower Risk Myelodysplastic Syndrome. Front. Oncol. 2020, 10, 610525. [Google Scholar] [CrossRef]
- Efficace, F.; Cottone, F.; Abel, G.; Niscola, P.; Gaidano, G.; Bonnetain, F.; Anota, A.; Caocci, G.; Cronin, A.; Fianchi, L.; et al. Patient-reported outcomes enhance the survival prediction of traditional disease risk classifications: An international study in patients with myelodysplastic syndromes. Cancer 2017, 124, 1251–1259. [Google Scholar] [CrossRef]
- Abel, G.A.; Efficace, F.; Buckstein, R.J.; Tinsley, S.; Jurcic, J.G.; Martins, Y.; Steensma, D.P.; Watts, C.D.; Raza, A.; Lee, S.J. Prospective international validation of the Quality of Life in Myelodysplasia Scale (QUALMS). Haematoligica 2016, 101, 781–788. [Google Scholar] [CrossRef]
- Jain, A.G.; Zhang, L.; Bennett, J.M.; Komrokji, R. Myelodysplastic Syndromes with Bone Marrow Fibrosis: An Update. Ann. Lab. Med. 2022, 42, 299–305. [Google Scholar] [CrossRef]
- Della Porta, M.G.; Malcovati, L.; Boveri, E.; Travaglino, E.; Pietra, D.; Pascutto, C.; Passamonti, F.; Invernizzi, R.; Castello, A.; Magrini, U.; et al. Clinical relevance of bone marrow fibrosis and CD34-positive cell clusters in primary myelodysplastic syndromes. J. Clin. Oncol. 2009, 27, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Steensma, D.P.; Hanson, C.A.; Letendre, L.; Tefferi, A. Myelodysplasia with fibrosis: A distinct entity? Leuk. Res. 2001, 25, 829–838. [Google Scholar] [CrossRef]
- Della Porta, M.G.; Malcovati, L. Myelodysplastic syndromes with bone marrow fibrosis. Haematologica 2011, 96, 180–183. [Google Scholar] [CrossRef]
- Buesche, G.; Teoman, H.; Wilczak, W.; Ganser, A.; Hecker, H.; Wilken, L.; Göhring, G.; Schlegelberger, B.; Bock, O.; Georgii, A.; et al. Marrow fibrosis predicts early fatal marrow failure in patients with myelodysplastic syndromes. Leukemia 2008, 22, 313–322. [Google Scholar] [CrossRef]
- Thiele, J.; Kvasnicka, H.M.; Facchetti, F.; Franco, V.; van der Walt, J.; Orazi, A. European consensus on grading bone marrow fibrosis and assessment of cellularity. Haematologica 2005, 90, 1128–1132. [Google Scholar]
- Lambertenghi-Deliliers, G.; Orazi, A.; Luksch, R.; Annaloro, C.; Soligo, D. Myelodysplastic syndrome with increased marrow fibrosis: A distinct clinico-pathological entity. Br. J. Haematol. 1991, 78, 161–166. [Google Scholar] [CrossRef]
- Sultan, C.; Sigaux, F.; Imbert, M.; Reyes, F. Acute myelodysplasia with myelofibrosis: A report of eight cases. Br. J. Haematol. 1981, 49, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Alessandrino, E.P.; Della Porta, M.G.; Bacigalupo, A.; Van Lint, M.T.; Falda, M.; Onida, F.; Bernardi, M.; Iori, A.P.; Rambaldi, A.; Cerretti, R.; et al. WHO classification and WPSS predict posttransplantation outcome in patients with myelodysplastic syndrome: A study from the Gruppo Italiano Trapianto di Midollo Osseo (GITMO). Blood 2008, 112, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Du, M.Y.; Xu, M.; Deng, J.; Liu, L.; Guo, T.; Xia, L.H.; Hu, Y.; Mei, H. Evaluation of different scoring systems and gene mutations for the prognosis of myelodysplastic syndrome (MDS) in Chinese population. J. Cancer 2020, 11, 508–519. [Google Scholar] [CrossRef]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar]
- Chen, T.; He, T.; Benesty, M. Xgboost: Extreme gradient boosting. R Package Version 2015, 0.4--2 1, 1--4. Available online: https://cran.microsoft.com/snapshot/2017-12-11/web/packages/xgboost/vignettes/xgboost.pdf (accessed on 12 January 2022).
- Kimura, K.; Tabe, Y.; Ai, T.; Takehara, I.; Fukuda, H.; Takahashi, H.; Naito, T.; Komatsu, N.; Uchihashi, K.; Ohsaka, A. A novel automated image analysis system using deep convolutional neural networks can assist to differentiate MDS and AA. Sci. Rep. 2019, 9, 13385. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, A.; Merino, A.; Boldú, L.; Molina, Á.; Alférez, S.; Rodellar, J. A new convolutional neural network predictive model for the automatic recognition of hypogranulated neutrophils in myelodysplastic syndromes. Comput. Biol. Med. 2021, 134, 104479. [Google Scholar] [CrossRef] [PubMed]
- Radakovich, N.; Meggendorfer, M.; Malcovati, L.; Hilton, C.B.; Sekeres, M.A.; Shreve, J.; Rouphail, Y.; Walter, W.; Hutter, S.; Galli, A.; et al. A geno-clinical decision model for the diagnosis of myelodysplastic syndromes. Blood Adv. 2021, 5, 4361–4369. [Google Scholar] [CrossRef]
- Orazi, A.; Milanesi, B. The technicon H6000 automated hematology analyzer in the diagnosis and classification of the myelodysplastic syndromes. Haematologica 1990, 75, 87–90. [Google Scholar]
- Cherian, S.; Moore, J.; Bantly, A.; Vergilio, J.A.; Klein, P.; Luger, S.; Bagg, A. Peripheral blood MDS score: A new flow cytometric tool for the diagnosis of myelodysplastic syndromes. Cytom. B Clin. Cytom. 2005, 64, 9–17. [Google Scholar] [CrossRef]
- Aires, A.; Teixeira, M.D.A.; Lau, C.; Moreira, C.; Spínola, A.; Mota, A.; Freitas, I.; Coutinho, J.; Lima, M. A pilot study on the usefulness of peripheral blood flow cytometry for the diagnosis of lower risk myelodysplastic syndromes: The “MDS thermometer”. BMC Hematol. 2018, 18, 6. [Google Scholar] [CrossRef]
- Miguel, A.; Teixeira, M.; Lau, C.; Moreira, C.; Spínola, A.; Mota, A.; Freitas, I.; Coutinho, J.; Lima, M. Automated neutrophil morphology and its utility in the assessment of neutrophil dysplasia. Lab. Hematol. 2007, 13, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Furundarena, J.R.; Araiz, M.; Uranga, M.; Sainz, M.R.; Agirre, A.; Trassorras, M.; Uresandi, N.; Montes, M.C.; Argoitia, N. The utility of the Sysmex XE-2100 analyzer’s NEUT-X and NEUT-Y parameters for detecting neutrophil dysplasia in myelodysplastic syndromes. Int. J. Lab. Hematol. 2010, 32, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Ku, Y.; Yoo, B.W.; Kim, J.A.; Lee, D.S.; Chai, Y.J.; Kong, H.J.; Kim, H.C. White blood cell differential count of maturation stages in bone marrow smear using dual-stage convolutional neural networks. PLoS ONE 2017, 12, e0189259. [Google Scholar] [CrossRef]
- Kainz, P.; Burgsteiner, H.; Asslaber, M.; Ahammer, H. Training echo state networks for rotation-invariant bone marrow cell classification. Neural. Comput. Appl. 2017, 28, 1277–1292. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; Huang, T.C.; Ye, R.H.; Fang, W.H.; Lai, S.W.; Chang, P.Y.; Liu, W.N.; Kuo, T.Y.; Lee, C.H.; Tsai, W.C.; et al. A Hematologist-Level Deep Learning Algorithm (BMSNet) for Assessing the Morphologies of Single Nuclear Balls in Bone Marrow Smears: Algorithm Development. JMIR Med. Inform. 2020, 8, e15963. [Google Scholar] [CrossRef]
- Mori, J.; Kaji, S.; Kawai, H.; Kida, S.; Tsubokura, M.; Fukatsu, M.; Harada, K.; Noji, H.; Ikezoe, T.; Maeda, T.; et al. Assessment of dysplasia in bone marrow smear with convolutional neural network. Sci. Rep. 2020, 10, 14734. [Google Scholar] [CrossRef]
- Rosenberg, C.A.; Bill, M.; Rodrigues, M.A.; Hauerslev, M.; Kerndrup, G.B.; Hokland, P.; Ludvigsen, M. Exploring dyserythropoiesis in patients with myelodysplastic syndrome by imaging flow cytometry and machine-learning assisted morphometrics. Cytom. B Clin. Cytom. 2021, 100, 554–567. [Google Scholar] [CrossRef]
- Ali, A.M.; Huang, Y.; Pinheiro, R.F.; Xue, F.; Hu, J.; Iverson, N.; Hoehn, D.; Coutinho, D.; Kayani, J.; Chernak, B.; et al. Severely impaired terminal erythroid differentiation as an independent prognostic marker in myelodysplastic syndromes. Blood Adv. 2018, 2, 1393–1402. [Google Scholar] [CrossRef]
- Goasguen, J.E.; Bennett, J.M.; Bain, B.J.; Brunning, R.; Vallespi, M.T.; Tomonaga, M.; Zini, G.; Renault, A. The International Working Group on Morphology of MDS Dyserythropoiesis in the diagnosis of the myelodysplastic syndromes and other myeloid neoplasms: Problem areas. Br. J. Haematol. 2018, 182, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Hellström-Lindberg, E.; Bowen, D.; Adès, L.; Cermak, J.; Del Cañizo, C.; Della Porta, M.G.; Fenaux, P.; Gattermann, N.; Germing, U.; et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: Recommendations from the European LeukemiaNet. Blood 2013, 122, 2943–2964. [Google Scholar] [CrossRef] [PubMed]
- Kieseberg, P.; Malle, B.; Frühwirt, P.; Weippl, E.; Holzinger, A. A tamper-proof audit and control system for the doctor in the loop. Brain Inform. 2016, 3, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Radhachandran, A.; Garikipati, A.; Iqbal, Z.; Siefkas, A.; Barnes, G.; Hoffman, J.; Mao, Q.; Das, R. A machine learning approach to predicting risk of myelodysplastic syndrome. Leuk. Res. 2021, 109, 106639. [Google Scholar] [CrossRef] [PubMed]
- Brück, O.E.; Lallukka-Brück, S.E.; Hohtari, H.R.; Ianevski, A.; Ebeling, F.T.; Kovanen, P.E.; Kytölä, S.I.; Aittokallio, T.A.; Ramos, P.M.; Porkka, K.V.; et al. Machine Learning of Bone Marrow Histopathology Identifies Genetic and Clinical Determinants in Patients with MDS. Blood Cancer Discov. 2021, 2, 238–249. [Google Scholar] [CrossRef]
- Macedo, L.C.; Lallukka-Brück, S.E.; Hohtari, H.R.; Ianevski, A.; Ebeling, F.T.; Kovanen, P.E.; Kytölä, S.I.; Aittokallio, T.A.; Ramos, P.M.; Porkka, K.V.; et al. Genetics factors associated with myelodysplastic syndromes. Blood Cells Mol. Dis. 2015, 55, 76–81. [Google Scholar] [CrossRef]
- Zhang, L.; Padron, E.; Lancet, J. The molecular basis and clinical significance of genetic mutations identified in myelodysplastic syndromes. Leuk. Res. 2015, 39, 6–17. [Google Scholar] [CrossRef]
- Malcovati, L.; Papaemmanuil, E.; Ambaglio, I.; Elena, C.; Gallì, A.; Della Porta, M.G.; Travaglino, E.; Pietra, D.; Pascutto, C.; Ubezio, M.; et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood 2014, 124, 1513–1521. [Google Scholar] [CrossRef]
- Nagata, Y.; Zhao, R.; Awada, H.; Kerr, C.K.; Mirzaev, I.; Kongkiatkamon, S.; Nazha, A.; Makishima, H.; Radivoyevitch, T.; Scott, J.G.; et al. Machine learning demonstrates that somatic mutations imprint invariant morphologic features in myelodysplastic syndromes. Blood 2020, 136, 2249–2262. [Google Scholar] [CrossRef]
- Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Matteuzzi, T.; Sala, C.; Mosca, E.; Chiereghin, C.; Di Nanni, N.; Gnocchi, M.; Zampini, M.; et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1223–1233. [Google Scholar] [CrossRef]
- Nazha, A.; Komrokji, R.; Meggendorfer, M.; Jia, X.; Radakovich, N.; Shreve, J.; Hilton, C.B.; Nagata, Y.; Hamilton, B.K.; Mukherjee, S.; et al. Personalized Prediction Model to Risk Stratify Patients with Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 3737–3746. [Google Scholar] [CrossRef] [PubMed]
- Nazha, A.; Sekeres, M.A.; Bejar, R.; Rauh, M.J.; Othus, M.; Komrokji, R.S.; Barnard, J.; Hilton, C.B.; Kerr, C.M.; Steensma, D.P.; et al. Genomic Biomarkers to Predict Resistance to Hypomethylating Agents in Patients With Myelodysplastic Syndromes Using Artificial Intelligence. JCO Precis. Oncol. 2019, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Arango Ossa, J.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognostic Scoring System. Blood 2021. Abstract #61. [Google Scholar]
- Pagliuca, S.; Gurnari, C.; Visconte, V. Molecular Targeted Therapy in Myelodysplastic Syndromes: New Options for Tailored Treatments. Cancers 2021, 13, 784. [Google Scholar] [CrossRef] [PubMed]
- Björnsson, B.; Borrebaeck, C.; Elander, N.; Gasslander, T.; Gawel, D.R.; Gustafsson, M.; Jörnsten, R.; Lee, E.J.; Li, X.; Lilja, S.; et al. Digital twins to personalize medicine. Genome Med. 2019, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020, 26, 1549–1556. [Google Scholar] [CrossRef]
- Abuhadra, N.; Mukherjee, S.; Al-Issa, K.; Adema, V.; Hirsch, C.M.; Advani, A.; Przychodzen, B.; Makhoul, A.; Awada, H.; Maciejewski, J.P.; et al. BCOR and BCORL1 mutations in myelodysplastic syndromes (MDS): Clonal architecture and impact on outcomes. Leuk. Lymphoma 2019, 60, 1587–1590. [Google Scholar] [CrossRef]
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern. Med. 2018, 178, 1544–1547. [Google Scholar] [CrossRef]
- Vokinger, K.N.; Feuerriegel, S.; Kesselheim, A.S. Mitigating bias in machine learning for medicine. Commun. Med. 2021, 1, 25. [Google Scholar] [CrossRef]
- Price, W.N.; Gerke, S.; Cohen, I.G. Potential Liability for Physicians Using Artificial Intelligence. JAMA 2019, 322, 1765–1766. [Google Scholar] [CrossRef]
| IPSS, IPSS-R, and WPSS | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 1.5 | 2 | 3 | 4 | ||
| % BM Blasts | IPSS | <5 | ≥5 to ≤10 | ≥11 to ≤20 | ≥21 to ≤30 | |||
| IPSS-R | ≤2 | >2 to <5 | ≥5 to ≤10 | >10 | ||||
| Karyotype | IPSS * | Good | Intermediate | Poor | ||||
| IPSS-R ** | Very Good | Good | Intermediate | Poor | Very Poor | |||
| WPSS * | Good | Intermediate | Poor | |||||
| Cytopenias *** | IPSS | <2 | ≥2 | |||||
| Hemoglobin | IPSS-R | ≥10 | ≥8 to <10 | <8 | ||||
| Platelets | IPSS-R | >100,000 | ≥50,000 to ≤100,000 | <50,000 | ||||
| ANC | IPSS-R | ≥800 | <800 | |||||
| WHO Classification | WPSS | Refractory anemia, Refractory Anemia with ringed sideroblasts, MDS with isolated 5q- | Refractory anemia with multilineage dysplasia, Refractory anemia with multilineage dysplasia and ring sideroblasts | Refractory anemia with excess blasts-1 | Refractory anemia with excess blasts-2 | |||
| RBC Transfusions | WPSS | Absent | Every 8 weeks for 4 months | |||||
| Risk Category | Very Low | Low | Intermediate (Intermediate 1) | Intermediate 2 | High | Very High | ||
| IPSS Score | 0 | ≥0.5 to ≤1 | ≥1.5 to ≤2 | ≥2.5 | ||||
| IPSS-R Score | ≤1.5 | 1.5 to ≤3 | >3 to ≤4.5 | >4.5 to ≤6 | >6 | |||
| WPSS Score | 0 | 1 | 2 | ≥3 to ≤4 | ≥5 to ≤6 | |||
| IPSS Overall Survival (years) | 5.7 | 3.5 | 1.2 | 0.4 | ||||
| IPSS-R Overall Survival (years) | 8.8 | 5.3 | 3 | 1.6 | 0.8 | |||
| WPSS Median Survival (years) | 11.75 | 5.5 | 4 | 2.17 | 0.75 | |||
| IPSS 25% AML Transformation (years) | 9.4 | 3.3 | 1.1 | 0.2 | ||||
| IPSS-R AML Transformation (years) | >14.5 | >10.8 | 3.2 | 1.4 | 0.7 | |||
| WPSS % AML Transformation at 5 years | 3 | 14 | 33 | 54 | 84 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awada, H.; Gurnari, C.; Durmaz, A.; Awada, H.; Pagliuca, S.; Visconte, V. Personalized Risk Schemes and Machine Learning to Empower Genomic Prognostication Models in Myelodysplastic Syndromes. Int. J. Mol. Sci. 2022, 23, 2802. https://doi.org/10.3390/ijms23052802
Awada H, Gurnari C, Durmaz A, Awada H, Pagliuca S, Visconte V. Personalized Risk Schemes and Machine Learning to Empower Genomic Prognostication Models in Myelodysplastic Syndromes. International Journal of Molecular Sciences. 2022; 23(5):2802. https://doi.org/10.3390/ijms23052802
Chicago/Turabian StyleAwada, Hussein, Carmelo Gurnari, Arda Durmaz, Hassan Awada, Simona Pagliuca, and Valeria Visconte. 2022. "Personalized Risk Schemes and Machine Learning to Empower Genomic Prognostication Models in Myelodysplastic Syndromes" International Journal of Molecular Sciences 23, no. 5: 2802. https://doi.org/10.3390/ijms23052802
APA StyleAwada, H., Gurnari, C., Durmaz, A., Awada, H., Pagliuca, S., & Visconte, V. (2022). Personalized Risk Schemes and Machine Learning to Empower Genomic Prognostication Models in Myelodysplastic Syndromes. International Journal of Molecular Sciences, 23(5), 2802. https://doi.org/10.3390/ijms23052802







