Extracellular Vesicles Secreted by Glioma Stem Cells Are Involved in Radiation Resistance and Glioma Progression
Abstract
:1. Introduction
2. Results and Discussion
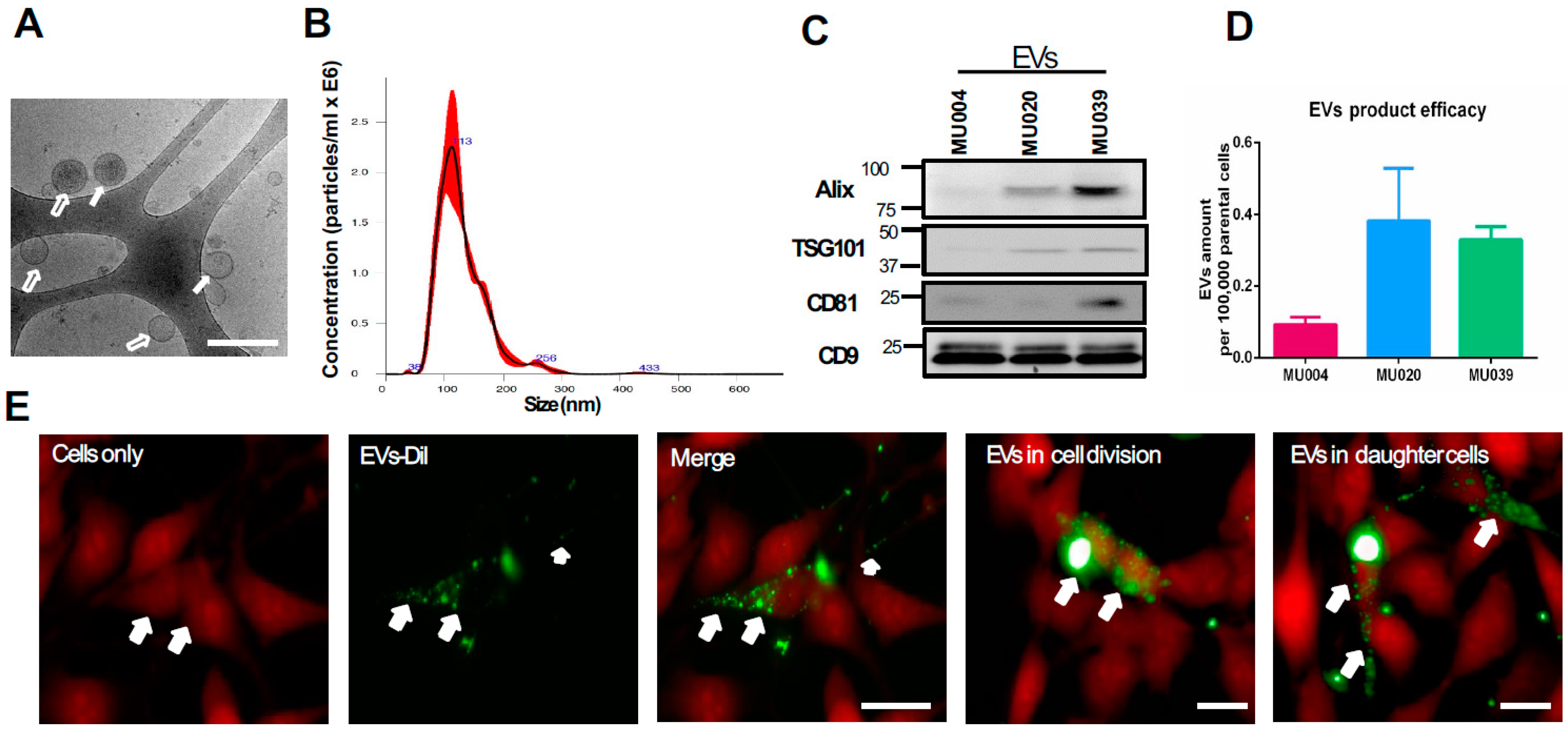
2.1. Exosomes Characterisation and Internalisation
2.2. GSC-EVs Enhance Radiation Resistance in Glioma Cells
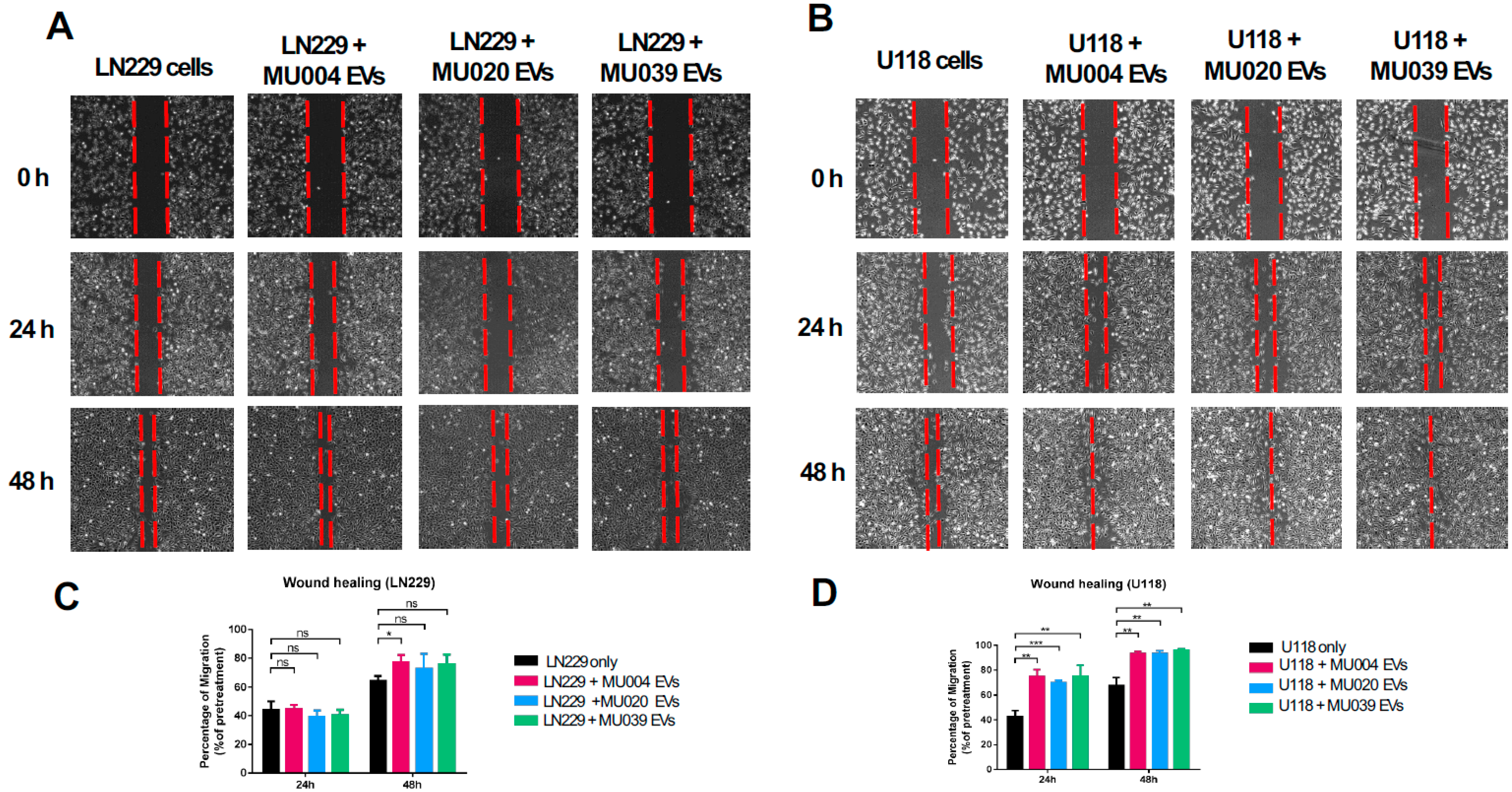
2.3. GSC-Derived Exosomes Enhanced Tumour Cell Migration and Spreading
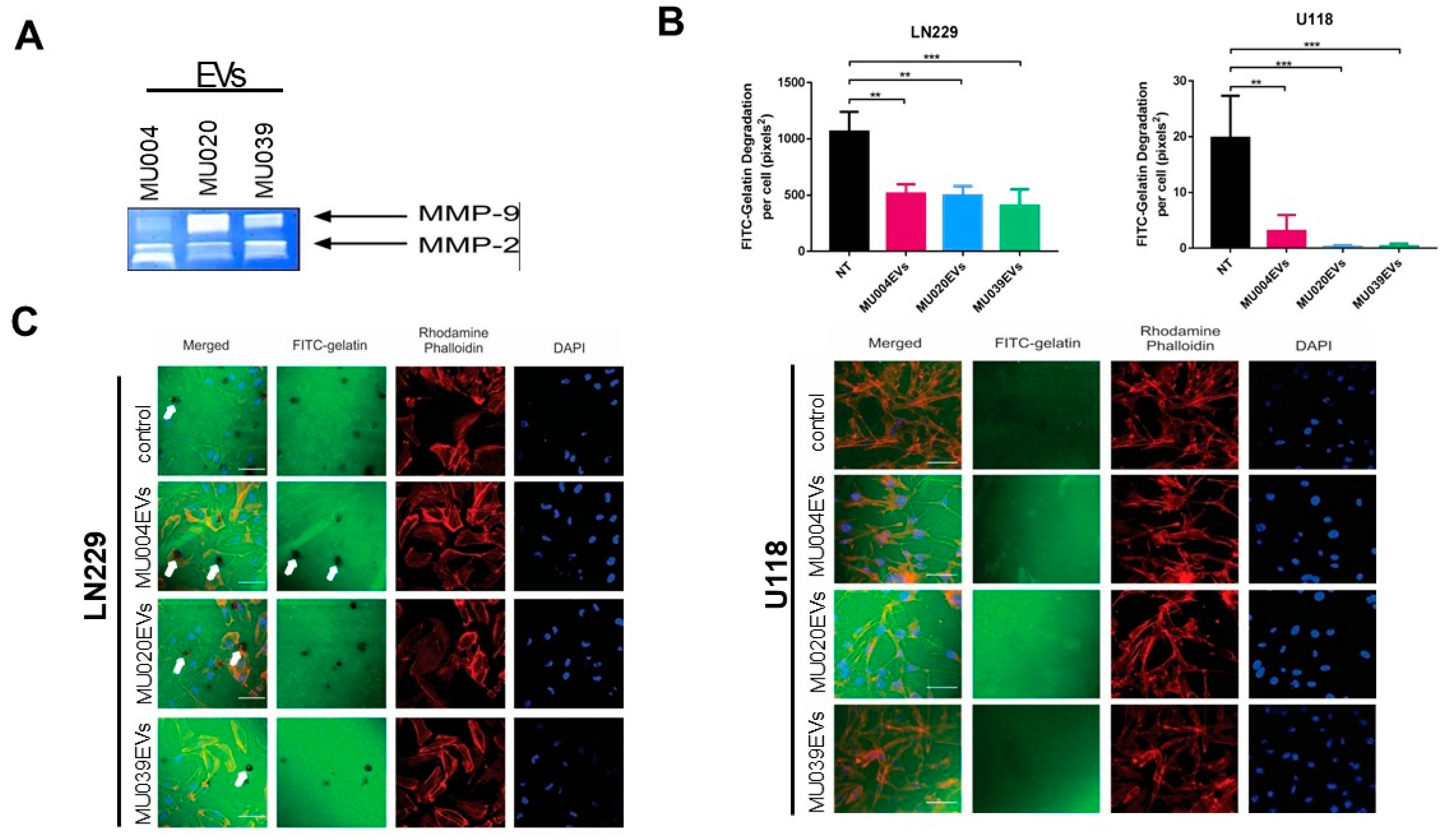
2.4. GSC-EVs Are Involved in Invadopodia and Proliferative Activity
2.5. miRNAs Cargoes in EVs Reflect GBM Phenotype
3. Discussion
4. Methods
4.1. Ethical Approval
4.2. Cell Culture
4.3. Exosome Isolation and Characterisation and Uptake
4.4. EVs Size and Morphology Quantification
4.5. Western Blot
4.6. Radiation Treatment
4.7. Cell Viability Assay
4.8. Clonogenic Assay
4.9. Cell Migration/Invasion
4.10. Zymography and Invadopodia Assay
4.11. Exosomes miRNA Profiling and Analysis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaye, A.H.; Morokoff, A. The Continuing Evolution: Biology and Treatment of Brain Tumors. Neurosurgery 2014, 61 (Suppl S1), 100–104. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.M.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavaré, S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef] [Green Version]
- Pasqualetti, F.; Montemurro, N.; Desideri, I.; Loi, M.; Giannini, N.; Gadducci, G.; Malfatti, G.; Cantarella, M.; Gonnelli, A.; Montrone, S.; et al. Impact of recurrence pattern in patients undergoing a second surgery for recurrent glioblastoma. Acta Neurol. Belg. 2021, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Bhamidipati, D.; Song, A.; Eldredge-Hindy, H.B.; Siglin, J.; Dan, T.D.; Champ, C.E.; Zhang, I.; Bar-Ad, V.; Kim, L.; et al. Bevacizumab and re-irradiation for recurrent high grade gliomas: Does sequence matter? J. Neurooncol. 2018, 140, 623–628. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Yu, T.-S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 2008, 455, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wang, Q.; Wang, Y.A.; Hua, S.; Sauvé, C.-E.G.; Ong, D.; Lan, Z.D.; Chang, Q.; Ho, Y.W.; Monasterio, M.M.; et al. Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell 2016, 167, 1281–1295.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.V.; Filiz, G.; Daniel, P.M.; Hollande, F.; Dworkin, S.; Amiridis, S.; Kountouri, N.; Ng, W.; Morokoff, A.P.; Mantamadiotis, T. Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS ONE 2017, 12, e0172791. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Dieppa, D.R.; Steinberg, J.; Gonda, D.; Cheung, V.J.; Carter, B.S.; Chen, C.C. Extracellular vesicles as a platform for ‘liquid biopsy’ in glioblastoma patients. Expert. Rev. Mol. Diagn. 2014, 14, 819–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godlewski, J.; Krichevsky, A.M.; Johnson, M.D.; Chiocca, E.A.; Bronisz, A. Belonging to a network—Micrornas, extracellular vesicles, and the glioblastoma microenvironment. Neuro-Oncology 2015, 17, 652–662. [Google Scholar] [CrossRef] [Green Version]
- Luga, V.; Wrana, J.L. Tumor–Stroma Interaction: Revealing Fibroblast-Secreted Exosomes as Potent Regulators of Wnt-Planar Cell Polarity Signaling in Cancer Metastasis. Cancer Res. 2013, 73, 6843–6847. [Google Scholar] [CrossRef] [Green Version]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastoma biomarker development. PLoS ONE 2013, 8, e78115. [Google Scholar] [CrossRef] [PubMed]
- Putz, U.; Howitt, J.; Doan, A.; Goh, C.-P.; Low, L.-H.; Silke, J.; Tan, S.-S. The Tumor Suppressor PTEN Is Exported in Exosomes and Has Phosphatase Activity in Recipient Cells. Sci. Signal. 2012, 5, ra70. [Google Scholar] [CrossRef] [PubMed]
- Li, C.C.Y.; Eaton, S.; Young, P.E.; Lee, M.; Shuttleworth, R.; Humphreys, D.; Grau, G.; Combes, V.; Bebawy, M.; Gong, J.; et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. 2013, 10, 1333–1344. [Google Scholar] [CrossRef] [Green Version]
- Ricklefs, F.; Mineo, M.; Rooj, A.K.; Nakano, I.; Charest, A.; Weissleder, R.; Breakefield, X.O.; Chiocca, E.A.; Godlewski, J.; Bronisz, A. Extracellular Vesicles from High-Grade Glioma Exchange Diverse Pro-oncogenic Signals That Maintain Intratumoral Heterogeneity. Cancer Res. 2016, 76, 2876–2881. [Google Scholar] [CrossRef] [Green Version]
- van der Vos, K.E.; Abels, E.R.; Zhang, X.; Lai, C.; Carrizosa, E.; Oakley, D.; Prabhakar, S.; Mardini, O.; Crommentuijn, M.H.W.; Skog, J.; et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro-Oncology 2016, 18, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef] [Green Version]
- Boelens, M.C.; Wu, T.J.; Nabet, B.Y.; Xu, B.; Qiu, Y.; Yoon, T.; Azzam, D.J.; Victor, C.T.-S.; Wiemann, B.Z.; Ishwaran, H.; et al. Exosome Transfer from Stromal to Breast Cancer Cells Regulates Therapy Resistance Pathways. Cell 2014, 159, 499–513. [Google Scholar] [CrossRef] [Green Version]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.-K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [Green Version]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.-F.; et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef]
- Yang, J.-K.; Yang, J.-P.; Tong, J.; Jing, S.-Y.; Fan, B.; Wang, F.; Sun, G.-Z.; Jiao, B.-H. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J. Neuro-Oncol. 2016, 131, 255–265. [Google Scholar] [CrossRef]
- Zhang, Z.; Yin, J.; Lu, C.; Wei, Y.; Zeng, A.; You, Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 166. [Google Scholar] [CrossRef]
- Yue, X.; Lan, F.; Xia, T. Hypoxic Glioma Cell-Secreted Exosomal miR-301a Activates Wnt/β-catenin Signaling and Promotes Radiation Resistance by Targeting TCEAL7. Mol. Ther. 2019, 27, 1939–1949. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Brown, D.V.; Daniel, P.M.; D’Abaco, G.M.; Gogos, A.; Ng, W.; Morokoff, A.; Mantamadiotis, T. Coexpression analysis of CD133 and CD44 identifies Proneural and Mesenchymal subtypes of glioblastoma multiforme. Oncotarget 2015, 6, 6267–6280. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, C.; Nguyen, H.P.T.; Morokoff, A.; Luwor, R.; Paradiso, L.; Kaye, A.H.; Mantamadiotis, T.; Stylli, S.S. Inhibition of Radiation and Temozolomide-Induced Invadopodia Activity in Glioma Cells Using FDA-Approved Drugs. Transl. Oncol. 2018, 11, 1406–1418. [Google Scholar] [CrossRef]
- Mao, L.; Whitehead, C.; Paradiso, L.; Kaye, A.H.; Morokoff, A.; Luwor, R.; Stylli, S.S. Enhancement of invadopodia activity in glioma cells by sublethal doses of irradiation and temozolomide. J. Neurosurg. 2017, 129, 598–610. [Google Scholar] [CrossRef]
- Stylli, S.S.; Kaye, A.H.; Lock, P. Invadopodia: At the cutting edge of tumour invasion. J. Clin. Neurosci. 2008, 15, 725–737. [Google Scholar] [CrossRef]
- Gourlay, J.; Morokoff, A.; Luwor, R.; Zhu, H.-J.; Kaye, A.H.; Stylli, S.S. The emergent role of exosomes in glioma. J. Clin. Neurosci. 2017, 35, 13–23. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Montemurro, N.; Fanelli, G.N.; Scatena, C.; Ortenzi, V.; Pasqualetti, F.; Mazzanti, C.M.; Morganti, R.; Paiar, F.; Naccarato, A.G.; Perrini, P. Surgical outcome and molecular pattern characterization of recurrent glioblastoma multiforme: A single-center retrospective series. Clin. Neurol. Neurosurg. 2021, 207, 106735. [Google Scholar] [CrossRef]
- Piryani, S.O.; Jiao, Y.; Kam, A.Y.F.; Liu, Y.; Vo-Dinh, T.; Chen, B.J.; Chao, N.J.; Doan, P.L. Endothelial Cell-Derived Extracellular Vesicles Mitigate Radiation-Induced Hematopoietic Injury. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 291–301. [Google Scholar] [CrossRef]
- Wen, S.; Dooner, M.; Cheng, Y.; Papa, E.; del Tatto, M.; Pereira, M.; Deng, Y.; Goldberg, L.; Aliotta, J.; Chatterjee, D.; et al. Mesenchymal stromal cell-derived extracellular vesicles rescue radiation damage to murine marrow hematopoietic cells. Leukemia 2016, 30, 2221–2231. [Google Scholar] [CrossRef]
- Wang, J.; Cazzato, E.; Ladewig, E.; Frattini, V.; Rosenbloom, D.I.S.; Zairis, S.; Abate, F.; Liu, Z.; Elliott, O.; Shin, Y.-J.; et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016, 48, 768–776. [Google Scholar] [CrossRef] [Green Version]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yan, C.; Mu, L.; Huang, K.; Li, X.; Tao, D.; Wu, Y.; Qin, J. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS ONE 2015, 10, e0125625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bronisz, A.; Godlewski, J.; Chiocca, E.A. Extracellular Vesicles and MicroRNAs: Their Role in Tumorigenicity and Therapy for Brain Tumors. Cell. Mol. Neurobiol. 2016, 36, 361–376. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Sheng, S.; Ye, L.; Xu, X.; Ma, Y.; Feng, X.; Qiu, L.; Fan, Z.; Wang, Y.; Xia, X.; et al. Extracellular vesicles derived from glioblastoma promote proliferation and migration of neural progenitor cells via PI3K-Akt pathway. Cell Commun. Signal. 2022, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Dalrymple, S.J.; Parisi, J.E.; Roche, P.C.; Ziesmer, S.C.; Scheithauer, B.W.; Kelly, P.J. Changes in Proliferating Cell Nuclear Antigen Expression in Glioblastoma Multiforme Cells along a Stereotactic Biopsy Trajectory. Neurosurgery 1994, 35, 1036–1045. [Google Scholar] [CrossRef]
- Schiffer, D.; Cavalla, P.; Dutto, A.; Borsotti, L. Cell proliferation and invasion in malignant gliomas. Anticancer Res. 1997, 17, 61–69. [Google Scholar] [PubMed]
- Giese, A.; Bjerkvig, R.; Berens, M.; Westphal, M. Cost of Migration: Invasion of Malignant Gliomas and Implications for Treatment. J. Clin. Oncol. 2003, 21, 1624–1636. [Google Scholar] [CrossRef] [PubMed]
- Garnier, D.; Ratcliffe, E.; Briand, J.; Cartron, P.-F.; Oliver, L.; Vallette, F.M. The Activation of Mesenchymal Stem Cells by Glioblastoma Microvesicles Alters Their Exosomal Secretion of miR-100-5p, miR-9-5p and let-7d-5p. Biomedicines 2022, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, D.; Kirkbride, K.C.; Costello, K.; Clark, E.S.; Sinha, S.; Grega-Larson, N.; Tyska, M.J.; Weaver, A.M. Exosome Secretion Is Enhanced by Invadopodia and Drives Invasive Behavior. Cell Rep. 2013, 5, 1159–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, L.; Regan, M.; Higashimori, H.; Ng, S.K.; Esau, C.; Vidensky, S.; Rothstein, J.; Yang, Y. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J. Biol. Chem. 2013, 288, 7105–7116. [Google Scholar] [CrossRef] [Green Version]
- Lessi, F.; Aretini, P.; Rizzo, M.; Morelli, M.; Menicagli, M.; Franceschi, S.; Mazzanti, C.M. Analysis of exosome-derived microRNAs reveals insights of intercellular communication during invasion of breast, prostate and glioblastoma cancer cells. Cell Adhes. Migr. 2021, 15, 180–201. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wei, B.; Peng, C.; Wang, L.; Li, C. Identification of serum exosomal miR-98-5p, miR-183-5p, miR-323-3p and miR-19b-3p as potential biomarkers for glioblastoma patients and investigation of their mechanisms. Curr. Res. Transl. Med. 2022, 70, 103315. [Google Scholar] [CrossRef]
- Movahedpour, A.; Khatami, S.H.; Khorsand, M.; Salehi, M.; Savardashtaki, A.; Mirmajidi, S.H.; Negahdari, B.; Khanjani, N.; Naeli, P.; Vakili, O.; et al. Exosomal noncoding RNAs: Key players in glioblastoma drug resistance. Mol. Cell. Biochem. 2021, 476, 4081–4092. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Pillai, P.P. Current insights on extracellular vesicle-mediated glioblastoma progression: Implications in drug resistance and epithelial-mesenchymal transition. Biochim. Biophys. Acta Gen. Subj. 2021, 1866, 130065. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef]
- Ma, C.; Nguyen, H.P.T.; Luwor, R.; Stylli, S.S.; Gogos, A.; Paradiso, L.; Kaye, A.H.; Morokoff, A.P. A comprehensive meta-analysis of circulation miRNAs in glioma as potential diagnostic biomarker. PLoS ONE 2018, 13, e0189452. [Google Scholar] [CrossRef]
- Morokoff, A.; Jones, J.; Nguyen, H.; Ma, C.; Lasocki, A.; Gaillard, F.; Bennett, I.; Luwor, R.; Stylli, S.; Paradiso, L.; et al. Serum microRNA is a biomarker for post-operative monitoring in glioma. J. Neuro-Oncol. 2020, 149, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Atai, N.A.; Balaj, L.; van Veen, H.; Breakefield, X.O.; Jarzyna, P.A.; van Noorden, C.J.; Skog, J.; Maguire, C.A. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J. Neurooncol. 2013, 115, 343–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef] [Green Version]
- Greening, D.W.; Nguyen, H.P.; Elgass, K.; Simpson, R.J.; Salamonsen, L.A. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions. Biol. Reprod. 2016, 94, 38. [Google Scholar] [CrossRef] [PubMed]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Stylli, S.S.; I, S.T.; Verhagen, A.M.; Xu, S.S.; Pass, I.; Courtneidge, S.A.; Lock, P. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J. Cell Sci. 2009, 122, 2727–2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celiku, O.; Johnson, S.; Zhao, S.; Camphausen, K.; Shankavaram, U. Visualizing molecular profiles of glioblastoma with GBM-BioDP. PLoS ONE 2014, 9, e101239. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Accession # | NanoString Raw Counts | Sequence | Genome Context | PTEN Position | ||
|---|---|---|---|---|---|---|---|
| MU004 EVs | MU020 EVs | MU039 EVs | |||||
| hsa-miR-451a | MIMAT0001631 | 15,460 | 23 | 14 | aaaccguuaccauuacugaguu | chr17: 28861369-28861440 [-] | |
| * hsa-miR-320e | MIMAT0015072 | 4936 | 31 | 30 | aaagcuggguugagaagg | chr19: 46709293-46709345 [-] | Position 4736–4742 of PTEN 3′ UTR |
| * hsa-miR-520f-3p | MIMAT0002830 | 726 | 1184 | 140 | aagugcuuccuuuuagaggguu | chr19: 53682159-53682245 [+] | Position 1149–1155 of PTEN 3′ UTR |
| hsa-miR-873-3p | MIMAT0022717 | 162 | 154 | 123 | ggagacugaugaguucccggga | chr9: 28888879-28888955 [-] | |
| hsa-miR-223-3p | MIMAT0000280 | 482 | 24 | 15 | ugucaguuugucaaauacccca | chrX: 66018870-66018979 [+] | |
| * hsa-miR-363-3p | MIMAT0000707 | 121 | 131 | 92 | aauugcacgguauccaucugua | chrX: 134169378-134169452 [-] | Position 2859–2866 of PTEN 3′ UTR |
| * hsa-miR-144-3p | MIMAT0000436 | 390 | 21 | 16 | uacaguauagaugauguacu | chr17: 28861533-28861618 [-] | Position 2917–2923 of PTEN 3′ UTR |
| hsa-miR-598-3p | MIMAT0003266 | 117 | 112 | 75 | uacgucaucguugucaucguca | chr8: 11035206-11035302 [-] | |
| hsa-miR-6721-5p | MIMAT0025852 | 124 | 103 | 63 | ugggcaggggcuuauuguaggag | chr6: 32170030-32170116 [-] | |
| * hsa-miR-16-5p | MIMAT0000069 | 214 | 48 | 30 | uagcagcacguaaauauuggcg | chr3: 160404745-160404825 [+] | Position 4318–4325 of PTEN 3′ UTR |
| hsa-miR-4443 | MIMAT0018961 | 79 | 91 | 56 | uuggaggcguggguuuu | chr3: 48196564-48196616 [+] | |
| # hsa-miR-612 | MIMAT0003280 | 67 | 65 | 83 | gcugggcagggcuucugagcuccuu | chr11: 65444458-65444557 [+] | |
| hsa-miR-513b-5p | MIMAT0005788 | 71 | 110 | 25 | uucacaaggaggugucauuuau | chrX: 147199044-147199127 [-] | |
| hsa-miR-1183 | MIMAT0005828 | 71 | 51 | 47 | cacuguaggugauggugagagugggca | chr7: 21471058-21471146 [+] | |
| * hsa-miR-495-3p | MIMAT0002817 | 60 | 75 | 41 | aaacaaacauggugcacuucuu | chr14: 101033755-101033836 [+] | Position 3221–3227 of PTEN 3′ UTR, Position 3232–3239 of PTEN 3′ UTR |
| * hsa-miR-23a-3p | MIMAT0000078 | 145 | 20 | 25 | aucacauugccagggauuucc | chr19: 13836587-13836659 [-] | Position 1608–1615 of PTEN 3′ UTR, Position 2279–2286 of PTEN 3′ UTR, Position 4753–4760 of PTEN 3′ UTR |
| hsa-miR-150-5p | MIMAT0000451 | 184 | 13 | 13 | cugguacaggccugggggacag | chr19: 49500785-49500868 [-] | |
| hsa-miR-761 | MIMAT0010364 | 68 | 63 | 37 | gcagcagggugaaacugacaca | chr1: 51836344-51836402 [-] | |
| hsa-miR-149-5p | MIMAT0000450 | 55 | 55 | 39 | ucuggcuccgugucuucacuccc | chr2: 240456001-240456089 [+] | |
| hsa-miR-548ah-5p | MIMAT0018972 | 49 | 42 | 39 | aaaagugauugcaguguuug | chr4: 76575551-76575626 [+] | |
| # hsa-miR-142-3p | MIMAT0000434 | 129 | 20 | 16 | uguaguguuuccuacuuuaugga | chr17: 58331232-58331318 [-] | |
| hsa-miR-2682-5p | MIMAT0013517 | 49 | 46 | 49 | caggcagugacuguucagacguc | chr1: 98045242-98045351 [-] | |
| hsa-miR-548al | MIMAT0019024 | 49 | 49 | 46 | aacggcaaugacuuuuguacca | chr11: 74399237-74399333 [+] | |
| # hsa-miR-551a | MIMAT0003214 | 50 | 57 | 40 | gcgacccacucuugguuucca | chr1: 3560695-3560790 [-] | |
| * hsa-miR-155-5p | MIMAT0000646 | 41 | 43 | 53 | uuaaugcuaaucgugauaggggu | chr21: 25573980-25574044 [+] | Position 6323–6329 of PTEN 3′ UTR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Nguyen, H.P.T.; Jones, J.J.; Stylli, S.S.; Whitehead, C.A.; Paradiso, L.; Luwor, R.B.; Areeb, Z.; Hanssen, E.; Cho, E.; et al. Extracellular Vesicles Secreted by Glioma Stem Cells Are Involved in Radiation Resistance and Glioma Progression. Int. J. Mol. Sci. 2022, 23, 2770. https://doi.org/10.3390/ijms23052770
Ma C, Nguyen HPT, Jones JJ, Stylli SS, Whitehead CA, Paradiso L, Luwor RB, Areeb Z, Hanssen E, Cho E, et al. Extracellular Vesicles Secreted by Glioma Stem Cells Are Involved in Radiation Resistance and Glioma Progression. International Journal of Molecular Sciences. 2022; 23(5):2770. https://doi.org/10.3390/ijms23052770
Chicago/Turabian StyleMa, Chenkai, Hong P. T. Nguyen, Jordan J. Jones, Stanley S. Stylli, Clarissa A. Whitehead, Lucy Paradiso, Rodney B. Luwor, Zammam Areeb, Eric Hanssen, Ellie Cho, and et al. 2022. "Extracellular Vesicles Secreted by Glioma Stem Cells Are Involved in Radiation Resistance and Glioma Progression" International Journal of Molecular Sciences 23, no. 5: 2770. https://doi.org/10.3390/ijms23052770
APA StyleMa, C., Nguyen, H. P. T., Jones, J. J., Stylli, S. S., Whitehead, C. A., Paradiso, L., Luwor, R. B., Areeb, Z., Hanssen, E., Cho, E., Putz, U., Kaye, A. H., & Morokoff, A. P. (2022). Extracellular Vesicles Secreted by Glioma Stem Cells Are Involved in Radiation Resistance and Glioma Progression. International Journal of Molecular Sciences, 23(5), 2770. https://doi.org/10.3390/ijms23052770







