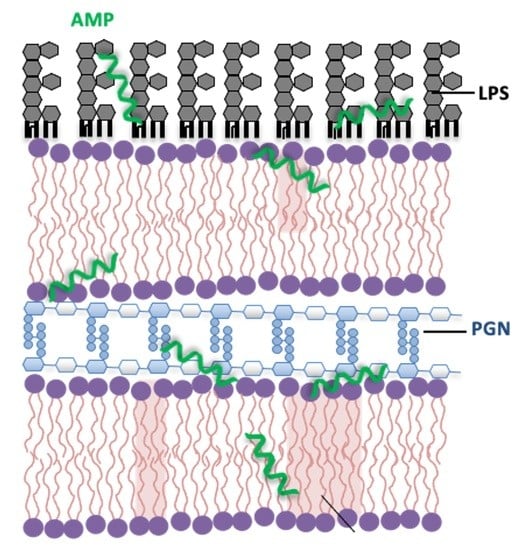
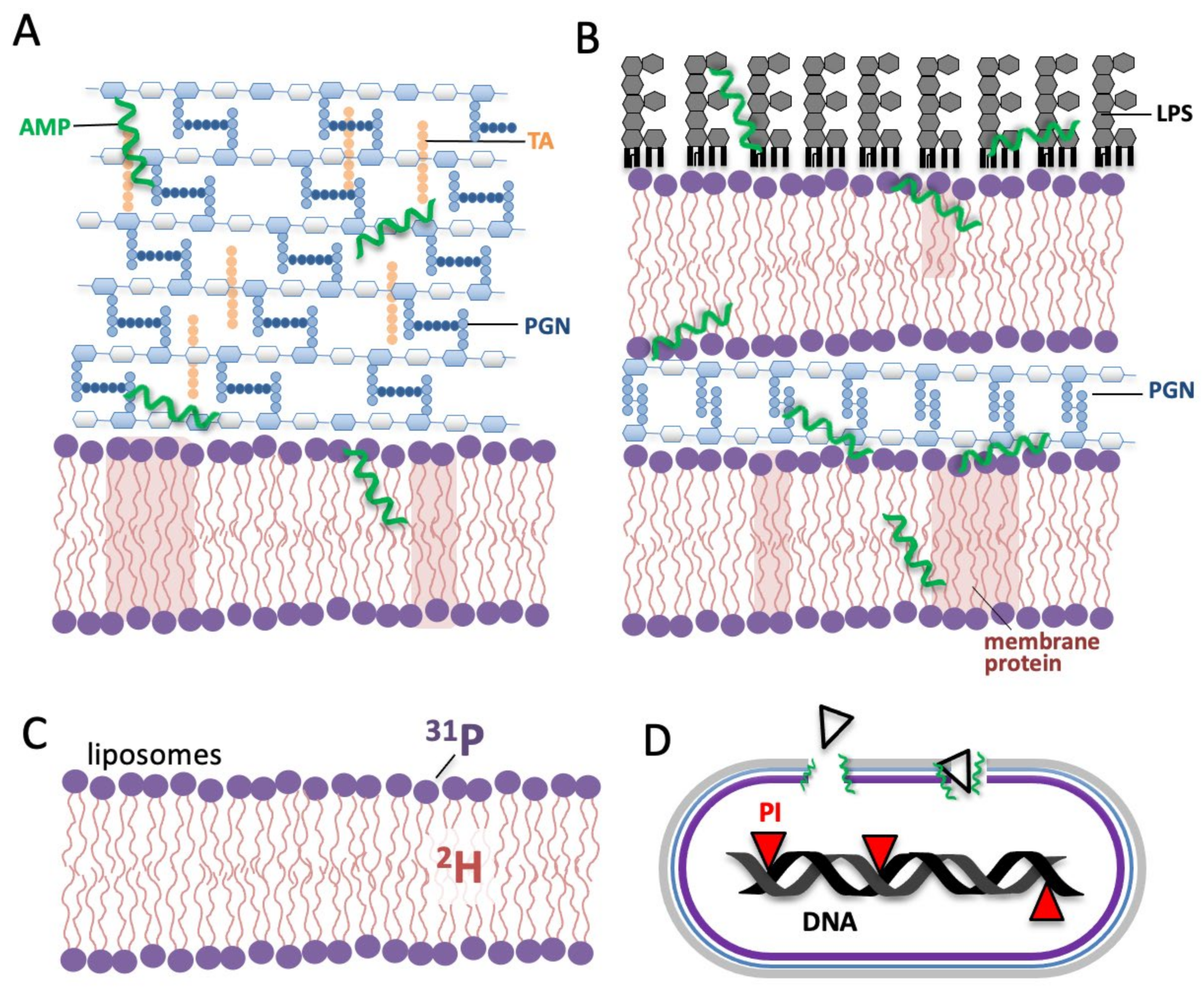
Antimicrobial Peptide Mechanisms Studied by Whole-Cell Deuterium NMR
Abstract
1. Introduction to Antimicrobial Peptides
2. Mode of Action of AMPs
2.1. Direct Killing of Target Cells by Permeabilizing the Membrane
2.2. Killing of Bacteria through Non-Membrane-Permeabilizing Mechanisms
2.3. AMPs vs. HDPs vs. CPPs
2.4. AMP Interactions with Non-Lipid Cell Envelope Components of Bacteria
3. Extending Biophysical Techniques That Probe AMP Mechanism from Model Membranes to Whole Cells
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.H. Tryptophan-rich and proline-rich antimicrobial peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef] [PubMed]
- Blondelle, S.E.; Lohner, K.; Aguilar, M.I. Lipid-induced conformation and lipid-binding properties of cytolytic and antimicrobial peptides: Determination and biological specificity. Biochim. Biophys. Acta-Biomembr. 1999, 1462, 89–108. [Google Scholar] [CrossRef]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engström, Å.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef]
- Hani, K.; Zairi, A.; Tangy, F.; Bouassida, K. Dermaseptins and magainins: Antimicrobial peptides from frogs’ skin-new sources for a promising spermicides microbicides-a mini review. J. Biomed. Biotechnol. 2009, 2009, 452567. [Google Scholar] [CrossRef]
- Aisenbrey, C.; Marquette, A.; Bechinger, B. The mechanisms of action of cationic antimicrobial peptides refined by novel concepts from biophysical investigations. In Antimicrobial Peptides, Advances in Experimental Medicine and Biology 1117; Matsuzaki, K., Ed.; Springer Nature: Singapore, 2019; pp. 33–64. [Google Scholar] [CrossRef]
- Mura, M.; Wang, J.; Zhou, Y.; Pinna, M.; Zvelindovsky, A.V.; Dennison, S.R.; Phoenix, D.A. The effect of amidation on the behaviour of antimicrobial peptides. Eur. Biophys. J. 2016, 45, 195–207. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef]
- Ting, D.S.J.; Beuerman, R.W.; Dua, H.S.; Lakshminarayanan, R.; Mohammed, I. Strategies in Translating the Therapeutic Potentials of Host Defense Peptides. Front. Immunol. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Gottler, L.M.; Ramamoorthy, A. Structure, Membrane Orientation, Mechanism, and Function of Pexiganan—A Highly Potent Antimicrobial Peptide Designed from Magainin. Biochim. Biophys. Acta 2009, 83, 1680–1686. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 546–566. [Google Scholar] [CrossRef]
- Murakami, M.; Ohtake, T.; Dorschner, R.A.; Schittek, B.; Garbe, C.; Gallo, R.L. Cathelicidin anti-microbial peptide expression in sweat, an innate defense system for the skin. J. Investig. Dermatol. 2002, 119, 1090–1095. [Google Scholar] [CrossRef]
- Falla, T.J.; Karunaratne, D.N.; Hancock, R.E.W. Mode of action of the antimicrobial peptide indolicidin. J. Biol. Chem. 1996, 271, 19298–19303. [Google Scholar] [CrossRef]
- Rozek, A.; Friedrich, C.L.; Hancock, R.E. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry 2000, 39, 15765–15774. [Google Scholar] [CrossRef]
- Kang, H.K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017, 55, 1–12. [Google Scholar] [CrossRef]
- Chen, C.H.; Starr, C.G.; Troendle, E.; Wiedman, G.; Wimley, W.C.; Ulmschneider, J.P.; Ulmschneider, M.B. Simulation-Guided Rational de Novo Design of a Small Pore-Forming Antimicrobial Peptide. J. Am. Chem. Soc. 2019, 141, 4839–4848. [Google Scholar] [CrossRef]
- Pérez-Peinado, C.; Dias, S.A.; Domingues, M.M.; Benfield, A.H.; Freire, J.M.; Rádis-Baptista, G.; Gaspar, D.; Castanho, M.A.; Craik, D.J.; Henriques, S.T.; et al. Mechanisms of bacterial membrane permeabilization by crotalicidin (Ctn) and its fragment Ctn(15–34), antimicrobial peptides from rattlesnake venom. J. Biol. Chem. 2018, 293, 1536–1549. [Google Scholar] [CrossRef]
- Melo, M.N.; Castanho, M.A. The mechanism of action of antimicrobial peptides: Lipid vesicles vs. bacteria. Front. Immunol. 2012, 3, 236. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta-Biomembr. 1999, 1462, 11–28. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Rapaport, D.; Shai, Y. Interaction of fluorescently labeled pardaxin and its analogues with lipid bilayers. J. Biol. Chem. 1991, 266, 23769–23775. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta-Biomembr. 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Matsuzaki, K. Membrane Permeabilization Mechanisms. In Antimicrobial Peptides, Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 9–16. [Google Scholar] [CrossRef]
- Kabelka, I.; Vácha, R. Advances in Molecular Understanding of α-Helical Membrane-Active Peptides. Acc. Chem. Res. 2021, 54, 2196–2204. [Google Scholar] [CrossRef]
- Cheng, J.T.; Hale, J.D.; Elliot, M.; Hancock, R.E.; Straus, S.K. Effect of membrane composition on antimicrobial peptides aurein 2.2 and 2.3 from australian southern bell frogs. Biophys. J. 2009, 96, 552–565. [Google Scholar] [CrossRef]
- Yang, L.; Harroun, T.A.; Weiss, T.M.; Ding, L.; Huang, H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–1485. [Google Scholar] [CrossRef]
- Sengupta, D.; Leontiadou, H.; Mark, A.E.; Marrink, S.J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim. Biophys. Acta-Biomembr. 2008, 1778, 2308–2317. [Google Scholar] [CrossRef]
- Nakajima, Y. Mode of Action and Resistance Mechanisms of Antimicrobial Macrolides, Macrolide Antibiot. In Macrolide Antibiotics, 2nd ed.; Academic Press: Cambridge, MA, USA, 2003; Volume 55, pp. 453–499. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Pept. Sci. Orig. Res. Biomol. 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Le, C.F.; Fang, C.M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Lu, W. Defensins: A Double-Edged Sword in Host Immunity. Front. Immunol. 2020, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2015, 16, 76–88. [Google Scholar] [CrossRef]
- Elmore, D.E. Insights into buforin II membrane translocation from molecular dynamics simulations. Peptides 2012, 38, 357–362. [Google Scholar] [CrossRef]
- Brannan, A.M.; Whelan, W.A.; Cole, E.; Booth, V. Differential scanning calorimetry of whole Escherichia coli treated with the antimicrobial peptide MSI-78 indicate a multi-hit mechanism with ribosomes as a novel target. PeerJ 2015, 3, e1516. [Google Scholar] [CrossRef][Green Version]
- Boman, H.G.; Agerberth, B.; Boman, A. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 1993, 61, 2978–2984. [Google Scholar] [CrossRef]
- Haney, E.F.; Straus, S.K.; Hancock, R.E. Reassessing the host defense peptide landscape. Front. Chem. 2019, 7, 43. [Google Scholar] [CrossRef]
- Fuselier, T.; Wimley, W.C. Spontaneous Membrane Translocating Peptides: The Role of Leucine-Arginine Consensus Motifs. Biophys. J. 2017, 113, 835–846. [Google Scholar] [CrossRef]
- Kauffman, W.B.; Guha, S.; Wimley, W.C. Synthetic molecular evolution of hybrid cell penetrating peptides. Nat. Commun. 2018, 9, 2568. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Melo, M.N.; Castanho, M.A. Cell-penetrating peptides and antimicrobial peptides: How different are they? Biochem. J. 2006, 399, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kalafatovic, D.; Giralt, E. Cell-penetrating peptides: Design strategies beyond primary structure and amphipathicity. Molecules 2017, 22, 1929. [Google Scholar] [CrossRef] [PubMed]
- Ruseska, I.; Zimmer, A. Internalization mechanisms of cell-penetrating peptides. Beilstein J. Nanotechnol. 2020, 11, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, K.; Helmfors, H.; Tudoran, O.; Juks, C.; Lindberg, S.; Padari, K.; El-Andaloussi, S.; Pooga, M.; Langel, Ü. Scavenger receptor-mediated uptake of cell-penetrating peptide nanocomplexes with oligonucleotides. FASEB J. 2012, 26, 1172–1180. [Google Scholar] [CrossRef]
- Battista, F.; Oliva, R.; Del Vecchio, P.; Winter, R.; Petraccone, L. Insights into the Action Mechanism of the Antimicrobial Peptide Lasioglossin III. Int. J. Mol. Sci. 2021, 22, 2857. [Google Scholar] [CrossRef]
- Oradd, G.; Schmidtchen, A.; Malmsten, M. Effects of peptide hydrophobicity on its incorporation in phospholipid membranes—An NMR and ellipsometry study. Biochim. Biophys. Acta 2011, 1808, 244–252. [Google Scholar] [CrossRef]
- Melo, M.N.; Ferre, R.; Castanho, M.A. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef]
- Roversi, D.; Luca, V.; Aureli, S.; Park, Y.; Mangoni, M.L.; Stella, L. How many antimicrobial peptide molecules kill a bacterium? The case of PMAP-23. ACS Chem. Biol. 2014, 9, 2003–2007. [Google Scholar] [CrossRef]
- Ebbensgaard, A.; Mordhorst, H.; Aarestrup, F.M.; Hansen, E.B. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front. Microbiol. 2018, 9, 2153. [Google Scholar] [CrossRef]
- Separovic, F.; Keizer, D.W.; Sani, M.A. In-cell Solid-State NMR Studies of Antimicrobial Peptides. Front. Med. Technol. 2020, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Navarro, S.; Moussaoui, M.; Nogués, M.V.; Boix, E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 2008, 47, 3544–3555. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Lohner, K. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 22, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Kwon, B.; Waring, A.J.; Hong, M. A 2H Solid-state nmr study of lipid clustering by cationic antimicrobial and Cell-penetrating peptides in model bacterial membranes. Biophys. J. 2013, 105, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, A.; Thennarasu, S.; Lee, D.K.; Tan, A.; Maloy, L. Solid-state NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophys. J. 2006, 91, 206–216. [Google Scholar] [CrossRef]
- Davis, J.H. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim. Biophys. Acta. 1983, 737, 117–171. [Google Scholar] [CrossRef]
- Lee, D.K.; Bhunia, A.; Kotler, S.A.; Ramamoorthy, A. Detergent-Type Membrane Fragmentation by MSI-78, MSI-367, MSI-594, and MSI-843 Antimicrobial Peptides and Inhibition by Cholesterol: A Solid-State Nuclear Magnetic Resonance Study. Biochemistry 2015, 54, 1897–1907. [Google Scholar] [CrossRef]
- Zamora-Carreras, H.; Strandberg, E.; Mühlhäuser, P.; Bürck, J.; Wadhwani, P.; Jiménez, M.Á.; Bruix, M.; Ulrich, A.S. Alanine scan and 2H NMR analysis of the membrane-active peptide BP100 point to a distinct carpet mechanism of action. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 1328–1338. [Google Scholar] [CrossRef]
- Luchinat, E.; Banci, L. In-cell NMR: A topical review. IUCrJ 2017, 4, 108–118. [Google Scholar] [CrossRef]
- Narasimhan, S.; Folkers, G.E.; Baldus, M. When Small becomes Too Big: Expanding the Use of In-Cell Solid-State NMR Spectroscopy. Chempluschem 2020, 85, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, R.; Romaniuk, J.A.; Rice, D.M.; Cegelski, L. Spectral snapshots of bacterial cell-wall composition and the influence of antibiotics by whole-cell NMR. Biophys. J. 2015, 108, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Cegelski, L.; O’Connor, R.D.; Stueber, D.; Singh, M.; Poliks, B.; Schaefer, J. Plant cell-wall cross-links by REDOR NMR spectroscopy. J. Am. Chem. Soc. 2010, 132, 16052–16057. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.H.; Nichol, C.P.; Weeks, G.; Bloom, M. Study of the cytoplasmic and outer membranes of Escherichia coli by deuterium magnetic resonance. Biochemistry 1979, 18, 2103–2112. [Google Scholar] [CrossRef]
- Pius, J.; Morrow, M.R.; Booth, V. 2H solid-state nuclear magnetic resonance investigation of whole Escherichia coli interacting with antimicrobial peptide MSI-78. Biochemistry 2012, 51, 118–125. [Google Scholar] [CrossRef]
- Santisteban, N.P.; Morrow, M.R.; Booth, V. Protocols for studying the interaction of MSI-78 with the membranes of Whole Gram-positive and Gram-negative Bacteria by NMR. In Antimicrobial Peptides, Methods and Protocols; Springer: New York, NY, USA, 2017; pp. 217–230. [Google Scholar] [CrossRef]
- Tardy-Laporte, C.; Arnold, A.A.; Genard, B.; Gastineau, R.; Morançais, M.; Mouget, J.L.; Tremblay, R.; Marcotte, I. A 2H solid-state NMR study of the effect of antimicrobial agents on intact Escherichia coli without mutating. Biochim. Biophys. Acta-Biomembr. 2013, 1828, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Booth, V. Deuterium Solid State NMR Studies of Intact Bacteria treated with Antimicrobial Peptides. Front. Med. Technol. 2020, 2, 26. [Google Scholar] [CrossRef]
- Santisteban, N.P.; Morrow, M.R.; Booth, V. Effect of AMPs MSI-78 and BP100 on the lipid acyl chains of 2H-labeled intact Gram positive bacteria. Biochim. Biophys. Acta-Biomembr. 2020, 1862, 183199. [Google Scholar] [CrossRef]
- Booth, V. Deuterium solid-state NMR of whole bacteria: Sample preparation and effects of cell envelope manipulation. In Solid-State NMR: Applications in Biomembrane Structure; Separovic, F., Sani, M.A., Eds.; IOP Publishing: Bristol, UK, 2020; pp. 1–121. [Google Scholar] [CrossRef]
- Laadhari, M.; Arnold, A.A.; Gravel, A.E.; Separovic, F.; Marcotte, I. Interaction of the antimicrobial peptides caerin 1.1 and aurein 1.2 with intact bacteria by 2H solid-state NMR. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 2959–2964. [Google Scholar] [CrossRef]
- Warnet, X.L.; Laadhari, M.; Arnold, A.A.; Marcotte, I.; Warschawski, D.E. A 2H magic-angle spinning solid-state NMR characterisation of lipid membranes in intact bacteria. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 146–152. [Google Scholar] [CrossRef]
- Overall, S.A.; Zhu, S.; Hanssen, E.; Separovic, F.; Sani, M.A. In situ monitoring of bacteria under antimicrobial stress using 31P solid-state NMR. Int. J. Mol. Sci. 2019, 20, 181. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakov, A.A.; Medeiros-Silva, J.; Tran, N.; Gelenter, M.D.; Hong, M. From Angstroms to Nanometers: Measuring Interatomic Distances by Solid-State NMR. Chem. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Sani, M.A.; Separovic, F. Antimicrobial Peptide Structures: From Model Membranes to Live Cells. Chem.-A Eur. J. 2018, 24, 286–291. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, K.M. Flow cytometry: An overview. Curr. Protoc. Immunol. 2018, 120, 1. [Google Scholar] [CrossRef]
- Freire, J.M.; Gaspar, D.; de la Torre, B.G.; Veiga, A.S.; Andreu, D.; Castanho, M.A. Monitoring antibacterial permeabilization in real time using time-resolved flow cytometry. Biochim. Biophys. Acta-Biomembr. 2015, 1848, 554–560. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, S.; Booth, V. Antimicrobial Peptide Mechanisms Studied by Whole-Cell Deuterium NMR. Int. J. Mol. Sci. 2022, 23, 2740. https://doi.org/10.3390/ijms23052740
Kumari S, Booth V. Antimicrobial Peptide Mechanisms Studied by Whole-Cell Deuterium NMR. International Journal of Molecular Sciences. 2022; 23(5):2740. https://doi.org/10.3390/ijms23052740
Chicago/Turabian StyleKumari, Sarika, and Valerie Booth. 2022. "Antimicrobial Peptide Mechanisms Studied by Whole-Cell Deuterium NMR" International Journal of Molecular Sciences 23, no. 5: 2740. https://doi.org/10.3390/ijms23052740
APA StyleKumari, S., & Booth, V. (2022). Antimicrobial Peptide Mechanisms Studied by Whole-Cell Deuterium NMR. International Journal of Molecular Sciences, 23(5), 2740. https://doi.org/10.3390/ijms23052740