Abstract
Mitochondria play key roles in cellular energy metabolism in eukaryotes. Mitochondria of most organisms contain their own genome and specific transcription and translation machineries. The expression of angiosperm mtDNA involves extensive RNA-processing steps, such as RNA trimming, editing, and the splicing of numerous group II-type introns. Pentatricopeptide repeat (PPR) proteins are key players in plant organelle gene expression and RNA metabolism. In the present analysis, we reveal the function of the MITOCHONDRIAL SPLICING FACTOR 2 gene (MISF2, AT3G22670) and show that it encodes a mitochondria-localized PPR protein that is crucial for early embryo development in Arabidopsis. Molecular characterization of embryo-rescued misf2 plantlets indicates that the splicing of nad2 intron 1, and thus respiratory complex I biogenesis, are strongly compromised. Moreover, the molecular function seems conserved between MISF2 protein in Arabidopsis and its orthologous gene (EMP10) in maize, suggesting that the ancestor of MISF2/EMP10 was recruited to function in nad2 processing before the monocot–dicot divergence ~200 million years ago. These data provide new insights into the function of nuclear-encoded factors in mitochondrial gene expression and respiratory chain biogenesis during plant embryo development.
Keywords:
group II intron; splicing; PPR; respiration; complex I; mitochondria; embryogenesis; Arabidopsis; angiosperms 1. Introduction
Mitochondria are key sites of cellular energy metabolism (i.e., ATP production), as well as of the biosynthesis of various essential metabolites. Most modern mitochondria contain vestigial genomes (mtDNA and mitogenome) derived from that of their ancestral bacterial progenitor, which vary quite widely in size between organisms. In plants, angiosperm mtDNAs are remarkably large and complex in structure [1], encoding rRNAs, tRNAs, ribosomal proteins, as well as various subunits of respiratory complexes (CI to CIV), the ATP synthase enzyme (CV), cofactors of the cytochrome c biogenesis (CCM) machinery, and at least one component of the twin-arginine protein translocation system [2].
In Arabidopsis, the oxidative phosphorylation (OXHPOS) machinery is composed of >100 different subunits, most of which are encoded by nuclear loci and about 20 of which are expressed from the mitogenome. Complex I (CI, or NADH-ubiquinone oxidoreductase), which catalyzes NADH dehydrogenation and electron transfer to coenzyme Q10 (CoQ10, or ubiquinone), is the largest and most complicated enzyme of the respiratory chain system [3]. In plants, CI is composed of more than 50 different subunits, 9 of which are encoded in the mitochondria (i.e., Nad1, Nad2, Nad3, Nad4, Nad4l, Nad5, Nad6, Nad7, and Nad9) [4]. These are assembled into two main sub-domains known as the ‘membrane arm’ and ‘peripheral arm’ of the holo-CI enzyme [5]. Nad1, Nad2, Nad3, Nad4, Nad4L, Nad5, and Nad6 belong to the membrane arm, which functions in proton pumping across the cristae membranes, while Nad7 and Nad9 are incorporated together with other nuclear-encoded subunits into the peripheral arm of CI, which is mainly associated with the transfer of electrons from NADH to ubiquinone. The biogenesis of the respiratory chain machinery involves various mechanisms for regulating the expression of subunits that are encoded by two physically separate genetic compartments [3,6,7,8]. Mutants affected in the expression of mitochondria-encoded CI subunits often show altered growth and developmental phenotypes, some of which contain developmentally arrested embryos [9,10]. Yet, the identity of the factors and pathways involved in these regulations still await further analysis.
The expression of mitochondrial genes in plants involves extensive RNA-processing steps. These include transcript trimming, RNA editing, and the removal of many group II intron sequences that interrupt the coding sequence of several essential genes [9,11,12,13]. These RNA processing steps are essential for mt-RNAs to synthesize the protein they encode. Group II introns are defined by a secondary structure formed by six stem-loop domains (D1 to D6). The excision of canonical group II introns relies on proteins encoded by the introns themselves (i.e., IEPs, or maturases) [14,15], whereas the splicing of group II introns in plant organelles involves a repertoire of nuclear-encoded factors that assist with the splicing reactions and which may serve as key control points in plant mitochondrial gene expression [1,16,17,18]. These belong to diverse families of RNA binding factors. A few are related to maturases [18,19], whereas others are identified as, for e.g., RNA helicases [20,21,22], PORR-related proteins [23], and relevantly to our study, pentatricopeptide repeat (PPR) proteins [24].
The PPR family constitutes a large protein family in land plants, with approximately 450 members identified in Arabidopsis and about 490 genes in maize [25,26,27]. PPR proteins are recognized by a degenerate 35 amino-acid motif folding into two antiparallel helices connected by a short loop/turn [28,29]. In association with the complexity of plant mitochondria gene expression, PPR proteins have been shown to play multifarious functions in organellar RNA metabolism, such as RNA stability and protection [12,30], C-to-U RNA editing [13], mRNA translation [31,32,33], and group II intron splicing [11,12,34,35,36]. Members of the PPR family are also linked to fertility restoration, where they regulate the expression of mitochondrial CMS-associated ORFs [37,38].
Genetic and biochemical studies indicated that PPRs are sequence-specific RNA-binding trans-factors, and that RNA recognition is mostly mediated by amino acids found at positions 5 and 35 in each PPR repeat. Association with each of the four RNA bases involves specific amino-acid combinations that are the basis of the PPR-RNA recognition code [39,40]. These data were further supported by the analysis of PPR protein-RNA crystal structures [28,29,41,42,43,44]. PPRs are classified into two main groups: P and PLS-type proteins, which in addition to canonical 35-amino acid PPR motifs (P) include long (L) or short (S) repeat variants [26,45]. While PLS-type proteins are almost exclusively associated with RNA editing [13,46], P-type PPR factors facilitate a wide range of organellar RNA expression steps going from stabilization to translation [34,47]. In this work, we analyzed the function of a mitochondrial P-type PPR factor that we named MITOCHONDRIAL INTRON SPLICING FACTOR 2 (MISF2), which is related to the PPR protein EMP10 in Zea mays [48]. As its ortholog in maize, the functions of MISF2 are essential for early embryo development. Embryo rescue techniques were used to investigate the molecular functions associated with two independent homozygous T-DNA insertional lines in MISF2. Loss-of-function mutants for MISF2 are strongly affected in the splicing of the first intron in nad2 gene (nad2 intron 1), which encodes a ~55 kDa core subunits of CI. Accordingly, the biogenesis of the respiratory CI is strongly affected in the misf2 mutants, while the transformation of the MISF2 gene restored the wild-type phenotype and the mtRNA metabolism defects detected in the homozygous mutants. The conserved molecular functions between MISF2 (in Arabidopsis) and EMP10 (in maize, [48]) suggest that the common ancestor MISF2/EMP10 was recruited to function in nad2 intron 1 splicing prior to the divergence of monocot and dicot plants, i.e., about ~200 million years ago [49].
2. Results
2.1. The Topology of MISF2 Protein
To better understand processes associated with mitochondrial RNA (mt-RNA) expression in plants, we assembled a collection of Arabidopsis T-DNA mutants affected in genes encoding mitochondria-targeted P-type PPR proteins and identified that heterozygous plants carrying insertions in the At3g22670 gene could not set homozygous mutants in their progeny. Domain search analysis using the PPR finder [50], PPRCODE [40], SMART [51] and CDD [52] algorithms indicated that the deduced product of AT3G22670 gene (Figure 1, Figure S1) encodes a 562 amino-acid PPR protein with a predicted topology of NH2-165-P-3-P-P-P-P-P-P-P-P-P-42-COOH (where ‘P’ designates P-type PPR motifs and amino acids not assigned to any defined domain are specified by numbers) (Figure 1 and Figure S1a).
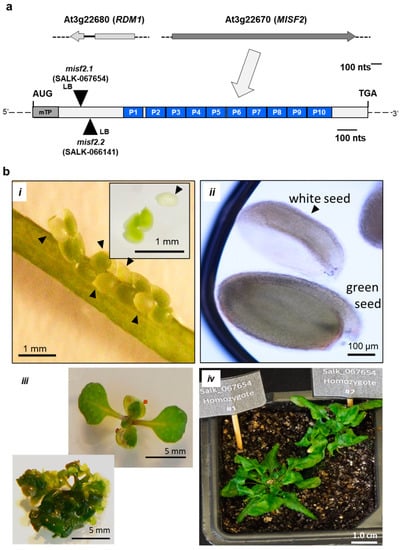
Figure 1.
MISF2 (At3g22670) gene topology and misf2 mutant phenotypes. (a) Scheme of the At3g22670 locus and gene structure. The large arrow points toward motif-structure arrangements of the MISF2 coding region. The position of the two T-DNA insertion sites in the coding region of MISF2 (i.e., SALK-line 067654, misf2.1, SALK-line 066141, and misf2.2) are located 324 and 350 nucleotides downstream to the ATG start codon, in a region that corresponds to the N-terminal domain of MISF2, upstream of the PPR motifs. (b) Morphologies of misf2 hetero- and homozygous mutants. Green and white seeds harboring wild-type/heterozygous and homozygous mutant embryos respectively were collected from surface-sterilized immature siliques of heterozygous misf2 plants (i) and sown on MS agar media supplemented with vitamins. Arrows point toward white seeds. Panel B (ii) shows differential interference contrast microscopy images (i.e., Nomarski) of embryos found in green or white seeds. Following germination, a few rescued homozygous misf2 plantlets (iii) were able to survive on soil, although failed to set flowers and viable seeds (iv).
Subcellular localization prediction algorithms, available at the ExPASy portal (https://www.expasy.org; accessed on 28 January 2022), UniProt [53] and the ‘SUBcellular location database for Arabidopsis proteins’ (SUBA4, http://suba.live; accessed on 28 January 2022) [54], indicated the presence of a predicted 24-amino acid mitochondrial targeting sequence in the N-terminal region of MISF2 (Figure S1a). In silico 3D structure prediction, using the AlphaFold server [55], suggested that MISF2 harbors a typical PPR helical fold (Figure S1a), with an inner basic core representing the RNA binding surface, as previously indicated from the analysis of the plant PPR10 protein [29].
2.2. MISF2 Encodes a Lowly-Expressed P-Type PPR Protein That Is Localized in Mitochondria
Expression analysis of MISF2 was performed using publicly available microarray and high-throughput sequencing databases. The Arabidopsis Information Resource (TAIR) (http://www.Arabidopsis.org; accessed on 28 January 2022) (Figure S2a) and ‘Genevestigator analysis toolbox’ [56] (Figure S2b) databases indicated differential expression of the MISF2 gene throughout development, with MISF2 expression being dominant in embryonic organs, young developing leaves, apical root tissues, flowers, and the shoot apex. To further investigate the intracellular location of MISF2, a fragment comprising the first 203 amino acids of MISF2 was fused in-frame to GFP (MISF2-GFP) expressed in Arabidopsis cells and the subcellular localization of the resulting fluorescence examined by confocal microscopy (Figure 2). In agreement with the in silico data, the MISF2-GFP signal was detected as round-shaped particles that co-localized with those of the MitoTracker® marker, a mitochondrion-specific fluorescent probe (Figure 2). These results are consistent with the predicted mitochondrial targeting of MISF2.
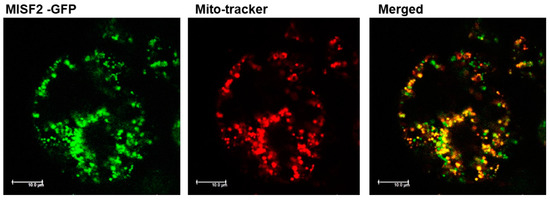
Figure 2.
MISF2 is localized to the mitochondria. Arabidopsis plant cells were transformed with a construct expressing the GFP fused in frame to the N-terminal region (i.e., 203 amino acids) of the MISF2 protein. The fluorescence corresponding to the GFP (green, left), the MitoTracker® marker (red, center), and the merged signals (right) are shown. Bars = 10 μm.
2.3. MISF2 Functions Are Required for Early Embryo Development in Arabidopsis thaliana
Several T-DNA insertion lines were identified within the MISF2 gene. These include two independent lines: SALK_067654 (misf2.1) and SALK_066141 (misf2.2), which contain T-DNA insertions located 324 and 350 nucleotides downstream of MISF2 translational start, respectively (Figure 1a and Figure S3a). Yet, no homozygous mutant plants could be recovered from the progeny of heterozygote misf2 lines, suggesting that the At3g22670 gene product is essential for embryogenesis. The heterozygous misf2.1 and misf2.2 plant lines did not show any obvious phenotypes under normal growth conditions (see Section 4, Material and Methods), suggesting that homozygous mutants could be embryonically lethal. To test this assumption, we compared the developmental phenotypes of embryos contained in immature seeds (10 days after pollination) of heterozygous misf2 with wild-type plants. Siliques of heterozygous misf2 plants contained about one-quarter of yellow to white seeds (Figure 1(bi)), which later degenerated into shrunken and brown mature seeds. Microscopy analyses further indicated that green seeds in siliques of heterozygous misf2 plants contained fully developed embryos, while white seeds had embryos arrested at the late torpedo/walking stick stages (Figure 1(bi,ii)).
2.4. Production of Embryo-Rescued misf2 Mutant Plants
Although misf2 is not found among the 32 Arabidopsis embryo-defective ppr mutants of the ‘SeedGene’ database [57], our genetic and microscopic analyses indicate that MISF2 is essential for proper embryo development (Figure 1b). Embryo rescue by in vitro culture allows to establish certain Arabidopsis mutants showing germination-defective phenotypes [58]. Among these are a few mutants affected in mitochondria biogenesis and function, such as the cod1 [59], ndufv1 [60] cal1/cal2 [61,62], nmat3 [63], or rfl8 [33] mutants. Therefore, white seeds contained in young siliques of heterozygous misf2 plants (i.e., 10~12 days post-anthesis, DPA), were sown on MS-agar plates supplemented with 1% sucrose and various vitamins (see Section 4, Materials and Methods) and then transferred to a controlled growth chamber. Indeed, under these conditions, 30% of the white seeds germinated after 3 months of culture and were then transferred to liquid culture using the same medium (see Section 4, Materials and Methods). PCR genotyping indicated that while green seeds derived from misf2.1 or misf2.2 heterozygote plants were either wild-type or heterozygous for the mutations, plantlets obtained from white seeds were all homozygous for either of the two misf2 mutant alleles.
The conditions used to rescue homozygous misf2.1 and misf2.2 seedlings were similar to those reported for the embryo rescue of Arabidopsis nmat3 [63] or cod1 mutant [59]. Phenotypical variations between individual homozygous-rescued misf2 plantlets were visible, with certain seedlings developing into slow-growing normal-looking plants with twisted leaves (Figure 1(biii)), while others produce miniature bushy-like structures (Figure 1(biii)). Similar observations were previously reported for several other emb mutants [64] affected in mitochondria biogenesis, including the rescued nmat3 or cod1 mutants [9,59,63]. A few homozygous-rescued misf2 plantlets (e.g., Figure 1(biv)) could be further transferred and cultivated on soil, but none of the plants could produce viable seeds.
2.5. MISF2 Is Essential for nad2 Pre-mRNAs Processing in Arabidopsis Mitochondria
For RNA and protein analyses, we used 3-week-old MS-grown homozygous misf2 mutant plantlets [64]. To further support the specific roles of MISF2 in mitochondria biogenesis, we also generated a functionally ‘complemented’ line (misf2.2/MISF2) by expressing the native MISF2 gene in homozygous misf2.2 plants (Figure S3b). Importantly, the expression of MISF2 in misf2.2 plants restored the growth and developmental defect phenotypes associated with the misf2.2 mutation. The complemented misf2.2/MISF2 mutant plants were able to complete their life cycle and set viable seeds (Figure S3c).
The steady-state levels of mitochondrial mRNAs in homozygous (embryo-rescued) misf2.1, misf2.2 and complemented misf2.2/MISF2 mutants were analyzed by RT-qPCR in comparison with wild-type (Col-0) plants. This analysis revealed a strong reduction (i.e., about 70 to 1200 folds) in the accumulation of mature nad2 transcripts spliced from their first intron in misf2.1 and misf2.2 mutants, respectively (i.e., nad2 exons a and b (nad2ab), Figure 3a). The steady-state levels of most other mitochondrial transcripts, including nad2 transcripts spliced from their other introns, were found to over-accumulate from 2 to 5 folds in both misf2 mutant lines (Figure 3a). As a control, we also analyzed the RNA profiles of plantlets derived from immature wild-type embryos (from the heart to torpedo stage) that were grown under the same conditions as the rescued misf2 mutants. The seedlings obtained from wild-type embryos did not show any significant reductions in the accumulation of mitochondrial transcripts, including nad2 (Figure 3b). Similarly, the accumulation of nad2 transcripts in functionally complemented plants were globally equivalent to those in wild-type plants (Figure 3b and Figure S3b). Based on these data, we concluded that the maturation defects observed for nad2 transcripts in misf2.1 and misf2.2 plants relate to the functions of MISF2 and not to physiological differences between the embryo-rescued plantlets and 3-week-old Arabidopsis seedlings germinated on MS-media plates.
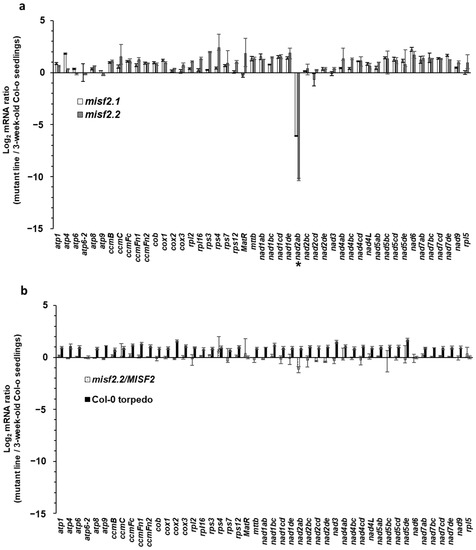
Figure 3.
Relative accumulation of mitochondrial mRNAs in misf2 mutants. Analysis of various mt-RNAs levels in Arabidopsis wild-type (Col-0), misf2 mutants, and complemented misf2.2/MISF2 plants by RT-qPCR. RNA extracted from 3-week-old wild-type seedlings (Col-0), 4-months-old rescued misf2 mutants, plantlets derived from immature Col-0 seeds (i.e., at the torpedo stage), and functionally complemented misf2.2/MISF2 mutants were reverse-transcribed and the relative steady-state levels of cDNAs corresponding to mitochondrial mRNAs evaluated by qPCR. Log2 ratios of mt mRNA abundances in misf2.1 and misf2.2 mutant lines (a), plantlets derived from immature Col-0 seeds, and complementation line (b) to those of 3-week-old MS-agar grown wild-type plants are shown. Asterisk indicates to reduced nad2ab transcript. The values are means of three biological replicates (error bars indicate one standard deviation).
2.6. MISF2 Is Required for Efficient Splicing of nad2 Intron 1
We reasoned that the reduced steady-state levels observed for the upstream region of mature nad2 transcripts (i.e., spliced exons ‘a’ and ‘b’) in the homozygous misf2 mutants likely relate to defects in the excision of the first intron in nad2. We thus determined the splicing efficiencies of nad2 intron 1 and that of the other 22 mitochondrial introns in wild-type plants and germinated embryos, as well as in misf2 mutants and functionally complemented misf2.2 plants by RT-qPCR. The obtained data revealed a strong reduction in the splicing efficiency of nad2 intron 1, with splicing reductions reaching about 360 and 11,000 folds in misf2.1 and misf2.2 plants, respectively, compared with the wild type (Figure 4a).
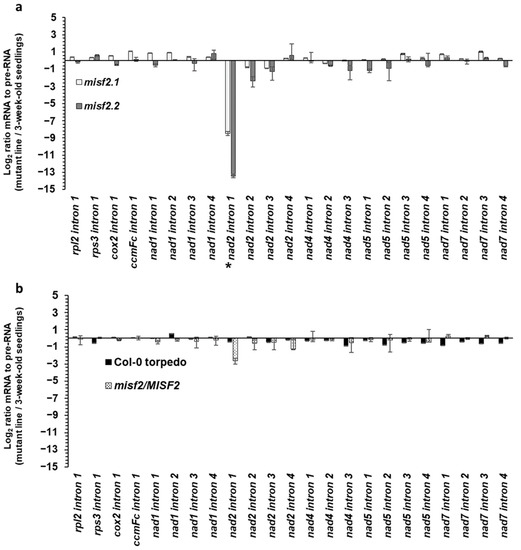
Figure 4.
Mitochondrial intron splicing efficiencies in misf2 mutants. The relative accumulation of mRNA and pre-RNA transcripts in wild-type, misf2 mutants, and complemented misf2.2/MISF2 plants, corresponding to the 23 group II intron sequences in Arabidopsis, was evaluated by RT-qPCR. The histogram shows the splicing efficiencies as indicated by the log2 ratios of pre-RNA to mature mRNA transcript abundance in misf2.1 and misf2.2 mutant lines compared with those in wild-type plants (a), as well as germinated wild-type seeds collected at the torpedo stage (Col-0-torpedo), and complemented line compared with those of wild-type plants (b). Asterisk indicates the altered splicing of nad2 intron 1. The values are means of three (misf2.1, misf2.2/MISF2) and five (misf2.2, Col-0) biological replicates (error bars indicate one standard deviation).
In contrast to nad2 intron 1, the splicing efficiency of other mitochondrial transcripts was not significantly affected in the homozygous misf2 mutants, although small reductions (i.e., from 2.5 to 6.7 folds) in the splicing efficiencies of nad2 introns 2 and 3 were seen in misf2.2 plants. The reduction in nad2 intron 1 splicing observed in misf2 mutants was largely corrected in complemented misf2.2 plants expressing the native MISF2 gene (misf2.2/MISF2), strongly supporting the role of MISF2 in the processing of nad2 intron 1 pre-mRNA (Figure 4b).
2.7. The MISF2 Protein Associates with nad2 Intron 1 In Vivo
A scheme of nad2 transcripts indicating the six typical stem-loop domains (D1–D6) within nad2 intron 1 (nad2 intron 1) is indicated in Figure 5a. PPR proteins are known to be sequence-specific RNA-binding factors [28,34,39,65,66,67]. A combinatorial code for RNA-recognition by PPR proteins was proposed, based on combinations of amino acids found at positions 5 and 35 of each PPR repeat [39,67,68]. The code applied to the 10 PPR repeats of MISF2 (Figure 1 and Figure S1a) indicated the following sequence: 5′-(C>U)-(G)-(U/C/G)-(U/C/G)-(A>G)-(G>>U)-(G>>A)-(C>U)-(G)-(?)-3′ (Figure 5b). Atomic structural model of MISF2 (Figure S1b) was predicted by the AlphaFold server [55]. A BLAST search along the updated Arabidopsis mtDNA (BK010421) revealed an eight-nucleotide matching sequence within the D1 stem-loop of nad2 intron 1 (Figure 5b).
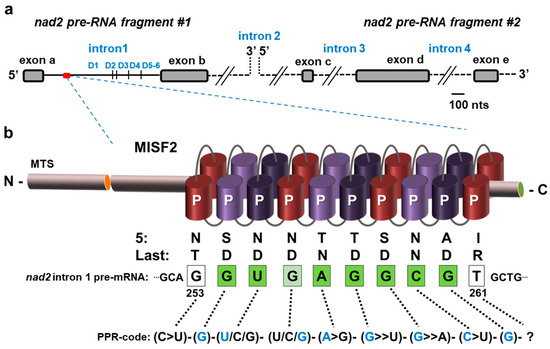
Figure 5.
The predicted MISF2 binding site in nad2 intron 1. (a) The expression of nad2 in Arabidopsis mitochondria involves the transcription of two precursor RNA transcripts, which are divided by the second intron. The maturation of nad2 requires the splicing of four introns found in cis (introns 1, 3, and 4) or trans (intron 2) configurations. The first pre-mRNA fragment consists of two exons separated by intron 1, while the second fragment harbors three exons separated by introns 3 and 4. The six typical stem-loop domains (D1–D6) are indicated for nad2 intron 1. (b) MISF2 is a P-type PPR protein, which harbors a mitochondrial targeting sequence (MTS) and 10 PPR motifs. The fifth and the last amino acids of each PPR repeat (Figure S1) are indicated below each PPR repeats. The best corresponding RNA binding site (i.e., 5′-GUGAGGCG-3′) is indicated within the first intron of nad2 pre-RNA fragment #1, with bases marked in green for perfect matches to the proposed binding site, in pale green for partial matches, and white for non-matching or unassigned nucleotides.
No other sequences of 10 bases long corresponding to the predicted MISF2 binding site could be identified elsewhere in the plant mitogenome. A model for the association of MISF2 with its predicted RNA binding site within nad2 intron 1 is illustrated in Figure 5b. The in silico data, therefore, correlated with the ‘genetically defined’ RNA target of MISF2, nad2 intron 1 (Figure 4 and Figure 5).
To further examine the in vivo RNA targets of MISF2, a cell line expressing an HA-tagged version of MISF2 was produced. After confirming the expression of the tagged protein in vivo (Figure 6a), the MISF2-3HA protein was immunoprecipitated from total extracts (Figure 6b) and co-purified RNAs were analyzed by RT-qPCR (Figure 6c).
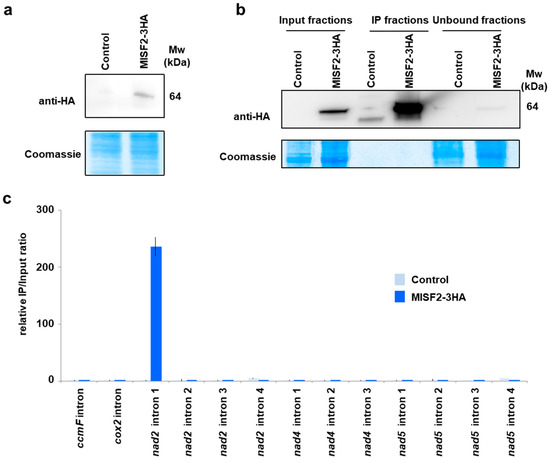
Figure 6.
The MISF2 protein associates with nad2 intron 1 in vivo. (a) Immunodetection of the MISF2-3HA fusion protein in protein extracts prepared form untransformed (control) and transformed (MISF2-3HA) Arabidopsis cell cultures. (b) Immunoprecipitation assays were conducted with the anti-HA antibody and the shown immunoblot analysis attests for the strong enrichment of the MISF2-3HA fusion in the immunoprecipitated (IP) fraction derived from the Arabidopsis transgenic cell line expressing the fusion. The weak MISF2-3HA signal in the unbound fraction demonstrates the efficiency of the immunoprecipitation. Parts of the blots stained with Coomassie blue are shown to display equal loading between samples. (c) Co-immunoprecipitated RNAs were analyzed by qRT-PCR using primer pairs specific to the indicated mitochondrial introns and relative enrichment ratios (immunoprecipitation fraction/input fraction) are shown.
Primers amplifying nad2 intron 1 were used in this analysis, along with other primers pairs targeting introns whose splicing was found to be slightly reduced in misf2 plants, plus a few additional controls. The obtained results reveal a very strong co-enrichment of nad2 intron 1, specifically in the co-IP ribonucleoprotein particle of MISF2-3HA. None of the other tested introns (i.e., the single introns within ccmF or cox2 mRNAs, nad2 introns 2, 3, and 4, nad4 introns 1 to 3, or nad5 introns 1 to 4) were co-enriched with MISF2-3HA, strongly supporting that nad2 intron 1 is the in vivo RNA target of this PPR protein, thereby confirming that MISF2 specifically associates with its genetically defined intron RNA.
2.8. Analysis of the Respiratory Chain Biogenesis in misf2 Mutants
The respiratory system of plant cells is made of five major protein complexes, termed as complex I (CI, about 1000 kDa in size), CII (160 kD), dimeric complex III (III2, 500 kDa), CIV (200 and 220 kDa forms), and the ATP synthase (CV, 660 kDa) [69]. Plant mitochondria also harbor various enzymes that belong to the ‘alternative electron transport’ pathway, involving alternative NADH dehydrogenases and the alternative cytochrome oxidase [70]. Genetic and biochemical studies showed that Nad2 is essential for complex I (CI) biogenesis and function [5,71,72,73,74,75,76,77]. The reduction in nad2 splicing (Figure 3 and Figure 4) suggests that the CI Nad2 subunit likely accumulates to very low levels in misf2 plants. Indeed, BN-PAGE analysis of Arabidopsis respiratory complexes indicated that CI is below detectable levels in misf2 mutant plants (Figure 7). Immunoblots made with antibodies against the carbonic anhydrase CA2 [78] further indicated the accumulation of several complex I assembly intermediates of about 610, 230 and 85 kDa in both misf2 mutants. While CI was considerably reduced in both misf2 mutants, BN-PAGE analyses indicated that other respiratory complexes, including CIII, CV, and particularly CIV, were rather upregulated in misf2 plants (Figure 7).
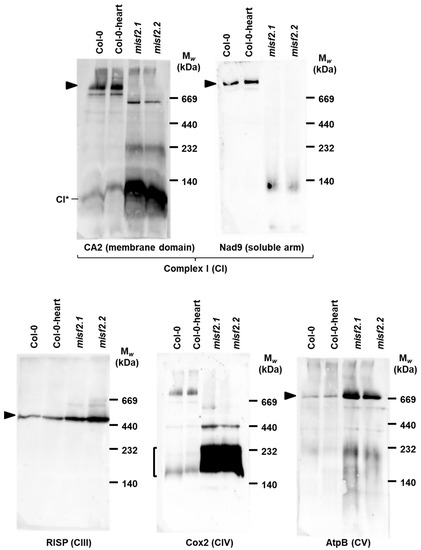
Figure 7.
Holo-complex I is below detectable levels in misf2 mutants. Blue native (BN)-PAGE analysis of crude organellar fractions was performed as described by [79]. Aliquots, equivalent to 40 mg of crude organellar membrane extracts, obtained from wild-type and misf2 plants, were solubilized with digitonin and resolved by BN-PAGE. For immunodetection, the proteins were transferred onto PVDF membranes and probed with the antibodies indicated below each blot (Table S2). Arrows point toward native complexes I (~1000 kDa), CIII dimer (III2, ~500 kDa), CIV (about 200 and 220 kDa forms), and CV (~660 kDa) [69]. CI* indicates the ~85 kDa sub-CI assembly intermediate [5].
We further analyzed the relative accumulation of various mitochondrial proteins in Col-0, misf2 mutants and the functionally complemented misf2.2/MISF2 line by immunoblotting analysis using various antibodies raised against different plant mitochondrial proteins. The data indicated that the CI-subunits CA2 and Nad9 accumulate in similar quantities in misf2 and wild-type plants. The levels of various other mitochondrial proteins, including the Rieske iron-sulfur protein (RISP) of CIII, the Cox2 subunit of CIV, the AtpB subunit of CV, and the mitochondrial outer-membrane voltage-dependent anion channel (VDAC or PORIN) proteins, were upregulated in misf2 mutants, as compared with wild-type plants (Figure 8a).
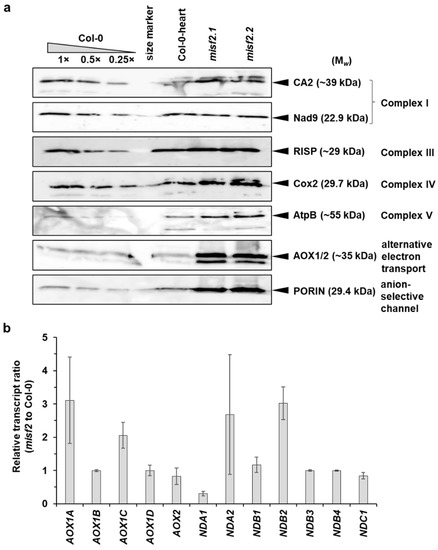
Figure 8.
Relative accumulation of different mitochondrial proteins and AOX or ND transcripts in wild-type and misf2 plants. (a) Immunoblots made with crude organellar fractions (equivalent to ~40 mg FW) extracted from 3-week-old MS-grown wild-type plants, in vitro germinated wild-type embryos at the heart to torpedo stages (Col-0 torpedo), and homozygous misf2 plantlets. The blots were probed with antibodies raised against the indicated mitochondrial proteins. (b) Analysis of the steady-state levels of various alternative oxidase (AOX) and rotenone-insensitive NAD(P)H dehydrogenase (ND) mRNAs by RT-qPCR. The histogram displays relative mRNAs levels in misf2 plantlets to 3-week-old MS-grown wild-type plants.
In contrast, the accumulation of all tested mitochondrial proteins was equivalent between the complemented line (misf2.2/MISF2) and wild-type plants (Figure S4).
Arabidopsis mutants affected in CI biogenesis undergo oxidative stress and often subsequently show a strong induction of the alternative respiratory pathways [10,60,62,77,80]. Accordingly, the relative accumulation of transcripts corresponding to various alternative oxidase (AOX) and rotenone-insensitive NAD(P)H dehydrogenase (NDs) mRNAs in misf2 was generally higher than in wild-type plants (Figure 8b). Similarly, immunoblot assays indicated that the steady-state levels of AOX1 or AOX2 proteins were higher in misf2 compared with the wild-type (Figure 8a and Figure S4).
3. Discussion
3.1. The MISF2 Gene Encodes a Mitochondria-Localized PPR Protein That Plays Essential Roles in Early Embryo-Development of Arabidopsis Plants
Mitochondria play key roles in energy metabolism and are thus vital organelle for plant life. During evolution, the mitochondrial genomes of land plants have undergone increased plasticity, showing substantial variations in genome size and structures and gene expression patterns between species (reviewed by e.g., [1]). Angiosperm mtDNAs are the largest and least gene-dense genomes among eukaryotes [2]. mRNA production and expression in land plant mitochondria involve extensive processing steps, which include endonucleolytic RNA cleavages, 5′ and 3′ mRNA trimming, extensive sequence editing and, relevantly to our study, the removal of intron (mostly group II-type) sequences that interrupt the coding regions of many mitochondrial genes (reviewed by e.g., [11]). These essential activities may serve as key control points of plant mitochondrial gene expression and are facilitated by numerous RNA binding cofactors [11,12].
In this study, we assigned a role to an Arabidopsis PPR protein, namely the MISF2 encoded by the AT3G22670 gene-locus in mt-RNA metabolism (i.e., nad2 maturation) and respiratory complex I assembly. MISF2, as its maize orthologue (i.e., EMP10 [48]), encodes a lowly expressed P-type PPR protein comprising 10 PPR motifs (Figure 1, Figures S2 and S3), which is located within the mitochondria (Figure 2). Figure S5 represents the sequence alignment [81,82] (Figure S5a) and phylogenetic analysis of MISF2/EMP10 paralogs in different angiosperms, including dicot (i.e., Arabidopsis, cauliflower, tobacco, and tomato) and monocot (i.e., barley, maize, rice, sorghum, and wheat) plant species (Figure S5b).
The Arabidopsis SeqViewer database (https://seqviewer.arabidopsis.org; accessed on 28 January 2022), which uses the outdated TIGR 4.0 version of the Arabidopsis genome, suggests that the 5′ untranslated region (UTR) of RDM1 (At3g22680) may overlap with the coding sequence of MISF2 (encoded on the opposite strand). Such different-strand overlapping of genes is especially untypical when considering the 2152 nucleotide-long 5′ UTR suggested for the RDM1 gene by the SeqViewer database in the compact genome of A. thaliana. However, this occurrence is not supported by the updated TAIR10 genome assembly that indicates a 76-nt-long 5′ UTR for RDM1 gene (Figures S6 and S7a). Likewise, the annotated RDM1 genes in other Arabidopsis species (i.e., A. lyrata LOC9321583 or A. suecica As03g023650) also harbor 5′ UTRs of about 80 and 200 nts, respectively, that do not overlap with MISF2. Rapid amplification of cDNA ends (RACE, Figure S3a) and RNA-seq data [83] (Figure S6) further showed that RDM1 harbors a 5′-UTR between 51 and 78 nucleotides long, which consequently does not extend to the MISF2 gene. This was also apparent by RT-PCRs with oligonucleotides designed to regions up- or down-stream of the 5′-UTR of RDM1 (Figure S7a,b), which further indicated that RDM1 is normally expressed in misf2 mutant plants (Figure S3b).
As for EMP10 in maize, downregulation of MISF2 expression results in premature arrest of Arabidopsis embryo development at the late torpedo stage (Figure 1b), whereas the function of RDM1 is regarded as non-essential for embryogenesis in A. thaliana plants [84]. Nevertheless, it was important to confirm that the developmental defect phenotypes and altered mt-RNA metabolism we see in misf2 mutants result directly from the downregulation of MISF2 expression. To this end, we analyzed the growth phenotypes (Figure S3c) and organellar RNA and protein profiles in a functionally complemented misf2 line (misf2/MISF2). These analyses revealed that the expression of MISF2 restored the embryogenesis defects and altered growth phenotypes associated with MISF2 gene disruption (Figure S3b) and that MISF2 is directly required for nad2 RNA maturation and respiratory CI biogenesis (Figure 3, Figure 4 and Figure S4). Co-IPs indicated that the MISF2 protein is specifically associated with its genetically defined intron RNA target (i.e., nad2 intron 1) in vivo.
3.2. MISF2 Is Required for the Splicing of nad2 Intron 1
Most mitochondrial introns in angiosperms are classified as group II type [16]. Model introns belonging to this class are large catalytic RNAs that are characterized by a conserved secondary structure consisting of six double-helical domains (D1 to D6), radiating from a central hub, with an internal ORF encoding a maturase in D4 [85,86]. The excision of group II introns in vivo in bacteria and in the organelles of eukaryotic cells requires the action of various RNA binding protein cofactors. In canonical group II introns, these at least include the maturase proteins (that are most often encoded by the introns themselves) [87]. In plant mitochondria, many additional proteinaceous splicing factors are required, which either derive from an ancient group of maturases [18], or from various other RNA binding cofactors that were recruited during evolution to facilitate mitochondrial intron splicing [17,88].
The PPR protein family is the largest RNA binding protein family known in plants, with about 400 to 600 members targeted to mitochondria or plastids [89]. PPR proteins bind their RNA substrates in a sequence specific manner and were shown to play pivotal roles in various aspects of posttranscriptional RNA processing, including the excision of group II introns in land plant organelles [11,13,34,47]. Here, we analyzed the molecular functions of the Arabidopsis MISF2 protein by characterizing loss-of-function mutants. As no homozygous mutant individuals could be identified among mature seeds of self-fertilized heterozygous misf2 progenies, we used embryo rescue approaches [59,63] to generate homozygous mutant plant material, which allowed us to analyze the role of MISF2 in mitochondrial RNA metabolism.
Analysis of mitochondrial RNA profiles in wild-type and misf2 plants showed a large reduction in the accumulation of mature nad2 mRNA in both mutant lines (Figure 3). The RT-qPCR analyses further revealed a strong reduction in the splicing efficiency of nad2 intron 1 in misf2 plants (Figure 4). The most probable RNA-binding site for MISF2 protein (i.e., GUGAGGCG) resides within the D1 stem-loop of nad2 intron 1 (Figure 5), which also corresponds to the genetic and biochemical RNA target of MISF2 (Figure 3, Figure 4 and Figure 6). In model group II introns, maturases were shown to bind with great affinity and specificity to their cognate intron-RNAs, in particular to regions of D1 and around the maturase coding sequences within the D4 stem-loop of canonical group II intron [90]. It will therefore be interesting to investigate whether sequence changes within plant nad2 intron 1 were accompanied by the recruitment of the PPR MISF2 factor to facilitate its splicing, possibly to stabilize or nucleate nad2 intron 1 folding into a catalytically active structure.
Taken together, our data provide strong evidence that MISF2 is specifically required for nad2 intron 1 splicing and that this RNA processing step is essential for early embryogenesis in Arabidopsis.
3.3. Embryo Development and Complex I Biogenesis
The electron transport chain is made of four major multi-subunit protein complexes, denoted as CI to CIV. Plants also possess several enzymes corresponding to non-phosphorylating bypasses of the electron transport chain (ETC), namely the alternative oxidase (AOX) and several rotenone-insensitive NAD(P)H dehydrogenases (NDs) [71,91,92,93,94]. The biogenesis of respiratory CI in angiosperms involves the incorporation of ~50 different subunits encoded by both mitochondrial (i.e., nad1, nad2, nad3, nad4, nad4l, nad5, nad6, nad7, and nad9) and nuclear gene loci [95]. These are incorporated into two main different CI domains, consisting of a membrane domain and a matrix (or peripheral) arm [3,5,10,96].
Nad2 is a pivotal subunit of CI, that is suggested to be incorporated very early during the assembly of the membrane arm [3,4,5,97]. The early steps of CI biogenesis involve the production of an ~85 kDa assembly intermediate of the membrane arm, which contains various gamma-type carbonic anhydrase subunits. Subsequently, Nad2 and a few other subunits are incorporated to form a ~200 kDa membrane-bound CI assembly intermediate [5]. It is therefore anticipated that a strong reduction in Nad2 would interfere with the assembly of the CI membrane arm, and hence, with the biogenesis of the ~1.0 MDa holo-CI. Consequently, BN-PAGE analysis of wild-type and mutant plants revealed a major reduction in CI abundance in both misf2 mutant lines (Figure 7). Immunoblot analysis with anti-CA2 antibodies further revealed the existence of various CI intermediates in misf2 mutants, among which a major particle of about 85 kDa, which was also observed in the abo5 mutant that is impaired in nad2 expression [74]. The CI particles of higher mass (i.e., 230 kDa and 610 kDa) detected in the mutants may correspond to Nad2-deprived assembly intermediates that are less stable than the ~85 kDa particles [5].
It has been demonstrated that the severity of CI deficiency correlates with the gravity of the phenotypes displayed by corresponding plant mutants [10,60,77]. Severe CI mutants are impaired in the storage of essential nutrients but not in the mobilization of stored reserves [60]. Accordingly, mutants affected in β-oxidation, a metabolic process by which fatty acids are broken down by various tissues to produce energy, contain embryos that are typically arrested at earlier developmental stages compared with CI mutants [98]. Embryo maturation is often incomplete in various CI mutants, leading to the production of seeds with reduced reserves and germination capacity. One can anticipate that altered respiration interferes with numerous essential metabolic activities, resulting in altered embryo development.
In our study, we noticed that a severe defect in the production of the Nad2 subunit of CI results in impaired embryogenesis and a loss of germination capacity of Arabidopsis mutant seeds. However, most characterized plant CI mutants are generally able to germinate under standard culture conditions (see e.g., [21,76,77,99,100,101]). The inability of misf2 mutants to germinate under normal conditions is expected to result from an early arrest of mutant embryo development, placing misf2 mutants among the most severe CI mutants reported so far. We currently do not know the role that the embryo-rescue medium plays in improving the seed germination of misf2 mutants. It may be due to the presence of certain important chemicals in the rescue medium, or simply to a weakening of the seed coat by the high sugar concentration of the medium. Once germination was induced, we could observe that misf2 mutants often showed growth phenotypes such as other Arabidopsis CI mutants (Figure 1(biv)). It was previously suggested that once photosynthesis is established, growth is to a lesser extent dependent on the application of external vitamins and/or sugars [60]. Subsequently, rescued misf2 mutants can slowly proceed with their vegetative growth phase but remain unable to complete their life cycle, flower, and produce viable seeds.
4. Materials and Methods
4.1. Plant Material and Growth Conditions
Arabidopsis thaliana of the Columbia (Col-0) accession was used in all experiments. The wild-type (Col-0 line), SALK-067654 (misf2.1), and SALK-066141 (misf2.2) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH, USA). Prior to germination, seeds obtained from wild-type and mutant lines were surface sterilized with Cl2 gas, generated by the addition of 1 mL HCl per 50 mL of bleach (sodium hypochlorite 4.7%), for 4 h at room temperature (RT). The seeds were then sown on MS-agar plates containing 1% (w/v) sucrose or rescued by a method described in detail below. For synchronized germinations, the seeds were kept in the dark for 5 days at 4 °C and then grown under long-day condition (LD, 16:8-h) in a growth chamber (Percival Scientific, Perry, IA, USA) at 22 °C and under light intensity of 300 µE m−2 s−1. PCR was used for genotyping the plants using specific oligonucleotides listed in Table S1. Sequencing of specific PCR products was used to check the T-DNA insertion site in both mutant lines.
4.2. GFP Localization Assay
The DNA region encoding the first 203 amino acids of MISF2 was PCR amplified with specific oligonucleotides (i.e., misf2-B1 and misf2-B2; Table S1. The 609 nts PCR DNA fragment was cloned into the pDONR207 vector using the Gateway BP clonase enzyme mix and verified by Sanger sequencing. The entry clone was then transferred into the pGWB5 vector by Gateway LR reaction to create a GFP translational fusion between the MISF2 N-terminal sequence and the GFP coding sequence. The vector was transformed into Agrobacterium tumefaciens (strain C58C51) and used to transform Arabidopsis plant cells, as previously described [102]. Transgenic cells were selected on hygromycin and GFP fluorescence was visualized by confocal microscopy Leica TCS SP8. To visualize mitochondria in vivo, plant cells were incubated with 1 µM MitoTracker® Red (Thermo Fisher, Scientific, Waltham, MA, USA) for 10 min at room temperature prior to observation under confocal microscopy.
4.3. Embryo-Rescue and Establishment of Homozygous misf2 Mutants
Siliques from wild-type and heterozygous misf2 plants were surfaced sterilized with 6% bleach solution for 10 min at RT. The seeds were then soaked in a 70% ethanol solution for 10 min at RT, washed briefly with sterile DDW, and opened in a sterile hood. Green and white seeds obtained from siliques of heterozygous misf2 plants 10 days after self-fertilization were sown on MS-agar plates supplemented with 1% (w/v) sucrose and 10 mg myoinositol, 100 μg thiamine, 100 μg pyridoxine, and 100 μg nicotinic acid. For DNA and RNA analysis, we used Arabidopsis wild-type and misf2 plantlets at stage R6 (i.e., 6 to 8 leaves) [64]. To obtain larger quantities of plant material, plantlets at stage R6 were grown on MS-agar plates and then transferred to MS-based liquid medium supplemented with 1% (w/v) sucrose and 10 mg myoinositol 100 μg Thiamine, 100 μg Pyridoxine, and 100 μg nicotinic acid and incubated at 22 °C under a light intensity of 300 µE m−2 s−1 with moderate (50~100 RPM) shaking.
4.4. Functional Complementation—Establishment of misf2.2/MISF2 Plants
For the complementation assay, the MISF2 gene and its predicted promoter region were amplified by PCR from Arabidopsis thaliana total DNA using the MISF2-promo-B1 and MISF2-Cpl-B2 primers, cloned into the pDONR207 vector by Gateway® BP reaction (Invitrogen, Waltham, MA, USA), and subsequently transferred into the pGWB13 expression vector [103] by LR reaction (Invitrogen, Waltham, MA, USA). The resulting vector was used to transform misf2 heterozygous plants by floral dip transformation. Transformed plants were selected on hygromycin and transgenic homozygous mutants were identified by PCR genotyping.
4.5. Expression of the 3XHA-Tagged MISF2 Protein in Arabidopsis Cell Cultures
For expressing a 3XHA-tagged version of MISF2 in Arabidopsis cell cultures, the MISF2 coding sequence was amplified by PCR using the MISF2-B1 and MISF2-Cpl-B2 primers, cloned into the pDONR207 vector by Gateway® BP reaction (Invitrogen, Waltham, MA, USA), and subsequently transferred into the pGWB14 expression vector [103]. The resulting construct was used to transform the PSBD Arabidopsis cell line as previously described [102].
4.6. Microscopic Analyses of Arabidopsis Wild-Type and Mutant Plants
Analysis of whole plant morphology, roots, leaves, siliques, and seeds of wild-type and mutant lines were examined under Stereoscopic (dissecting) microscope or light microscope at the bio-imaging unit of the Institute of Life Sciences (The Hebrew University of Jerusalem, Jerusalem, Israel). Seeds were incubated with Hoyer solution for 30 min and the cleared samples were analyzed by differential interference contrast (Nomarski) microscopy.
4.7. RNA Extraction and Analysis
RNA extraction and analysis was performed essentially as previously described [21,23,104,105,106]. Total RNA was prepared from 200 mg seedlings grown on MS-agar plates supplemented with 1% sucrose using the RNAzol RT reagent (Sigma-Aldrich, St. Louis, MO, USA). The RNA was then treated with RNase-free DNase I prior to its use in the assays. RT-qPCR was performed with specific oligonucleotides designed to exon-exon (mRNAs) regions corresponding to mitochondrial genes and intron-exon regions (pre-mRNAs) within each of the 23 group II introns in Arabidopsis thaliana (Table S1). cDNA was synthesized by reverse transcription with the Superscript III reverse transcriptase (Invitrogen, Waltham, MA, USA), using 1–2 µg of total RNA and 250 ng of a mixture of random hexanucleotides (Promega, Mannheim, Germany) and incubated for 50 min at 50 °C. Reactions were stopped by 15 min incubation at 70 °C and the RT samples served directly for real-time PCR on a LightCycler 480 (Roche, Penzberg, Germany) using 2.5 μL of LightCycler 480 SYBR Green I Master mix and 2.5 μM of primers in a final volume of 5 µL. Reactions were performed in triplicate in the following conditions: pre-heating at 95 °C for 10 min followed by 40 cycles of 10 s at 95 °C, 10 s at 58 °C, and 10 s at 72 °C. The nucleus-encoded 18S rRNA (At3g41768) and the mitochondrial 26S ribosomal rRNA subunit (ArthMr001) were used as reference genes.
4.8. Rapid Amplification of Complementary End (RACE) Analyses
Poly-A+ cDNA libraries were obtained from total RNA extracted from 3-week-old MS-grown Arabidopsis plants, using the Dynabeads™ mRNA Purification Kit (Thermo-Fisher, Thermo Fisher, Kiryat Shmona, Israel). The 5’ and 3′ ends of RDM1 were established by RACE analysis, using the SMARTer® RACE 5′/3′ Kit (Takara Bio Inc., Kusatsu, Shiga, Japan). For the analysis of the 5′ UTR of MISF2, we performed an ‘inverse single strand RACE’ analysis. First, a cDNA corresponding to MISF2 mRNA was generated by RT-PCR with a primer phosphorylated by T4 Polinucleotide Kinase (Promega). The cDNA was self-ligated with T4 RNA Ligase (Promega, Mannheim, Germany) overnight at 25 °C. The 5′ end of the MISF2 gene was generated by PCR with primers designed near the ends of the gene (i.e., MISF2-RACE_S1 and MISF2-RACE_AS2) and analyzed by sequencing.
4.9. Crude Mitochondria Preparations
Crude mitochondria extracts were prepared essentially as described previously [79]. To this end, 200 mg of plantlets grown in liquid culture were harvested and homogenized in 2 mL of 75 mM MOPS-KOH, pH 7.6, 0.6 M sucrose, 4 mM EDTA, 0.2% polyvinylpyrrolidone-40, 8 mM L-cysteine, 0.2% bovine serum albumin, and protease inhibitor cocktail ‘complete Mini’ from Roche Diagnostics GmbH (Mannheim, Germany). The lysate was filtrated through one layer of Miracloth and centrifuged at 1300× g for 4 min at 4 °C (to remove cell debris). The supernatant was then centrifuged at 22,000× g for 10 min at 4 °C. The resulting pellet containing thylakoid and mitochondrial membranes were washed twice with 1 mL of wash buffer 37.5 mM MOPS-KOH, 0.3 M sucrose, and 2 mM EDTA, with pH 7.6 prior to use.
4.10. Blue Native PAGE Analysis of Respiratory Complexes
Blue native (BN)-PAGE of crude organellar membranous fractions was performed according to the method described in Ref. [79]. An aliquot equivalent to 40 mg of crude Arabidopsis mitochondria extracts was solubilized with 5% (w/v) digitonin in BN-solubilization buffer (30 mM HEPES, pH 7.4, 150 mM potassium acetate, 10% (v/v) glycerol) and then incubated on ice for 30 min. The samples were centrifuged for 8 min at 20,000× g to pellet non-solubilized material and 0.2% (v/v) of Serva Blue G was added to the supernatant. The samples were then loaded onto a native 4% to 16% linear gradient gel. For ‘non-denaturing-PAGE’ immunoblotting, the gel was transferred to a PVDF membrane (Bio-Rad) in Cathode buffer (50 mM Tricine and 15 mM Bis-Tris-HCl, pH 7.0) for 16 h at 4 °C at constant current of 40 mA. The blots where then incubated with antibodies against mitochondrial proteins (Table S2) and hybridization signals were identified by chemiluminescence assay after incubation with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody.
4.11. RNA Co-Immunoprecipitation Assays
Immunoprecipitation of MISF2-3HA were performed using the μMACS HA-Tagged Protein Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) following a procedure previously described in Wang et al., 2020 [88].
5. Conclusions
Angiosperms encode numerous PPR proteins that are predominantly localized in plastids and mitochondria, which carry essential roles in organellar RNA metabolism. These include the EMP10 protein, which regulates the maturation of nad2 in maize mitochondria [48]. Analysis of the protein and RNA profiles of mutants affected in the Arabidopsis orthologous gene, designated MITOCHONDRIAL SPLICING FACTOR 2 (MISF2, encoded by At3g22670 gene), indicates that MISF2 also functions specifically in the excision of the first intron of nad2. Plant mutants affected in MISF2 accumulate high levels of nad2 pre-RNA due to a strong defect in nad2 intron 1 splicing. The altered splicing found in misf2 (or emp10) is tightly associated with CI biogenesis defects and arrested embryonic development. Together, these data show that the molecular functions are conserved between the Arabidopsis MISF2 protein and its related EMP10 homolog in maize [48], which suggests that the common PPR ancestor of MISF2 and EMP10 has been recruited to act in nad2 intron 1 splicing prior to the divergence of monocot and dicot plant species [49]. Our results provide important insights into the roles of nuclear-encoded PPR factors in mitochondria gene expression and the biogenesis of the respiratory system during early plant life.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23052670/s1.
Author Contributions
Methodology, T.-T.N., C.B., S.S., M.Z., M.Q., R.M., H.Z., H.M. and O.O.-B.; formal analysis, T.-T.N., C.B., S.S., M.Z., M.Q., R.M., H.Z., H.M. and O.O.-B.; data curation, T.-T.N., H.M., R.M. and O.O.-B.; writing—original draft preparation, H.M. and O.O.-B.; writing—review and editing, H.M., T.-T.N., R.M. and O.O.-B.; supervision, H.M. and O.O.-B.; funding acquisition, H.M. and O.O.-B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants to O.O.B from the ‘Israeli Science Foundation’ ISF grants No. 1834/20 and 3254-2020 and by grants to H. M. from the French National Research Agency No. ANR-16-CE11-0024-01. The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007). This work has benefited from the support of IJPB’s Plant Observatory technological platforms.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in FigShare at 10.6084/m9.figshare.19244277, accessed on 28 January 2022.
Acknowledgments
We thank Eduardo Zabaleta (UNMDP) for providing us with anti-CA2 antibodies. The authors also wish to thank Ariel Chipman (HUJI) for his help with microscopy analyses and Omer Ben-Dor (HUJI) for his help with misf2 mutant screening.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Best, C.; Mizrahi, R.; Ostersetzer-Biran, O. Why so complex? The intricacy of genome structure and gene expression, associated with angiosperm mitochondria, may relate to the regulation of embryo quiescence or dormancy—Intrinsic blocks to early plant life. Plants 2020, 9, 598. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Braun, H.-P.; Binder, S.; Brennicke, A.; Eubel, H.; Fernie, A.R.; Finkemeier, I.; Klodmann, J.; König, A.-C.; Kühn, K.; Meyer, E.; et al. The life of plant mitochondrial complex I. Mitochondrion 2014, 19, 295–313. [Google Scholar] [CrossRef] [PubMed]
- Soufari, H.; Parrot, C.; Kuhn, L.; Waltz, F.; Hashem, Y. Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM. Nat. Commun. 2020, 11, 5195. [Google Scholar] [CrossRef] [PubMed]
- Ligas, J.; Pineau, E.; Bock, R.; Huynen, M.A.; Meyer, E.H. The assembly pathway of complex I in Arabidopsis thaliana. Plant J. 2019, 97, 447–459. [Google Scholar] [CrossRef]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef]
- Kleine, T.; Leister, D. Retrograde signaling: Organelles go networking. Biochim. Biophys. Acta 2016, 1857, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, P.; Rugen, N.; Carrie, C.; Elsasser, M.; Finkemeier, I.; Giese, J.; Hildebrandt, T.M.; Kuhn, K.; Maurino, V.G.; Ruberti, C.; et al. Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J. 2020, 101, 420–441. [Google Scholar] [CrossRef]
- Colas des Francs-Small, C.; Small, I. Surrogate mutants for studying mitochondrially encoded functions. Biochimie 2014, 100, 234–242. [Google Scholar] [CrossRef]
- Ostersetzer-Biran, O. Respiratory complex I and embryo development. J. Exp. Bot. 2016, 67, 1205–1207. [Google Scholar] [CrossRef] [PubMed]
- Zmudjak, M.; Ostersetzer-Biran, O. RNA Metabolism and Transcript Regulation; Chichester John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; Volume 50, pp. 143–184. [Google Scholar]
- Hammani, K.; Giege, P. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014, 19, 380–389. [Google Scholar] [CrossRef]
- Small, I.D.; Schallenberg-Rudinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef] [PubMed]
- Michel, F.; Lang, B.F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature 1985, 316, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.A. On the origin of RNA splicing and introns. Cell 1985, 42, 397–400. [Google Scholar] [CrossRef]
- Bonen, L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 2008, 8, 26–34. [Google Scholar] [CrossRef]
- Brown, G.G.; Colas des Francs-Small, C.; Ostersetzer-Biran, O. Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 2014, 5, 35. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Lampe, M.-K.; Sultan, L.D.; Ostersetzer-Biran, O. Organellar maturases: A window into the evolution of the spliceosome. BBA-Bioenerg. 2015, 1847, 798–808. [Google Scholar] [CrossRef]
- Mohr, G.; Lambowitz, A.M. Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res. 2003, 31, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Köhler, D.; Schmidt-Gattung, S.; Binder, S. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol. Biol. 2010, 72, 459–467. [Google Scholar] [CrossRef]
- Zmudjak, M.; Shevtsov, S.; Sultan, L.D.; Keren, I.; Ostersetzer-Biran, O. Analysis of the roles of the Arabidopsis nMAT2 and PMH2 proteins provided with new insights into the regulation of group II intron splicing in land-plant mitochondria. Int. J. Mol. Sci. 2017, 18, 2428. [Google Scholar] [CrossRef]
- He, J.; Duan, Y.; Hua, D.; Fan, G.; Wang, L.; Liu, Y.; Chen, Z.; Han, L.; Qu, L.-J.; Gong, Z. DEXH box RNA helicase–mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 2012, 24, 1815–1833. [Google Scholar] [CrossRef]
- Colas des Francs-Small, C.; Kroeger, T.; Zmudjak, M.; Ostersetzer-Biran, O.; Rahimi, N.; Small, I.; Barkan, A. A PORR domain protein required for rpl2 and ccmFc intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 2012, 69, 996–1005. [Google Scholar] [CrossRef]
- Peeters, N.; Small, I. Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 2001, 1541, 54–63. [Google Scholar] [CrossRef]
- Chen, L.; Li, Y.-X.; Li, C.; Shi, Y.; Song, Y.; Zhang, D.; Li, Y.; Wang, T. Genome-wide analysis of the pentatricopeptide repeat gene family in different maize genomes and its important role in kernel development. BMC Plant Biol. 2018, 18, 366. [Google Scholar] [CrossRef] [PubMed]
- Lurin, C.; Andres, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyere, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008, 13, 663–670. [Google Scholar] [CrossRef]
- Coquille, S.; Filipovska, A.; Chia, T.; Rajappa, L.; Lingford, J.P.; Razif, M.F.M.; Thore, S.; Rackham, O. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 2014, 5, 5729. [Google Scholar] [CrossRef]
- Gully, B.S.; Cowieson, N.; Stanley, W.A.; Shearston, K.; Small, I.D.; Barkan, A.; Bond, C.S. The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 2015, 43, 1918–1926. [Google Scholar] [CrossRef] [PubMed]
- Binder, S.; Hölzle, A.; Jonietz, C. RNA processing and RNA stability in plant mitochondria. In Plant Mitochondria; Springer: Berlin/Heidelberg, Germany, 2011; pp. 107–130. [Google Scholar]
- Haili, N.; Planchard, N.; Arnal, N.; Quadrado, M.; Vrielynck, N.; Dahan, J.; des Francs-Small, C.C.; Mireau, H. The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH DEHYDROGENASE SUBUNIT7 mRNA in Arabidopsis. Plant Physiol. 2016, 170, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Waltz, F.; Nguyen, T.T.; Arrive, M.; Bochler, A.; Chicher, J.; Hammann, P.; Kuhn, L.; Quadrado, M.; Mireau, H.; Hashem, Y.; et al. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 2019, 5, 106–117. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Planchard, N.; Dahan, J.; Arnal, N.; Balzergue, S.; Benamar, A.; Bertin, P.; Brunaud, V.; Dargel-Graffin, C.; Macherel, D.; et al. A Case of Gene Fragmentation in Plant Mitochondria Fixed by the Selection of a Compensatory Restorer of Fertility-Like PPR Gene. Mol. Biol. Evol. 2021, 38, 3445–3458. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T.; Fujii, S. Function of PPR proteins in plastid gene expression. RNA Biol. 2013, 10, 1446–1456. [Google Scholar] [CrossRef]
- Geddy, R.; Brown, G.G. Genes encoding pentatricopeptide repeat (PPR) proteins are not conserved in location in plant genomes and may be subject to diversifying selection. BMC Genom. 2007, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Dahan, J.; Mireau, H. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 2013, 10, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Wang, Y.; Hua, J. Genome wide identification of PPR gene family and prediction analysis on restorer gene in Gossypium. J. Genet. 2018, 97, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.; Bond, C.; Small, I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012, 8, e1002910. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhang, D.; Yan, J.; Zhang, Q.; Hong, S.; Yang, Y.; Yao, Y.; Yin, P.; Zou, T. Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 2019, 47, 3728–3738. [Google Scholar]
- Yin, P.; Li, Q.; Yan, C.; Liu, Y.; Liu, J.; Yu, F.; Wang, Z.; Long, J.; He, J.; Wang, H.-W.; et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 2013, 504, 168. [Google Scholar] [CrossRef]
- Ke, J.; Chen, R.Z.; Ban, T.; Zhou, X.E.; Gu, X.; Tan, M.H.; Chen, C.; Kang, Y.; Brunzelle, J.S.; Zhu, J.K.; et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 2013, 20, 1377–1382. [Google Scholar] [CrossRef]
- Gully, B.S.; Shah, K.R.; Lee, M.; Shearston, K.; Smith, N.M.; Sadowska, A.; Blythe, A.J.; Bernath-Levin, K.; Stanley, W.A.; Small, I.D.; et al. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhang, D.; Guan, Z.; Liu, Y.; Yang, Z.; Yang, Y.; Wang, X.; Wang, Q.; Zhang, Q.; Fan, S.; et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016, 7, 11285. [Google Scholar] [CrossRef] [PubMed]
- Brehme, N.; Zehrmann, A.; Verbitskiy, D.; Hartel, B.; Takenaka, M. Mitochondrial RNA editing PPR proteins can tolerate protein tags at E as well as at DYW domain termini. Front. Plant Sci. 2014, 5, 127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takenaka, M.; Jörg, A.; Burger, M.; Haag, S. RNA editing mutants as surrogates for mitochondrial SNP mutants. Plant Physiol. Biochem. 2019, 135, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Rovira, A.G.; Smith, A.G. PPR proteins—Orchestrators of organelle RNA metabolism. Physiol. Plant 2019, 166, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Li, S.; Sun, F.; Sun, Q.; Zhao, H.; Ren, X.; Zhao, Y.; Tan, B.C.; Zhang, Z.; Qiu, F. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J. 2017, 91, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Gouy, M.; Yang, Y.W.; Sharp, P.M.; Li, W.H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 1989, 86, 6201–6205. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Royan, S.; Schallenberg-Rüdinger, M.; Lenz, H.; Castleden, I.R.; McDowell, R.; Vacher, M.A.; Tonti-Filippini, J.; Bond, C.S.; Knoop, V.; et al. The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol. Plant 2020, 13, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Anderson, J.B.; DeWeese-Scott, C.; Fedorova, N.D.; Geer, L.Y.; He, S.; Hurwitz, D.I.; Jackson, J.D.; Jacobs, A.R.; Lanczycki, C.J.; et al. CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003, 31, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Apweiler, R.; Bairoch, A.; Natale, D.A.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; et al. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006, 34, D187–D191. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747–420751. [Google Scholar] [CrossRef] [PubMed]
- Meinke, D.W. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2020, 226, 306–325. [Google Scholar] [CrossRef]
- Franzmann, L.; Patton, D.A.; Meinke, D.W. In vitro morphogenesis of arrested embryos from lethal mutants of Arabidopsis thaliana. Theor. Appl. Genet. 1989, 77, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Dahan, J.; Tcherkez, G.; Macherel, D.; Benamar, A.; Belcram, K.; Quadrado, M.; Arnal, N.; Mireau, H. Disruption of the CYTOCHROME C OXIDASE DEFICIENT1 gene leads to cytochrome c oxidase depletion and reorchestrated respiratory metabolism in Arabidopsis. Plant Physiol. 2014, 166, 1788–1802. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, K.; Obata, T.; Feher, K.; Bock, R.; Fernie, A.R.; Meyer, E.H. Complete mitochondrial complex I deficiency induces an up-regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol. 2015, 168, 1537–1549. [Google Scholar] [CrossRef]
- Cordoba, J.P.; Marchetti, F.; Soto, D.; Martin, M.V.; Pagnussat, G.C.; Zabaleta, E. The CA domain of the respiratory complex I is required for normal embryogenesis in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Fromm, S.; Going, J.; Lorenz, C.; Peterhansel, C.; Braun, H.P. Depletion of the ”gamma-type carbonic anhydrase-like“ subunits of complex I affects central mitochondrial metabolism in Arabidopsis thaliana. Biochim. Biophys. Acta 2016, 1857, 60–71. [Google Scholar] [CrossRef]
- Shevtsov-Tal, S.; Best, C.; Matan, R.; Chandran, S.A.; Brown, G.G.; Ostersetzer-Biran, O. nMAT3 is an essential maturase splicing factor required for holo-complex I biogenesis and embryo development in Arabidopsis thaliana plants. Plant J. 2021, 106, 1128–1147. [Google Scholar] [CrossRef] [PubMed]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth stage–based phenotypic analysis of Arabidopsis. A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar]
- Yagi, Y.; Hayashi, S.; Kobayashi, K.; Hirayama, T.; Nakamura, T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 2013, 8, e57286. [Google Scholar]
- Yagi, Y.; Nakamura, T.; Small, I. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 2014, 78, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Zehrmann, A.; Brennicke, A.; Graichen, K. Improved computational target site prediction for pentatricopeptide repeat rna editing factors. PLoS ONE 2013, 8, e65343. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Gutmann, B.; Zhong, X.; Ye, Y.; Fisher, M.F.; Bai, F.; Castleden, I.; Song, Y.; Song, B.; Huang, J.; et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016, 85, 532–547. [Google Scholar] [CrossRef] [PubMed]
- Klodmann, J.; Senkler, M.; Rode, C.; Braun, H.-P. Defining the “protein complex proteome” of plant mitochondria. Plant Physiol. 2011, 157, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Whelan, J.; Soole, K.L.; Day, D.A. Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 2011, 62, 79–104. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, F.; Li, N.; Shi, D.-Q.; Yang, W.-C. Pentatricopeptide repeat protein MID1 modulates nad2 intron 1 splicing and Arabidopsis development. Sci. Rep. 2020, 10, 2008. [Google Scholar] [CrossRef]
- Marchetti, F.; Cainzos, M.; Shevtsov, S.; Cordoba, J.P.; Sultan, L.D.; Brennicke, A.; Takenaka, M.; Pagnussat, G.; Ostersetzer-Biran, O.; Zabaleta, E. Mitochondrial Pentatricopeptide Repeat Protein, EMB2794, plays a pivotal role in NADH dehydrogenase subunit nad2 mRNA maturation in Arabidopsis thaliana. Plant Cell Physiol. 2020, 61, 1080–1094. [Google Scholar] [CrossRef]
- Xiu, Z.; Sun, F.; Shen, Y.; Zhang, X.; Jiang, R.; Bonnard, G.; Zhang, J.; Tan, B.C. EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J. 2016, 85, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, J.; Chen, Z.; Ren, X.; Hong, X.; Gong, Z. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J. 2010, 63, 749–765. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.W.; Wang, H.J.; Hsieh, M.H.; Hsieh, H.L.; Jauh, G.Y. Arabidopsis mTERF15 Is Required for Mitochondrial nad2 Intron 3 Splicing and Functional Complex I Activity. PLoS ONE 2014, 9, e112360. [Google Scholar] [CrossRef]
- Nakagawa, N.; Sakurai, N. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II Intron maturase, causes impaired splicing of mitochondrial nad4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 772–783. [Google Scholar] [CrossRef]
- Keren, I.; Tal, L.; Colas des Francs-Small, C.; Araújo, W.L.; Shevtsov, S.; Shaya, F.; Fernie, A.R.; Small, I.; Ostersetzer-Biran, O. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 2012, 71, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Perales, M.; Parisi, G.; Fornasari, M.S.; Colaneri, A.; Villarreal, F.; Gonzalez-Schain, N.; Echave, J.; Gomez-Casati, D.; Braun, H.P.; Araya, A.; et al. Gamma carbonic anhydrase like complex interact with plant mitochondrial complex I. Plant Mol. Biol. 2004, 56, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Pineau, B.; Layoune, O.; Danon, A.; De Paepe, R. L-galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 2008, 283, 32500–32505. [Google Scholar] [CrossRef] [PubMed]
- Karpova, O.V.; Kuzmin, E.V.; Elthon, T.E.; Newton, K.J. Differential Expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 2002, 14, 3271–3284. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, K.B. GeneDoc: Analysis and visualization of genetic variation. EMBNEW News 1997, 4, 14. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Best, C.; Sultan, L.; Murik, O.; Ostersetzer-Biran, O. Insights into the mitochondrial transcriptome landscapes of two Brassicales plant species, Arabidopsis thaliana (var. Col-0) and Brassica oleracea (var. botrytis). Endocyto Cell Res. 2020, 30, 16–38. [Google Scholar]
- Gao, Z.; Liu, H.-L.; Daxinger, L.; Pontes, O.; He, X.; Qian, W.; Lin, H.; Xie, M.; Lorkovic, Z.J.; Zhang, S.; et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 2010, 465, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Michel, F.; Umesono, K.; Ozeki, H. Comparative and functional anatomy of group II catalytic introns—A review. Gene 1989, 82, 5–30. [Google Scholar] [CrossRef]
- Michel, F.; Ferat, J.L. Structure and activities of group-II introns. Annu. Rev. Biochem. 1995, 64, 435–461. [Google Scholar] [CrossRef]
- Lazowska, J.; Jacq, C.; Slonimski, P.P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell 1980, 22, 333–348. [Google Scholar] [CrossRef]
- Wang, C.; Fourdin, R.; Quadrado, M.; Dargel-Graffin, C.; Tolleter, D.; Macherel, D.; Mireau, H. Rerouting of ribosomal proteins into splicing in plant organelles. Proc. Natl. Acad. Sci. USA 2020, 117, 29979–29987. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Small, I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011, 191, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, M.; Noah, J.W.; Lambowitz, A.M. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001, 20, 7259–7270. [Google Scholar] [CrossRef]
- Schertl, P.; Braun, H.P. Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 2014, 5, 163. [Google Scholar] [CrossRef] [PubMed]
- Senkler, J.; Senkler, M.; Eubel, H.; Hildebrandt, T.; Lengwenus, C.; Schertl, P.; Schwarzländer, M.; Wagner, S.; Wittig, I.; Braun, H.-P. The mitochondrial complexome of Arabidopsis thaliana. Plant J. 2017, 89, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Subrahmanian, N.; Remacle, C.; Hamel, P.P. Plant mitochondrial complex I composition and assembly: A review. BBA-Bioenerg. 2016, 1857, 1001–1014. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Li, L.; Huang, S.; Pong Lee, C.; Millar, A.H.; Taylor, N.L. Mitochondrial composition, function and stress response in plants. J. Integr. Plant Biol. 2012, 54, 887–906. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Taylor, N.L.; Millar, A.H. Recent advances in the composition and heterogeneity of the Arabidopsis mitochondrial proteome. Front. Plant Sci. 2013, 4, 4. [Google Scholar] [CrossRef]
- Klodmann, J.; Sunderhaus, S.; Nimtz, M.; Jänsch, L.; Braun, H.-P. Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 2010, 22, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, M.; Padavannil, A.; Zhou, L.; Guo, F.; Letts, J.A. Atomic structure of a mitochondrial complex I intermediate from vascular plants. eLife 2020, 9, e56664. [Google Scholar] [CrossRef] [PubMed]
- Pinfield-Wells, H.; Rylott, E.L.; Gilday, A.D.; Graham, S.; Job, K.; Larson, T.R.; Graham, I.A. Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J. 2005, 43, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Koprivova, A.; des Francs-Small, C.C.; Calder, G.; Mugford, S.T.; Tanz, S.; Lee, B.R.; Zechmann, B.; Small, I.; Kopriva, S. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 2010, 285, 32192–32199. [Google Scholar] [CrossRef] [PubMed]
- Colas des Francs-Small, C.; Falcon de Longevialle, A.; Li, Y.; Lowe, E.; Tanz, S.K.; Smith, C.; Bevan, M.W.; Small, I. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiol. 2014, 165, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Weissenberger, S.; Soll, J.; Carrie, C. The PPR protein SLOW GROWTH 4 is involved in editing of nad4 and affects the splicing of nad2 intron 1. Plant Mol. Biol. 2017, 93, 355–368. [Google Scholar] [CrossRef]
- Van Leene, J.; Eeckhout, D.; Persiau, G.; Van De Slijke, E.; Geerinck, J.; Van Isterdael, G.; Witters, E.; De Jaeger, G. Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol. Biol. 2011, 754, 195–218. [Google Scholar]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Zmudjak, M.; Colas des Francs-Small, C.; Keren, I.; Shaya, F.; Belausov, E.; Small, I.; Ostersetzer-Biran, O. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytol. 2013, 199, 379–394. [Google Scholar] [CrossRef]
- Cohen, S.; Zmudjak, M.; Colas des Francs-Small, C.; Malik, S.; Shaya, F.; Keren, I.; Belausov, E.; Many, Y.; Brown, G.G.; Small, I.; et al. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 2014, 78, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Sultan, L.D.; Mileshina, D.; Grewe, F.; Rolle, K.; Abudraham, S.; Głodowicz, P.; Khan Niazi, A.; Keren, I.; Shevtsov, S.; Klipcan, L.; et al. The reverse-transcriptase/RNA-maturase protein MatR is required for the splicing of various group II introns in Brassicaceae mitochondria. Plant Cell 2016, 28, 2805–2829. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).