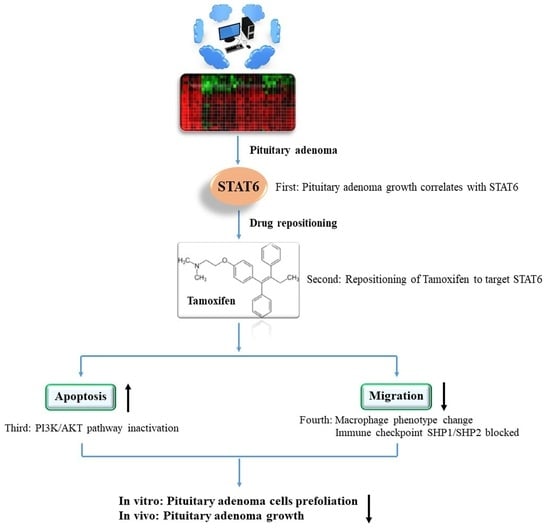
Tamoxifen Exerts Anticancer Effects on Pituitary Adenoma Progression via Inducing Cell Apoptosis and Inhibiting Cell Migration
Abstract
1. Introduction
2. Results
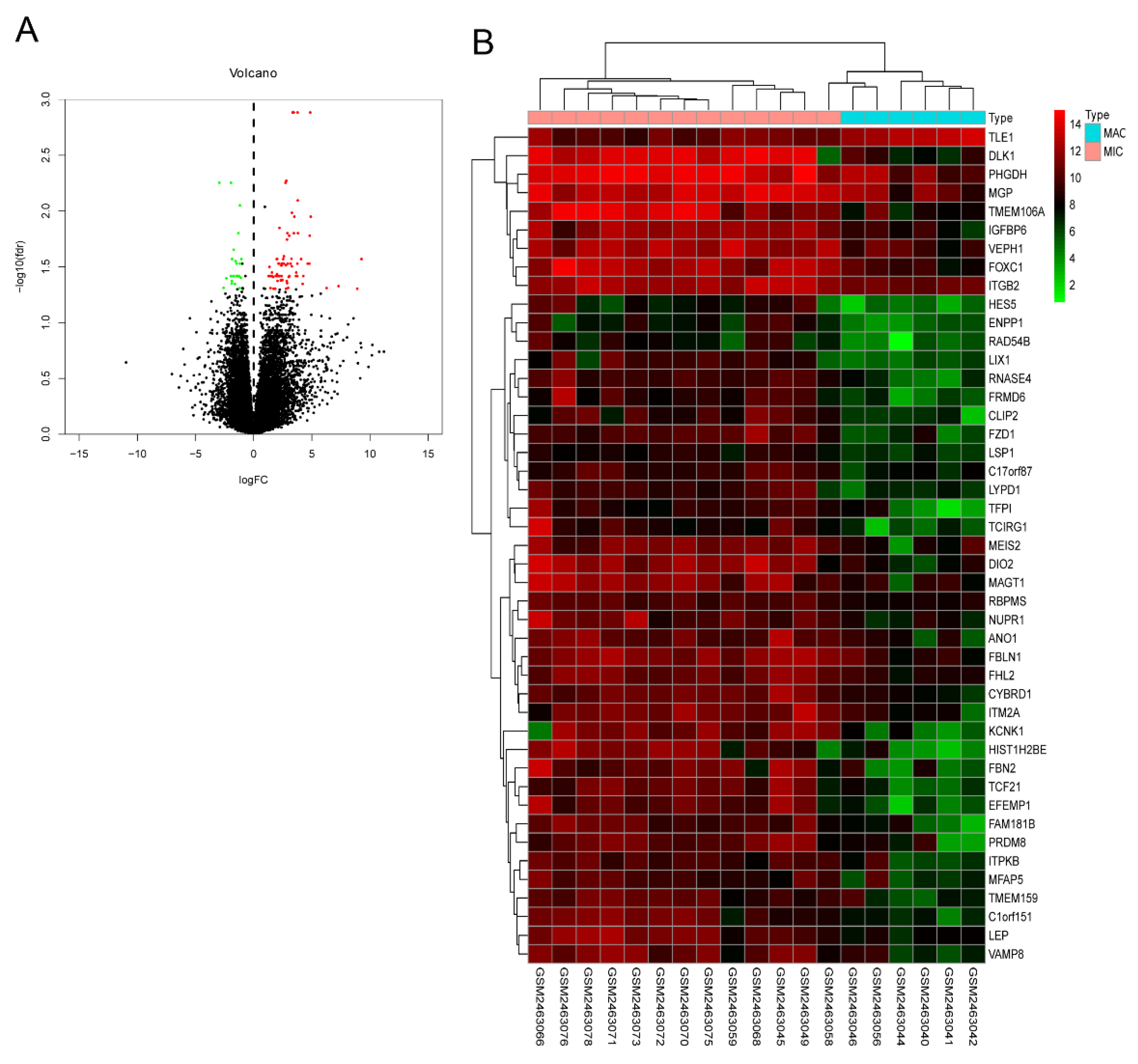
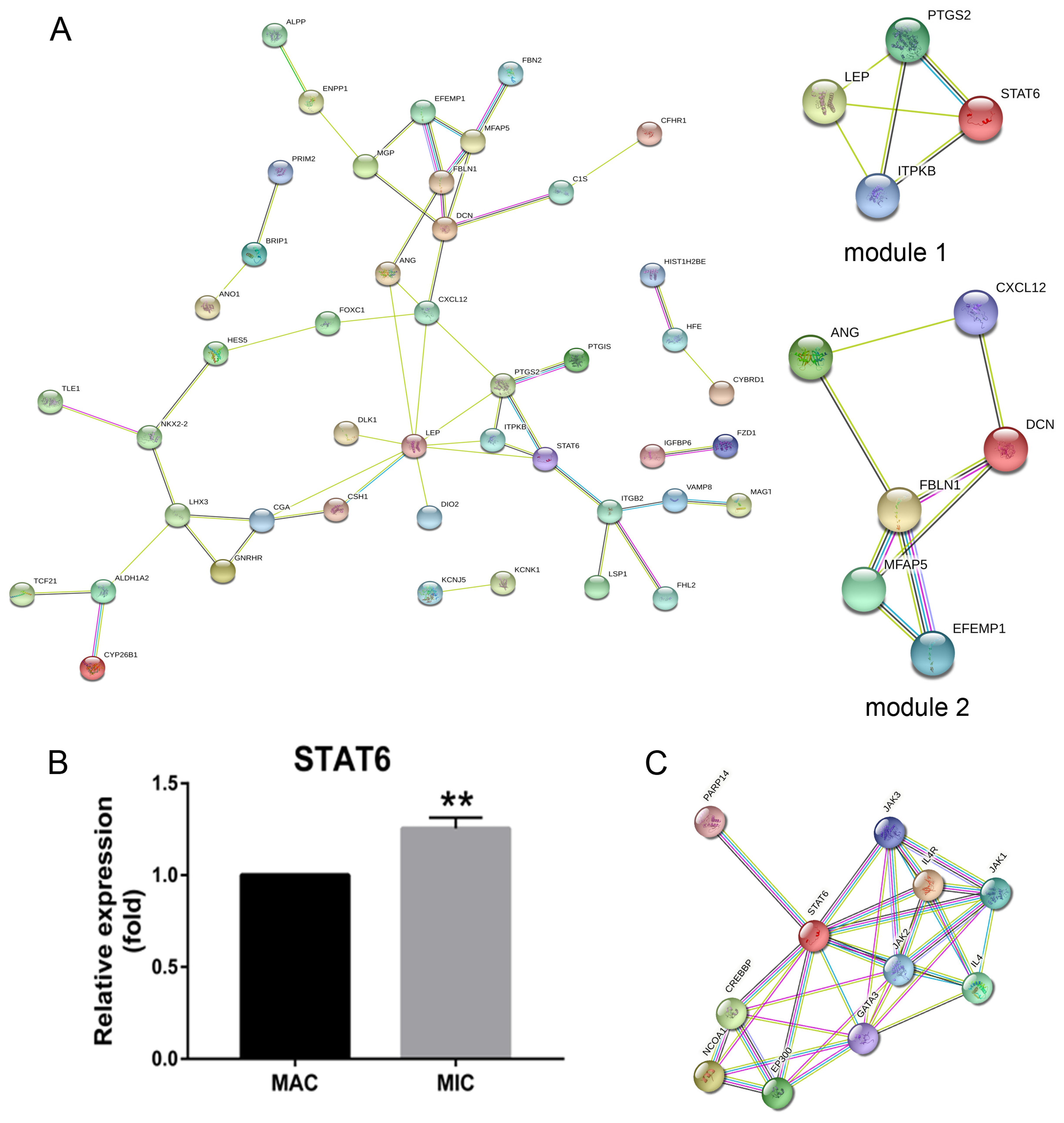
2.1. Pituitary Adenomas Progression Correlates with STAT6 Expression Levels
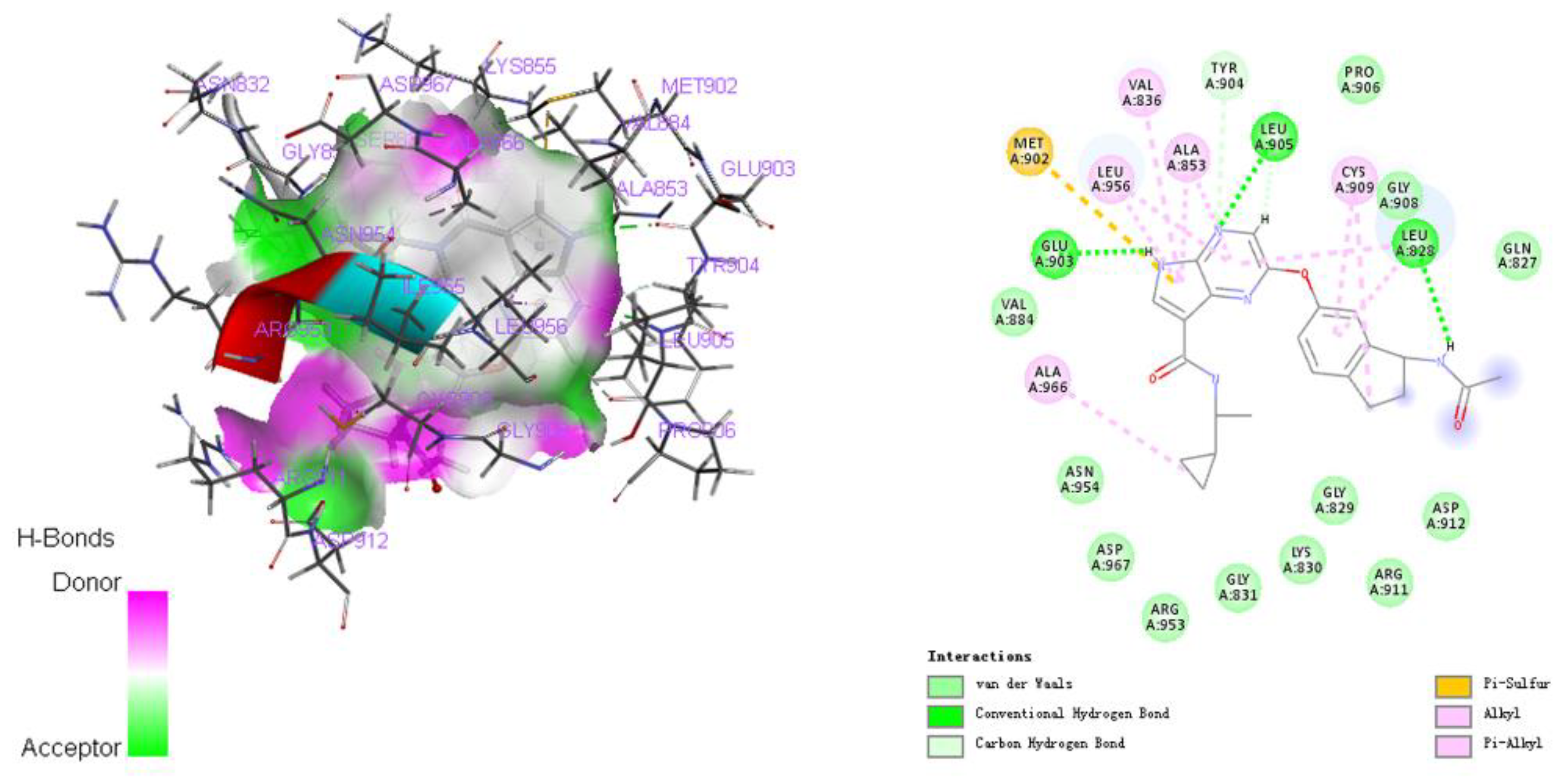
2.2. Repositioning of Tamoxifen to Target STAT6 to Control the Progression of Pituitary Adenomas
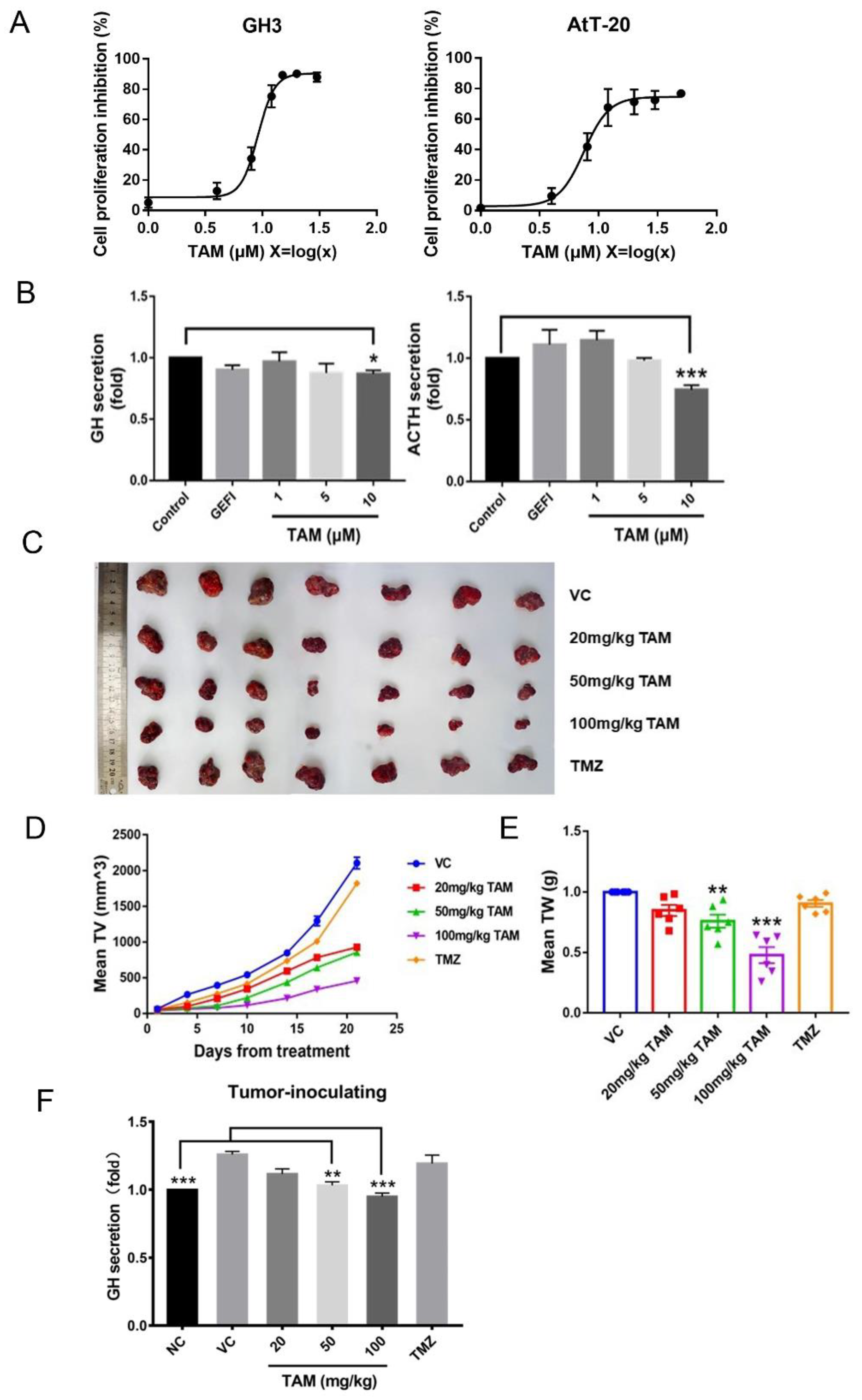
2.3. Tamoxifen Inhibited Pituitary Adenomas Progression Both In Vitro and In Vivo
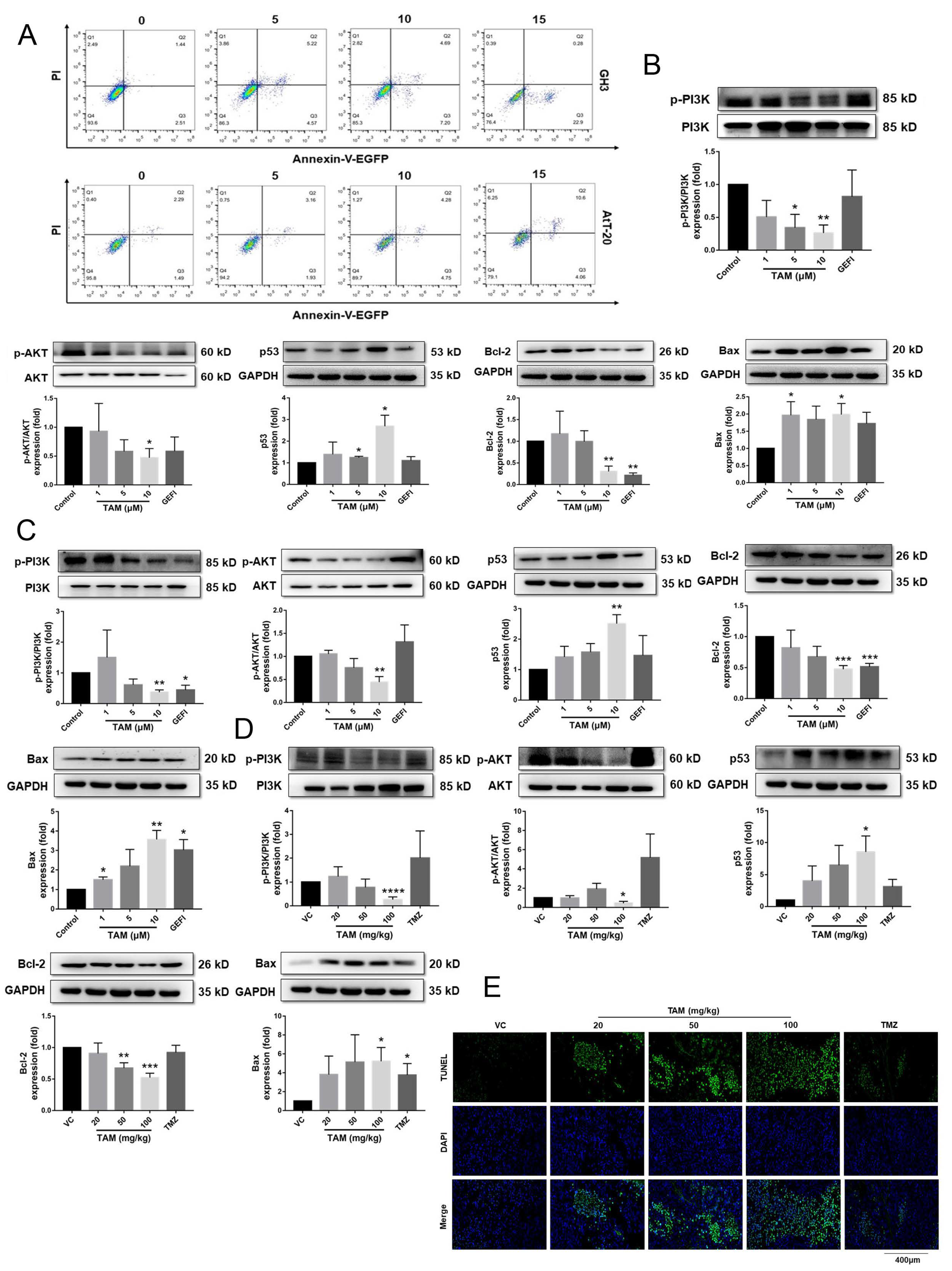
2.4. Tamoxifen Induced Apoptosis of the Pituitary Adenomas and Changed the Expression Levels of Apoptosis-Related Proteins in Pituitary Adenomas
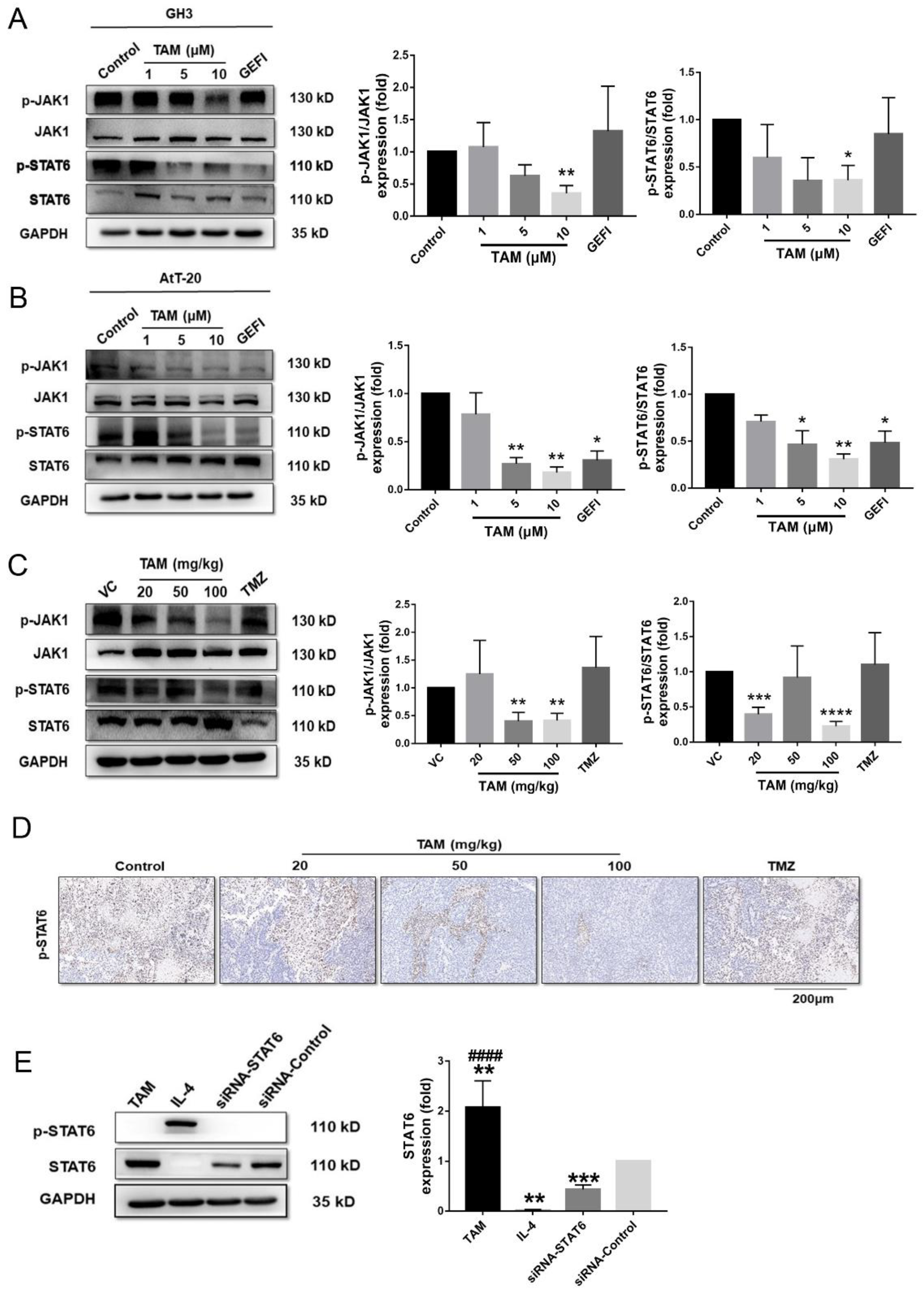
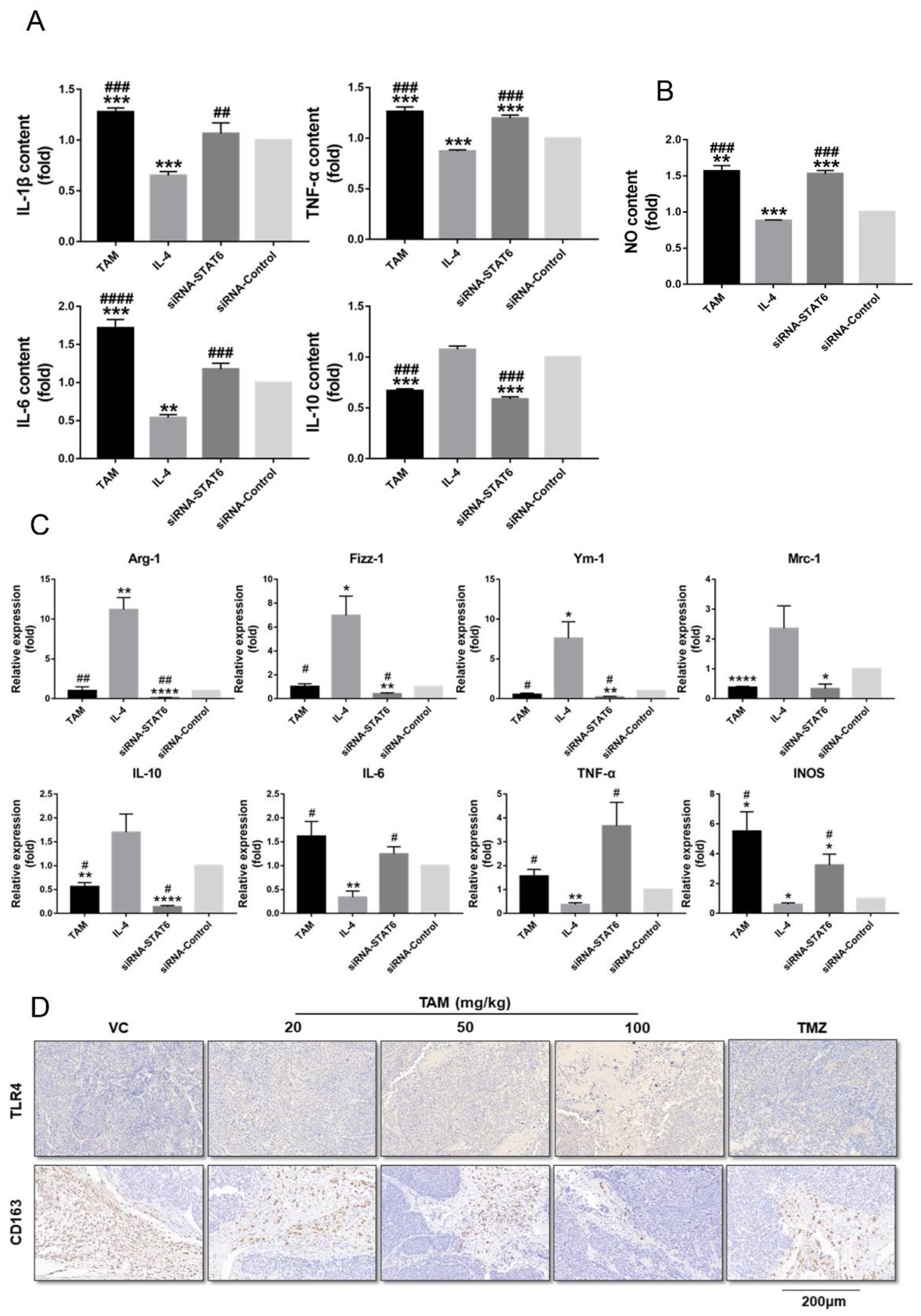
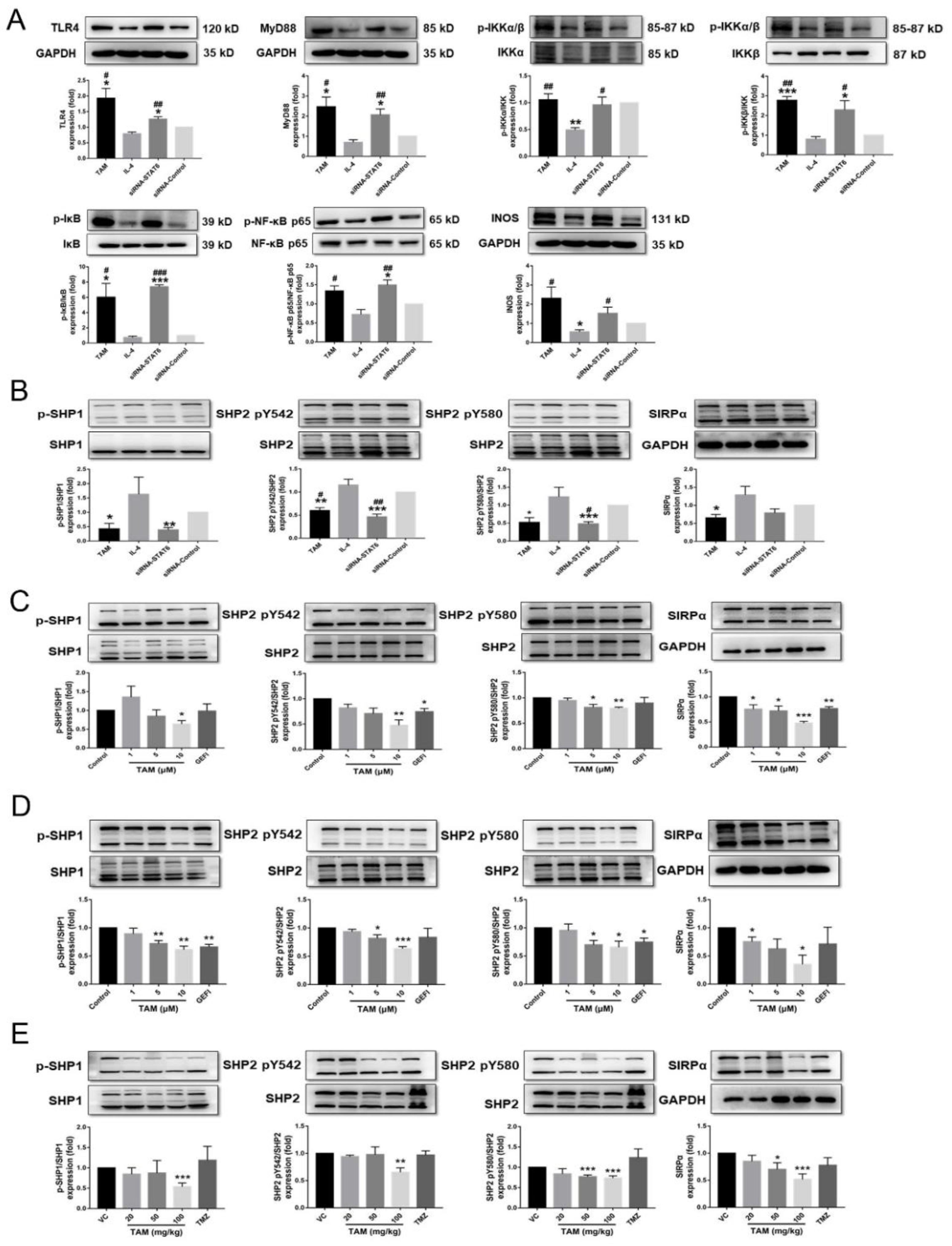
2.5. Tamoxifen Inhibited the Migration of Pituitary Adenomas Cells by Inducing M1 Polarization of Tumor-Associated Macrophages via STAT6 Inactivation and SHP1/SHP2 Blockade
3. Discussion
4. Materials and Methods
4.1. Gene Expression Profiles Analyses and Validation of the Hub Genes
4.2. Drug Repositioning and MOLECULAR Docking
4.3. Cell Culture and Proliferation Assay
4.4. Measurement of Hormone Levels
4.5. Annexin V-EGFP/PI Staining
4.6. SiRNA Transfection
4.7. Detection of Macrophage Polarization
4.8. Transwell Assay
4.9. Western Blotting
4.10. RNA Isolation and qRT-PCR
4.11. In Vivo Assessment
4.12. Immunohistochemistry, TUNEL Assay and Imaging
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kopczak, A.; Renner, U.; Karl Stalla, G. Advances in understanding pituitary tumors. F1000Prime Rep. 2014, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Mete, O.; Lopes, M.B. Overview of the 2017 WHO Classification of Pituitary Tumors. Endocr. Pathol. 2017, 28, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Nie, D.; Li, B.; Gui, S.; Li, C.; Zhang, Y.; Zhao, P. CircNFIX promotes progression of pituitary adenoma via CCNB1 by sponging miR-34a -5p. Mol. Cell. Endocrinol. 2021, 525, 111140. [Google Scholar] [CrossRef] [PubMed]
- Lenders, N.; McCormack, A. Malignant transformation in non-functioning pituitary adenomas (pituitary carcinoma). Pituitary 2018, 21, 217–229. [Google Scholar] [CrossRef]
- Losa, M.; Bogazzi, F.; Cannavo, S.; Ceccato, F.; Curtò, L.; De Marinis, L.; Iacovazzo, D.; Lombardi, G.; Mantovani, G.; Mazza, E.; et al. Temozolomide therapy in patients with aggressive pituitary adenomas or carcinomas. J. Neuro-Oncol. 2015, 126, 519–525. [Google Scholar] [CrossRef]
- El-Sharkawy, E.R.; Mahmoud, K. Cytotoxity of two new coumarin derivatives isolated from Launaea mucronata. Nat. Prod. Res. 2016, 30, 394–398. [Google Scholar] [CrossRef]
- Zhou, Y.; Zheng, J.; Li, Y.; Xu, D.-P.; Li, S.; Chen, Y.-M.; Li, H.-B. Natural Polyphenols for Prevention and Treatment of Cancer. Nutrients 2016, 8, 515. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F.; Atashpour, S.; Haghighat, S. Effects of Genistein and Synergistic Action in Combination with Tamoxifen on the HepG2 Human Hepatocellular Carcinoma Cell Line. Asian Pac. J. Cancer Prev. 2017, 18, 2381–2385. [Google Scholar] [CrossRef]
- Porta, C.; Riboldi, E.; Ippolito, A.; Sica, A. Molecular and epigenetic basis of macrophage polarized activation. Semin. Immunol. 2015, 27, 237–248. [Google Scholar] [CrossRef]
- Zitvogel, L.; Galluzzi, L.; Smyth, M.J.; Kroemer, G. Mechanism of Action of Conventional and Targeted Anticancer Therapies: Reinstating Immunosurveillance. Immunity 2013, 39, 74–88. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, T.; Li, L.; Sun, L.; Hou, Y.; Hu, X.; Zhang, L.; Tian, H.; Zhao, Q.; Peng, J.; et al. TSC1 controls macrophage polarization to prevent inflammatory disease. Nat. Commun. 2014, 5, 4696. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.C. Innate immune response triggers lupus-like autoimmune disease. Cell 2007, 130, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Fiset, P.O.; Tulic, M.K.; Skrablin, P.S.A.; Grover, S.M.; Létuvé, S.; Mazer, B.D.; Hamid, Q. Signal transducer and activator of transcription 6 down-regulates toll-like receptor-4 expression of a monocytic cell line. Clin. Exp. Allergy 2006, 36, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, W.; Wang, S.; Yang, T.; Zhang, G.; Wang, D.; Ju, R.; Lu, Y.; Wang, H.; Wang, L. Tumor microenvironment remodeling and tumor therapy based on M2-like tumor associated macrophage-targeting nano-complexes. Theranostics 2021, 11, 2892–2916. [Google Scholar] [CrossRef]
- Tariq, M.; Zhang, J.Q.; Liang, G.K.; He, Q.J.; Ding, L.; Yang, B. Gefitinib inhibits M2-like polarization of tumor-associated macrophages in Lewis lung cancer by targeting the STAT6 signaling pathway. Acta Pharmacol. Sin. 2017, 38, 1501–1511. [Google Scholar] [CrossRef]
- Kong, X.N.; Yan, H.X.; Chen, L.; Dong, L.W.; Yang, W.; Liu, Q.; Yu, L.X.; Huang, D.D.; Liu, S.Q.; Liu, H.; et al. LPS-induced down-regulation of signal regulatory protein {alpha} contributes to innate immune activation in macrophages. J. Exp. Med. 2007, 204, 2719–2731. [Google Scholar] [CrossRef]
- Huang, H.; Zheng, Y.; Li, L.; Shi, W.; Zhang, R.; Liu, H.; Chen, Z.; Wu, L. The roles of post-translational modifications and coactivators of STAT6 signaling in tumor growth and progression. Futur. Med. Chem. 2020, 12, 1945–1960. [Google Scholar] [CrossRef]
- Liu, D.; Tao, T.; Xu, B.; Chen, S.; Liu, C.; Zhang, L.; Lu, K.; Huang, Y.; Jiang, L.; Zhang, X.; et al. MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6). Biochem. Biophys. Res. Commun. 2014, 445, 151–156. [Google Scholar] [CrossRef]
- Xu, B.; Lu, X.; Zhao, Y.; Liu, C.; Huang, X.; Chen, S.; Zhu, W.; Zhang, L.; Chen, M. MicroRNA-135a induces prostate cancer cell apoptosis via inhibition of STAT6. Oncol. Lett. 2018, 17, 1889–1895. [Google Scholar] [CrossRef]
- Wang, N.; Tao, L.; Zhong, H.; Zhao, S.; Yu, Y.; Yu, B.; Chen, X.; Gao, J.; Wang, R. miR-135b inhibits tumour metastasis in prostate cancer by targeting STAT6. Oncol. Lett. 2015, 11, 543–550. [Google Scholar] [CrossRef]
- Cheng, S.; Xie, W.; Miao, Y.; Guo, J.; Wang, J.; Li, C.; Zhang, Y. Identification of key genes in invasive clinically non-functioning pituitary adenoma by integrating analysis of DNA methylation and mRNA expression profiles. J. Transl. Med. 2019, 17, 407. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.; Tait, S.W. Mitochondrial apoptosis: Killing cancer using the enemy within. Br. J. Cancer 2015, 112, 957–962. [Google Scholar] [CrossRef]
- Xie, F.; Feng, Y.; Huang, J.; Ding, Y.; Shen, X. Tamoxifen-induced apoptosis of rat C6 glioma cells via PI3K/Akt, JNK and ERK activation. Oncol. Rep. 2010, 24, 1561–1567. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.J.; Cantley, L.C. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 2006, 441, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Jiang, X.; Shen, Y.; Li, M.; Ma, H.; Xing, M.; Lu, Y. Frequent mutations and amplifications of the PIK3CA gene in pituitary tumors. Endocr.-Relat. Cancer 2009, 16, 301–310. [Google Scholar] [CrossRef]
- Muşat, M.; Korbonits, M.; Kola, B.; Borboli, N.; Hanson, M.R.; Nanzer, A.M.; Grigson, J.; Jordan, S.; Morris, D.G.; Gueorguiev, M.; et al. Enhanced protein kinase B/Akt signalling in pituitary tumours. Endocr.-Relat. Cancer 2005, 12, 423–433. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; Watt, A.C.; BrinJones, H.; Sceneay, J.; Li, B.B.; Khan, N.; Ubellacker, J.M.; Xie, S.; Metzger-Filho, O.; et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017, 548, 471–475. [Google Scholar] [CrossRef]
- Wu, S.Y.; Sharma, S.; Wu, K.; Tyagi, A.; Zhao, D.; Deshpande, R.P.; Watabe, K. Tamoxifen suppresses brain metastasis of estrogen receptor-deficient breast cancer by skewing microglia polarization and enhancing their immune functions. Breast Cancer Res. 2021, 23, 35. [Google Scholar] [CrossRef]
- Fridman, J.S.; Lowe, S.W. Control of apoptosis by p53. Oncogene 2003, 22, 9030–9040. [Google Scholar] [CrossRef]
- Budram-Mahadeo, V.; Morris, P.J.; Latchman, D.S. The Brn-3a transcription factor inhibits the pro-apoptotic effect of p53 and enhances cell cycle arrest by differentially regulating the activity of the p53 target genes encoding Bax and p21(CIP1/Waf1). Oncogene 2002, 21, 6123–6131. [Google Scholar] [CrossRef][Green Version]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Evan, G.I.; Vousden, K.H. Proliferation, cell cycle and apoptosis in cancer. Nature 2001, 411, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef] [PubMed]
- Stout, R.D.; Jiang, C.; Matta, B.; Tietzel, I.; Watkins, S.K.; Suttles, J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J. Immunol. 2005, 175, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Azimian, H.; Dayyani, M.; Toossi, M.T.B.; Mahmoudi, M. Bax/Bcl-2 expression ratio in prediction of response to breast cancer radiotherapy. Iran. J. Basic Med. Sci. 2018, 21, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.; Bossy-Wetzel, E.; Goldstein, J.C.; Fitzgerald, P.; Green, D.R. p53 Induces Apoptosis by Caspase Activation through Mitochondrial Cytochrome c Release. J. Biol. Chem. 2000, 275, 7337–7342. [Google Scholar] [CrossRef]
- Walford, H.H.; Doherty, T.A. STAT6 and lung inflammation. JAKSTAT 2013, 2, e25301. [Google Scholar] [CrossRef]
- Miyashita, T.; Reed, J.C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995, 80, 293–299. [Google Scholar]
- Wang, Y.; Zhang, X.; Xie, X.; Chen, W.; Li, M.; Diao, D.; Dang, C. Obesity and metabolic syndrome related macrophage promotes PD-L1 expression in TNBC through IL6/JAK/STAT pathway and can be reversed by telmisartan. Cancer Biol. Ther. 2020, 21, 1179–1190. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Y.; Xu, Y.; Xu, L.; Zheng, W.; Wu, Y.; Li, L.; Shen, P. Estrogen Represses Hepatocellular Carcinoma (HCC) Growth via Inhibiting Alternative Activation of Tumor-associated Macrophages (TAMs). J. Biol. Chem. 2012, 287, 40140–40149. [Google Scholar] [CrossRef] [PubMed]
- Schroder, A.J.; Pavlidis, P.; Arimura, A.; Capece, D.; Rothman, P.B. Cutting edge: STAT6 serves as a positive and negative regulator of gene expression in IL-4-stimulated B lymphocytes. J. Immunol. 2002, 168, 996–1000. [Google Scholar] [CrossRef] [PubMed]
- Ladoire, S.; Mignot, G.; Dabakuyo, S.; Arnould, L.; Apetoh, L.; Rébé, C.; Coudert, B.; Martin, F.; Bizollon, M.H.; Vanoli, A.; et al. In situ immune response after neoadjuvant chemotherapy for breast cancer predicts survival. J. Pathol. 2011, 224, 389–400. [Google Scholar] [CrossRef] [PubMed]
- A Lien, E.; Wester, K.; E Lønning, P.; Solheim, E.; Ueland, P.M. Distribution of tamoxifen and metabolites into brain tissue and brain metastases in breast cancer patients. Br. J. Cancer 1991, 63, 641–645. [Google Scholar] [CrossRef]
- Zarate CAJr Singh, J.B.; Carlson, P.J.; Quiroz, J.; Jolkovsky, L.; Luckenbaugh, D.A.; Manji, H.K. Efficacy of a protein kinase C inhibitor (tamoxifen) in the treatment of acute mania: A pilot study. Bipolar. Disord. 2007, 9, 561–570. [Google Scholar] [CrossRef]
- Lamberts, S.W.J.; De Quijada, M.; Klijn, J.G.M. The effect of tamoxifen on GH and PRL secretion by human pituitary tumors. J. Endocrinol. Investig. 1980, 3, 343–347. [Google Scholar] [CrossRef]
- Lamberts, S.W.; Verleun, T.; Oosterom, R. Effect of Tamoxifen Administration on Prolactin Release by Invasive Prolactin-Secreting Pituitary Adenomas. Neuroendocrinology 1982, 34, 339–342. [Google Scholar] [CrossRef]
- Gollard, R.; Kosty, M.; Cheney, C.; Copeland, B.; Bordin, G. Prolactin-secreting pituitary carcinoma with implants in the cheek pouch and metastases to the ovaries. A case report and literature review. Cancer 1995, 76, 1814–1820. [Google Scholar] [CrossRef]
- Koizumi, K.; Aono, T. Pregnancy after combined treatment with bromocriptine and tamoxifen in two patients with pituitary prolactinomas. Fertil. Steril. 1986, 46, 312–314. [Google Scholar] [CrossRef]
- Yu, H.; Ren, S.; Wang, J.; Lv, T.; Sun, L.; Du, G. Bexarotene combined with lapatinib for the treatment of Cushing’s disease: Evidence based on drug repositioning and experimental confirmation. Signal Transduct. Target. Ther. 2020, 5, 175. [Google Scholar] [CrossRef]
- Sbiera, S.; Deutschbein, T.; Weigand, I.; Reincke, M.; Fassnacht, M.; Allolio, B. The New Molecular Landscape of Cushing’s Disease. Trends Endocrinol. Metab. 2015, 26, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, H.; Cooper, O.; Ben-Shlomo, A.; Mamelak, A.; Ren, S.-G.; Bruyette, D.; Melmed, S. EGFR as a therapeutic target for human, canine, and mouse ACTH-secreting pituitary adenomas. J. Clin. Investig. 2011, 121, 4712–4721. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Shahinian, H.; Maya, M.M.; Yong, W.; Heaney, A.P. Temozolomide: A novel treatment for pituitary carcinoma. Lancet Oncol. 2006, 7, 518–520. [Google Scholar] [CrossRef]
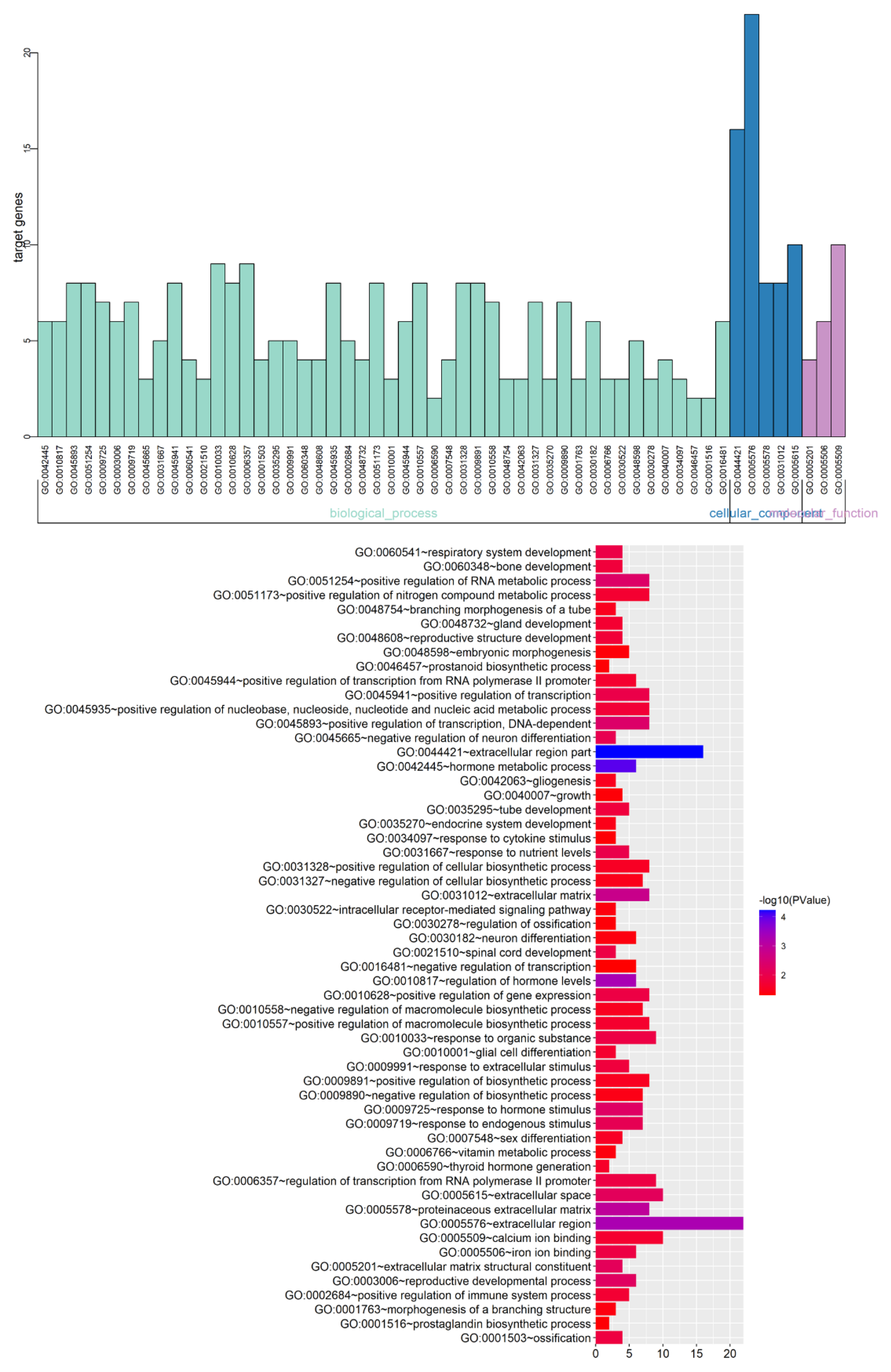

| a, BP | |||
|---|---|---|---|
| GO Term | Function | Count | p-Value |
| GO:0042445 | Hormone metabolic process | 6 | 9.61 × 10−5 |
| GO:0010817 | Regulation of hormone levels | 6 | 5 × 10−4 |
| GO:0045893 | Regulation of transcription, DNA-dependent | 8 | 0.004535 |
| GO:0051254 | Positive regulation of RNA metabolic process | 8 | 0.004745 |
| GO:0009725 | Response to hormone stimulus | 7 | 0.005137 |
| GO:0003006 | Reproductive developmental process | 6 | 0.005603 |
| GO:0009719 | Response to endogenous stimulus | 7 | 0.008202 |
| GO:0045665 | Negative regulation of neuron differentiation | 3 | 0.009053 |
| GO:0031667 | Response to nutrient levels | 5 | 0.010641 |
| GO:0045941 | Positive regulation of transcription | 8 | 0.011004 |
| GO:0060541 | Respiratory system development | 4 | 0.011637 |
| GO:0021510 | Spinal cord development | 3 | 0.012494 |
| GO:0010033 | Response to organic substance | 9 | 0.012515 |
| GO:0010628 | Positive regulation of gene expression | 8 | 0.012807 |
| GO:0006357 | Regulation of transcription from RNA polymerase | 9 | 0.013109 |
| GO:0001503 | Ossification | 4 | 0.013776 |
| GO:0035295 | Tube development | 5 | 0.015426 |
| GO:0009991 | Response to extracellular stimulus | 5 | 0.015426 |
| GO:0060348 | Bone development | 4 | 0.016477 |
| GO:0048608 | Reproductive structure development | 4 | 0.017561 |
| GO:0045935 | Regulation of nucleic acid metabolic process | 8 | 0.018314 |
| GO:0002684 | Positive regulation of immune system process | 5 | 0.01999 |
| GO:0048732 | Gland development | 4 | 0.021049 |
| GO:0051173 | Regulation of nitrogen compound metabolic | 8 | 0.021377 |
| GO:0010001 | Glial cell differentiation | 3 | 0.022349 |
| GO:0045944 | Regulation of transcription from RNA polymerase | 6 | 0.022552 |
| GO:0010557 | Regulation of macromolecule biosynthetic | 8 | 0.023036 |
| GO:0006590 | Thyroid hormone generation | 2 | 0.025889 |
| GO:0007548 | Sex differentiation | 4 | 0.028124 |
| GO:0031328 | Positive regulation of cellular biosynthetic | 8 | 0.028748 |
| GO:0009891 | Positive regulation of biosynthetic process | 8 | 0.03078 |
| GO:0010558 | Regulation of macromolecule biosynthetic | 7 | 0.031329 |
| GO:0048754 | Branching morphogenesis of a tube | 3 | 0.032633 |
| GO:0042063 | Gliogenesis | 3 | 0.032633 |
| GO:0031327 | Negative regulation of cellular biosynthetic | 7 | 0.034828 |
| GO:0035270 | Endocrine system development | 3 | 0.036402 |
| GO:0009890 | Negative regulation of biosynthetic process | 7 | 0.038022 |
| GO:0001763 | Morphogenesis of a branching structure | 3 | 0.041338 |
| GO:0030182 | Neuron differentiation | 6 | 0.041736 |
| GO:0006766 | ViTamoxifenin metabolic process | 3 | 0.042354 |
| GO:0030522 | Intracellular receptor-mediated signaling pathway | 3 | 0.042354 |
| GO:0048598 | Embryonic morphogenesis | 5 | 0.044701 |
| GO:0030278 | Regulation of ossification | 3 | 0.045457 |
| GO:0040007 | Growth | 4 | 0.0456 |
| GO:0034097 | Response to cytokine stimulus | 3 | 0.04651 |
| GO:0046457 | Prostanoid biosynthetic process | 2 | 0.046959 |
| GO:0001516 | Prostaglandin biosynthetic process | 2 | 0.046959 |
| GO:0016481 | Negative regulation of transcription | 6 | 0.049316 |
| b, CC | |||
| GO term | Function | Count | p-value |
| GO:0044421 | Extracellular region part | 16 | 5.93 × 10−5 |
| GO:0005576 | Extracellular region | 22 | 5.27 × 10−4 |
| GO:0005578 | Proteinaceous extracellular matrix | 8 | 0.001055 |
| GO:0031012 | Extracellular matrix | 8 | 0.001628 |
| GO:0005615 | Extracellular space | 10 | 0.006775 |
| c, MF | |||
| GO term | Function | Count | p-value |
| GO:0005201 | Extracellular matrix structural constituent | 4 | 0.006984 |
| GO:0005506 | Iron ion binding | 6 | 0.012809 |
| GO:0005509 | Calcium ion binding | 10 | 0.022046 |
| Module | Function Description | p-Value | Nodes | Genes |
|---|---|---|---|---|
| 1 | Negative regulation of response to stimulus | 0.007499 | 4 | LEP, PTGS2, STAT6, ITPKB |
| T cell differentiation | 0.00243 | 4 | ||
| 2 | Angiogenesis | 0.016832 | 6 | ANG, FBLN1, MFAP5, EFEMP1, DCN, CXCL12 |
| Gene | RMSD | -Docking Energy original | -Docking Energy TAM |
|---|---|---|---|
| STAT6 | 0.3199 | 14.3921 | 6.93868 |
| Sequence Number of Chips | GSE93825 |
|---|---|
| Platform | GPL18281 |
| Sample type | Pituitary human tissues |
| Sample | MACs and MICs |
| Reference | Cassarino et al. (2018) |
| Genes | Primer Sequence (5′→3′) |
|---|---|
| β-Actin | Forward primer: TCTGTGTGGATTGGTGGCTCTA |
| Reverse primer: CTGCTTGCTGATCCACATCTG | |
| Mrc-1 | Forward primer: GACTGCTGCTGAGTCCAGTT |
| Reverse primer: AGGGATCGCCTGTTTTCCAG | |
| Arg-1 | Forward primer: ACATTGGCTTGCGAGACGTA |
| Reverse primer: ATCACCTTGCCAATCCCCAG | |
| Ym1 | Forward primer: GGGCCCTTATTGAGAGGAGC |
| Reverse primer: CCAGCTGGTACAGCAGACAA | |
| Fizz1 | Forward primer: CCTGCTGGGATGACTGCTAC |
| Reverse primer: CAGTGGTCCAGTCAACGAGT | |
| IL-10 | Forward primer: CCAAGGTGTCTACAAGGCCA |
| Reverse primer: GCTCTGTCTAGGTCCTGGAGT | |
| IL-6 | Forward primer: CCGGAGAGGAGACTTCACAG |
| Reverse primer: CAGAATTGCCATTGCACAAC | |
| TNF-α | Forward primer: AGCCGATGGGTTGTACCT |
| Reverse primer: TGAGTTGGTCCCCCTTC | |
| INOS | Forward primer: CTCTACAACATCCTGGAGCAAGTG |
| Reverse primer: ACTATGGAGCACAGCCACATTGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, T.; Zhang, Z.; Yu, H.; Ren, S.; Wang, J.; Li, S.; Sun, L. Tamoxifen Exerts Anticancer Effects on Pituitary Adenoma Progression via Inducing Cell Apoptosis and Inhibiting Cell Migration. Int. J. Mol. Sci. 2022, 23, 2664. https://doi.org/10.3390/ijms23052664
Lv T, Zhang Z, Yu H, Ren S, Wang J, Li S, Sun L. Tamoxifen Exerts Anticancer Effects on Pituitary Adenoma Progression via Inducing Cell Apoptosis and Inhibiting Cell Migration. International Journal of Molecular Sciences. 2022; 23(5):2664. https://doi.org/10.3390/ijms23052664
Chicago/Turabian StyleLv, Tingting, Zirui Zhang, Haoying Yu, Shuyue Ren, Jingrong Wang, Shang Li, and Lan Sun. 2022. "Tamoxifen Exerts Anticancer Effects on Pituitary Adenoma Progression via Inducing Cell Apoptosis and Inhibiting Cell Migration" International Journal of Molecular Sciences 23, no. 5: 2664. https://doi.org/10.3390/ijms23052664
APA StyleLv, T., Zhang, Z., Yu, H., Ren, S., Wang, J., Li, S., & Sun, L. (2022). Tamoxifen Exerts Anticancer Effects on Pituitary Adenoma Progression via Inducing Cell Apoptosis and Inhibiting Cell Migration. International Journal of Molecular Sciences, 23(5), 2664. https://doi.org/10.3390/ijms23052664