Cyclin-Dependent Kinases (CDKs) and the Human Cytomegalovirus-Encoded CDK Ortholog pUL97 Represent Highly Attractive Targets for Synergistic Drug Combinations
Abstract
1. Introduction
2. Results
2.1. Evaluation of Antiviral Efficacy and Drug-Induced Cytotoxicity of Selected CDK Inhibitors and Drug Combinations
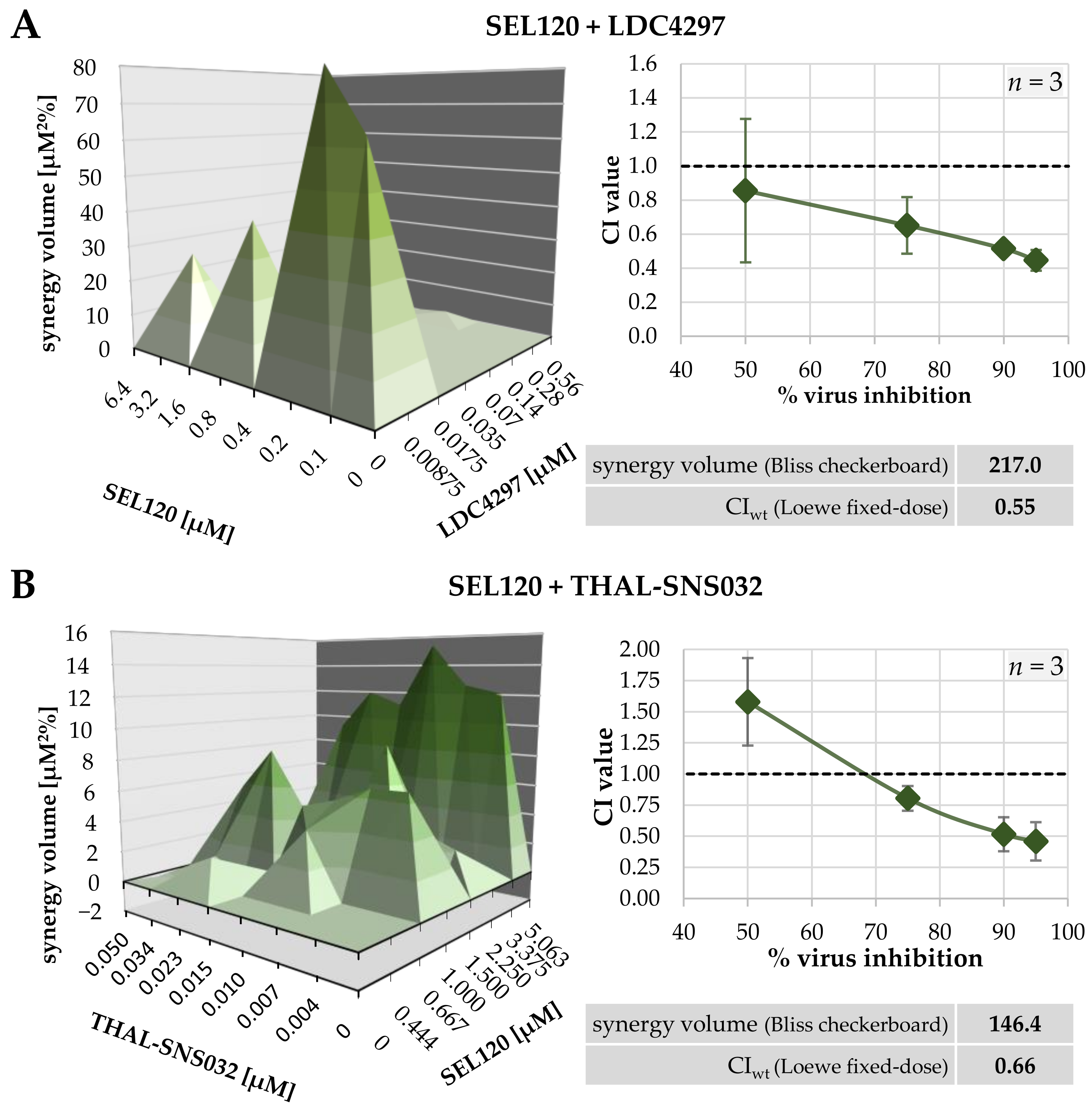
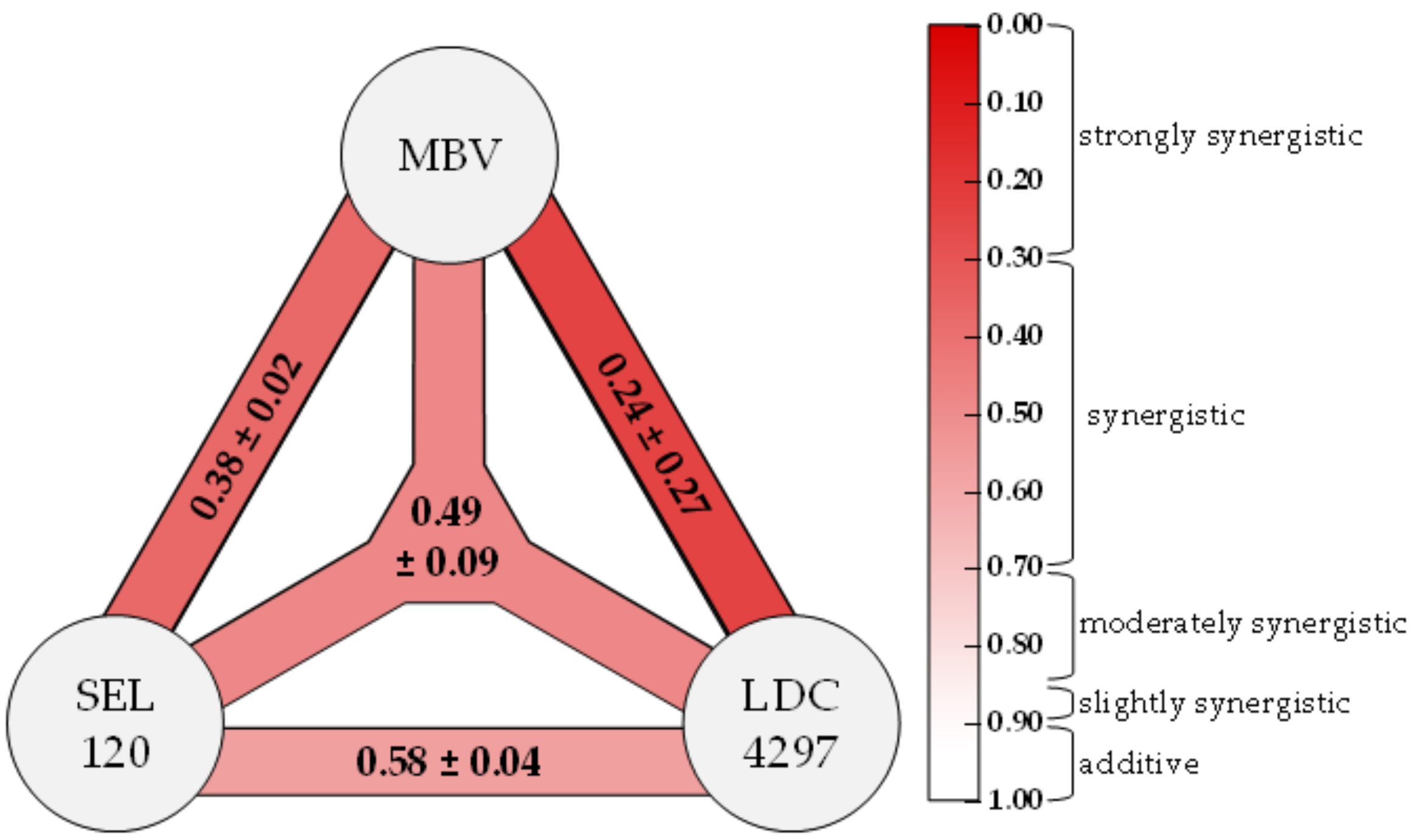
2.2. Assessment of Drug–Drug Interactions of Purely Host-Directed PKI Combinations
| Drug Combination | 95% Confidence Interval Synergy Volume [µM2%] b | Drug Interaction Type | ||
|---|---|---|---|---|
| Replicates a | Positive | Negative | ||
| SEL120 + LDC4297 | 1 | 217.0 | 0 | strongly synergistic |
| SEL120 + THAL-SNS032 | 1 | 148.4 | −2.0 | strongly synergistic |
| CDK2 Inh II + LDC4297 | 1 | 0 | −251.3 | antagonistic |
| Drug Combination | Ratio | Replicates a | CI Values Extrapolated at x Virus Inhibition | CIwt b | |||
|---|---|---|---|---|---|---|---|
| 50% | 75% | 90% | 95% | ||||
| SEL120 + LDC4297 | 10:1 | 3 | 0.86 ± 0.42 | 0.65 ± 0.17 | 0.51 ± 0.03 | 0.45 ± 0.06 | 0.55 ± 0.06 |
| SEL120 + THAL-SNS032 | 50:1 | 3 | 1.58 ± 0.35 | 0.80 ± 0.10 | 0.52 ± 0.14 | 0.46 ± 0.15 | 0.66 ± 0.10 |
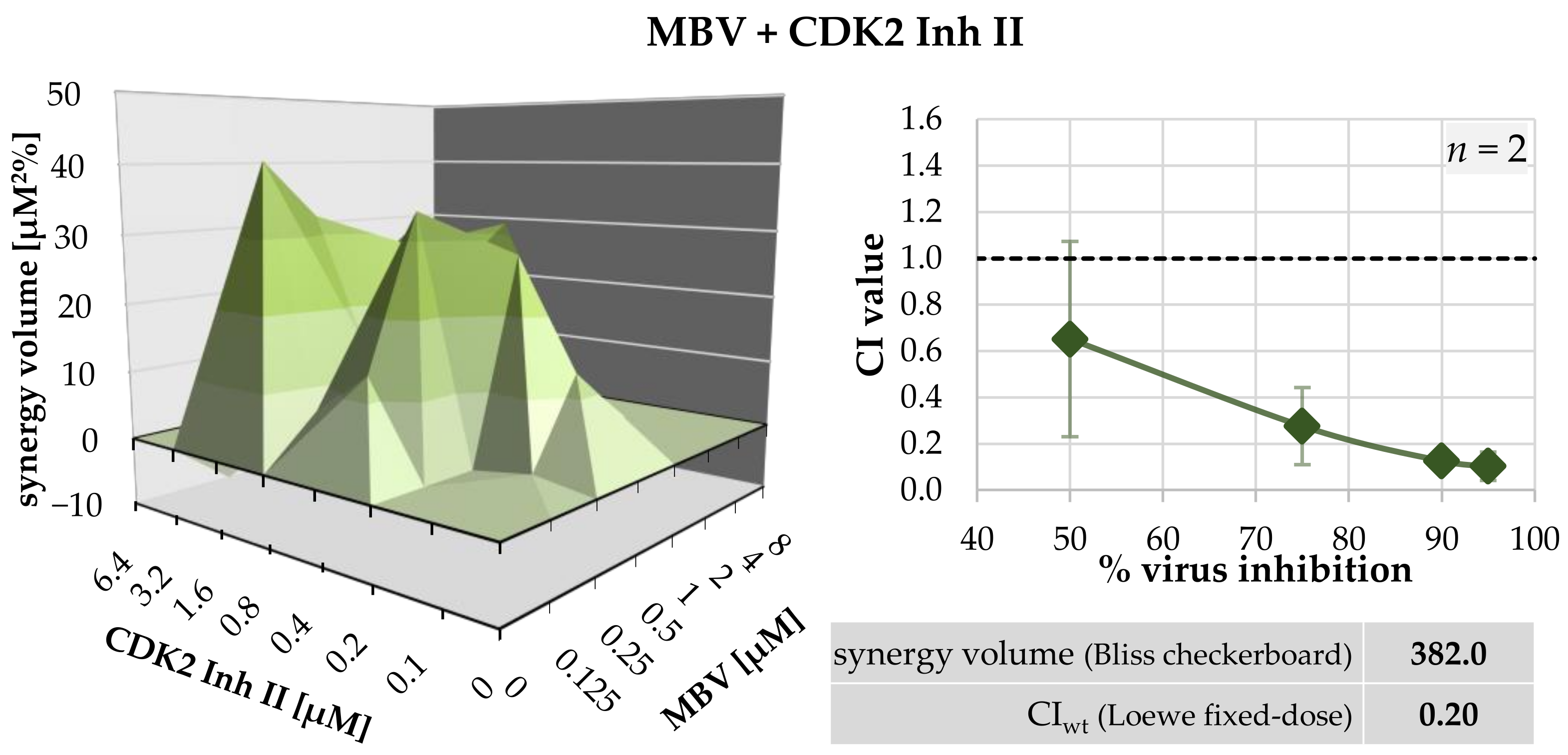
2.3. Assessment of Drug–Drug Interactions of PKI Combinations Directed to Both Host CDKs and vCDK/pUL97
| Drug Combination | Ratio | Replicates a | CI Values Extrapolated at x Virus Inhibition | CIwt b | |||
|---|---|---|---|---|---|---|---|
| 50% | 75% | 90% | 95% | ||||
| MBV + LDC4297 c | 100:1 | 4 | 0.37 ± 0.41 | 0.29 ± 0.23 | 0.30 ± 0.18 | 0.44 ± 0.36 | 0.36 ± 0.22 |
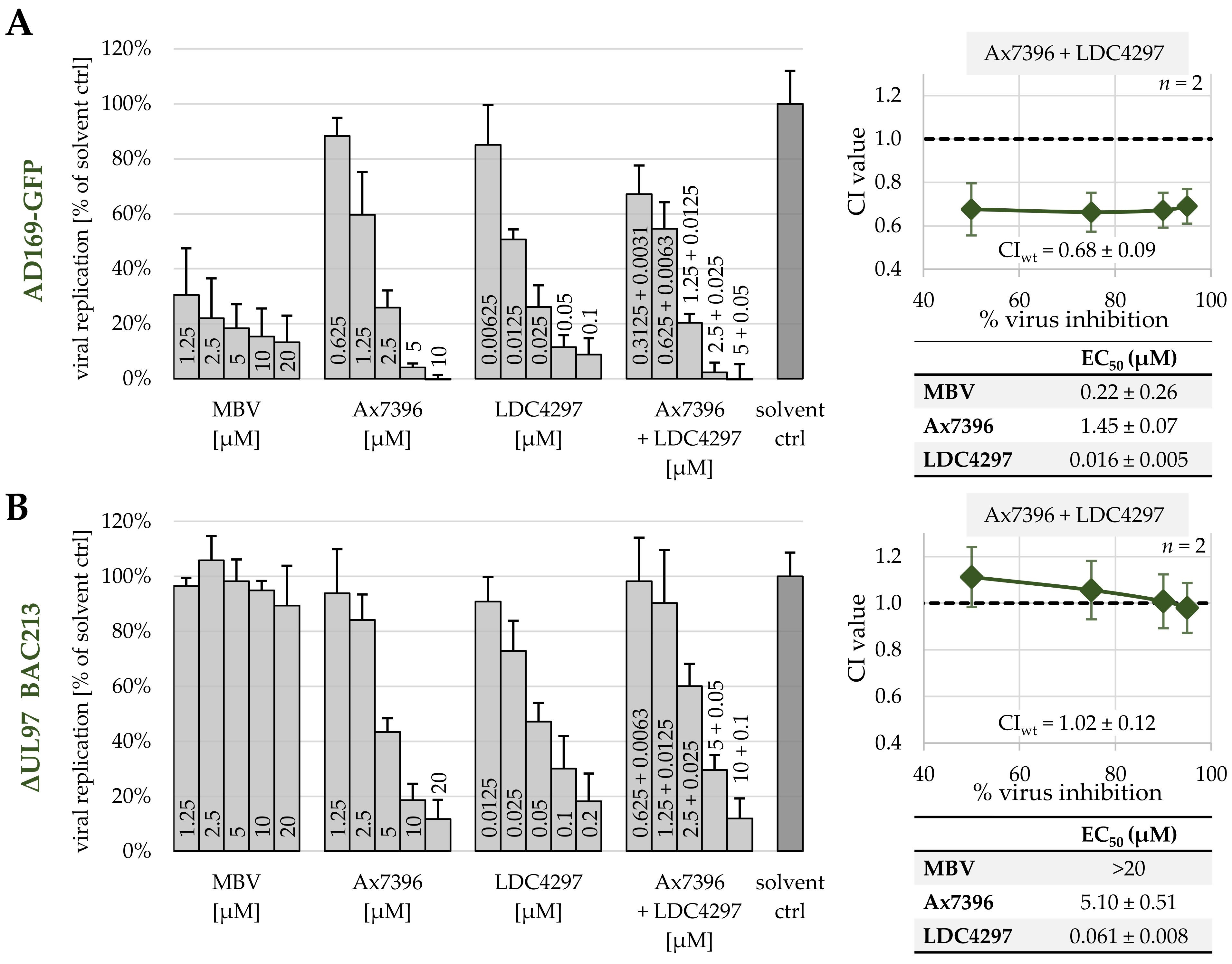
| Ax7396 + LDC4297 | 100:1 | 3 | 0.94 ± 0.47 | 0.68 ± 0.07 | 0.57 ± 0.18 | 0.55 ± 0.26 | 0.62 ± 0.12 |
| Gö6976 + LDC4297 | 100:1 | 3 | 0.70 ± 0.66 | 0.34 ± 0.14 | 0.37 ± 0.18 | 0.42 ± 0.23 | 0.42 ± 0.14 |
| Vi7392 + LDC4297 | 100:1 | 3 | 0.49 ± 0.12 | 0.51 ± 0.11 | 0.55 ± 0.14 | 0.59 ± 0.17 | 0.55 ± 0.13 |
| MBV + CDK2 Inh II | 1:1 | 2 | 0.65 ± 0.56 | 0.28 ± 0.24 | 0.13 ± 0.09 | 0.10 ± 0.01 | 0.20 ± 0.13 |
| Drug Combination | Cell type/Virus | Replicates a | 95% Confidence Interval Synergy Volume [µM2%] b | Drug Interaction Type | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| MBV + CDK2 Inh II | HFF/HCMV | 1 | 394.3 | −12.3 | strongly synergistic |
| MBV [µM] | LDC4297 [µM] | ||
|---|---|---|---|
| EC50 | single drugs | 0.35 ± 0.42 | 0.009 ± 0.002 |
| in combination (100:1) | 0.043 | 0.0004 | |
| dosage reduction | 8 × reduced | 23 × reduced | |
| EC90 | single drugs | 8.43 ± 3.68 | 0.124 ± 0.025 |
| in combination (100:1) | 2.76 | 0.028 | |
| dosage reduction | 3 × reduced | 4 × reduced | |
| Ax7396[µM] | LDC4297[µM] | ||
| EC50 | single drugs | 1.93 ± 0.84 | 0.013 ± 0.007 |
| in combination (100:1) | 0.570 ± 0.124 | 0.006 ± 0.001 | |
| dosage reduction | 3 × reduced | 2 × reduced | |
| EC90 | single drugs | 8.93 ± 9.72 | 0.097 ± 0.047 |
| in combination (100:1) | 2.09 ± 1.01 | 0.021 ± 0.010 | |
| dosage reduction | 4 × reduced | 5 × reduced | |
| Gö6976[µM] | LDC4297[µM] | ||
| EC50 | single drugs | 1.40 ± 1.97 | 0.009 ± 0.002 |
| in combination (100:1) | 0.228 ± 0.195 | 0.002 ± 0.002 | |
| dosage reduction | 6 × reduced | 5 × reduced | |
| EC90 | single drugs | 280.3 ± 468.6 | 0.147 ± 0.090 |
| in combination (100:1) | 3.13 ± 2.69 | 0.031 ± 0.027 | |
| dosage reduction | 90 × reduced | 5 × reduced | |
| Vi7392[µM] | LDC4297[µM] | ||
| EC50 | single drugs | 2.67 ± 0.36 | 0.016 ± 0.010 |
| in combination (100:1) | 6 × reduced | 3 × reduced | |
| dosage reduction | 0.451 ± 0.157 | 0.005 ± 0.002 | |
| EC90 | single drugs | 9.08 ± 2.70 | 0.088 ± 0.051 |
| in combination (100:1) | 2.16 ± 0.98 | 0.022 ± 0.010 | |
| dosage reduction | 4 × reduced | 4 × reduced | |
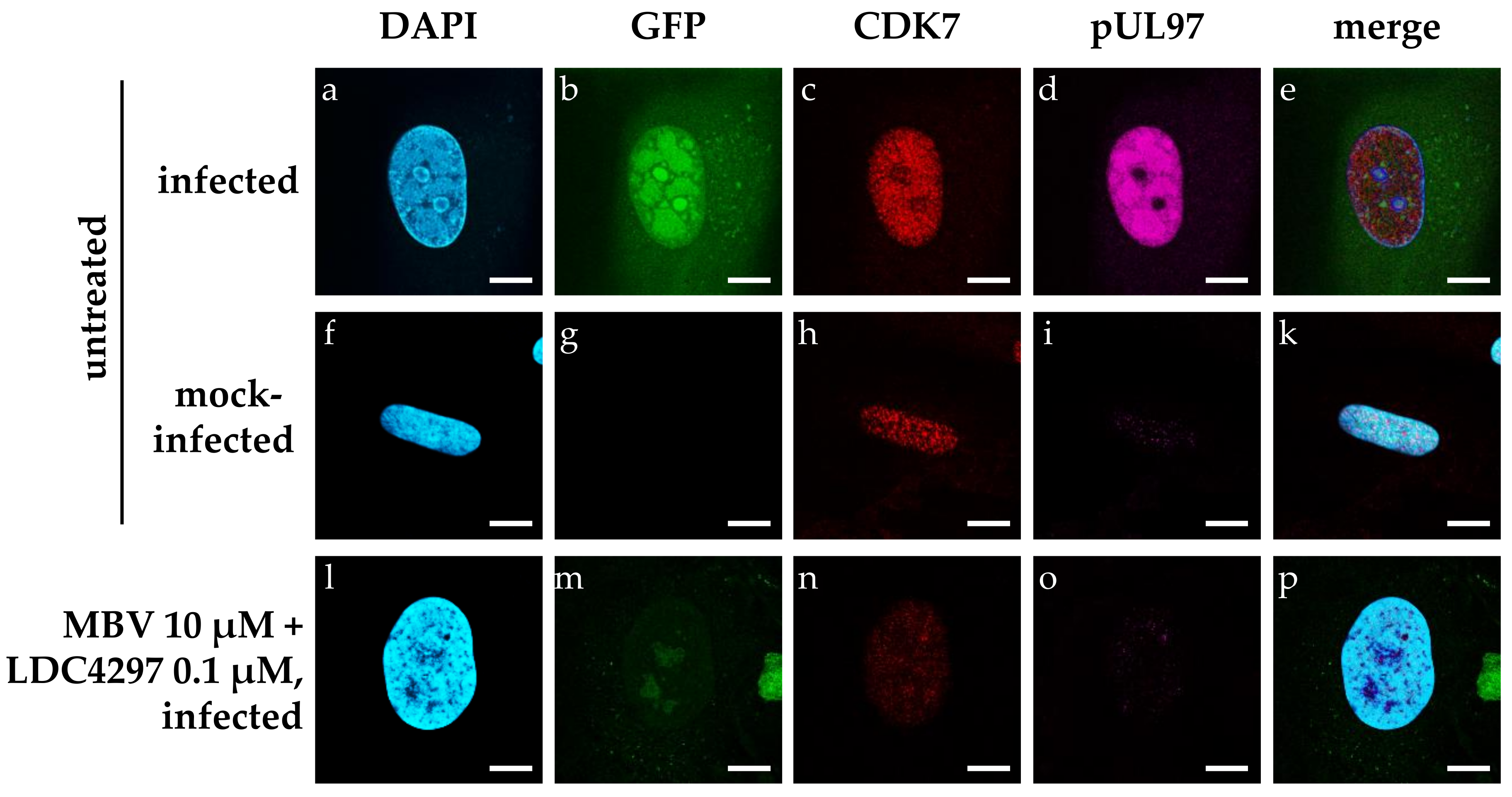
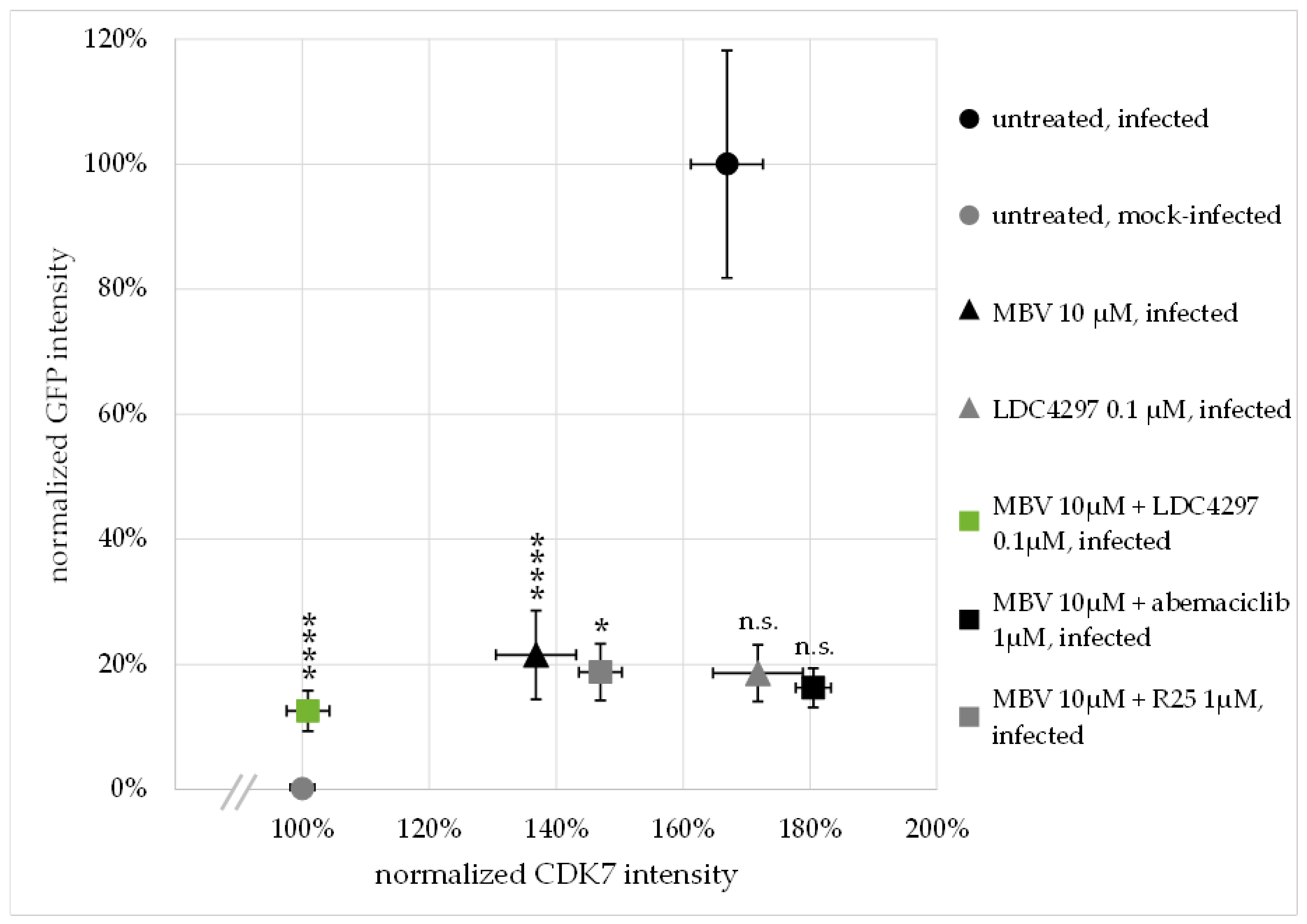
2.4. Confocal Imaging-Based Analysis: The HCMV-Induced Upregulation of CDK7 Expression Levels Is Completetly Abrogated by Synergistic Drug Treatment
2.5. Synergy between PKIs Ax7392 and LDC4297 Is pUL97-Dependent and pUL97-Specific, as Assessed by a HCMV ΔUL97 Deletion Mutant
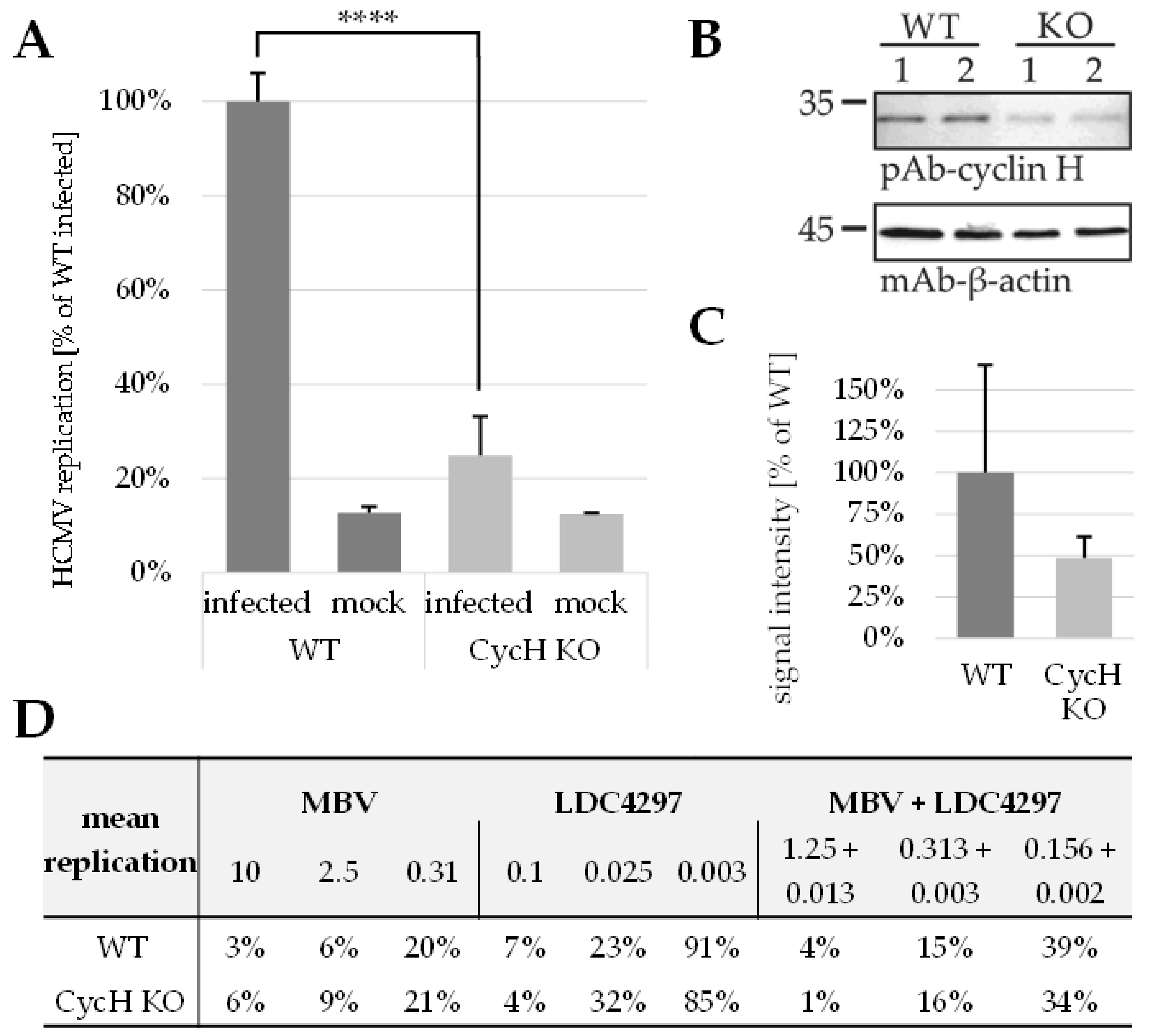
2.6. PKI Synergy Specifically Reinvestigated Using Host Cell Populations with Cyclin H Knock-Out
2.7. Triple Drug Combination Treatment Additionally Optimizes the Synergistic Potency of Anti-HCMV Activity of PKIs
2.8. Conclusions
3. Discussion
4. Materials and Methods
4.1. Cells and Viruses
4.2. Antiviral Compounds
4.3. Antibodies
4.4. Determination of Cell Viability by Neutral Red Uptake Assay
4.5. Drug Interaction Assessment via Bliss Independence Checkerboard Assay Adapted to HCMV-GFP In Vitro Infection
4.6. Drug Interaction Assessment via Loewe Additivity Fixed-Dose Assay Adapted to HCMV-GFP In Vitro Infection
4.7. Indirect Immunofluorescence Assay Utilizing Confocal Laser-Scanning Microscopy
4.8. Quantitation of Intracellular Immunofluorescence Signals
4.9. Generation of Lentiviral Transfer Constructs
4.10. Transient Cyclin H Knock-Out of HFFs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIDS | acquired immunodeficiency syndrome |
| app. | approximately |
| CAK | CDK-activating kinase |
| cCMV | congenital cytomegalovirus infection |
| CDK | cyclin-dependent kinase |
| CDV | cidofovir |
| CI | combination index |
| CIwt | weighted combination index |
| CTD | C-terminal domain |
| CycH | cyclin H |
| d | day(s) |
| DAA | direct-acting antiviral(s) |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DMSO | dimethyl sulfoxide |
| EGFR | epithelial growth factor receptor kinase |
| FBS | fetal bovine serum |
| FOS | foscarnet |
| GCV | ganciclovir |
| GFP | green fluorescent protein |
| gRNA | guide RNA |
| HCMV | human cytomegalovirus |
| HDA | host-directed antiviral(s) |
| HFF | human foreskin fibroblast |
| HIV | human immunodeficiency virus |
| IE | immediate early |
| KO | knock-out |
| LMV | letermovir |
| MBV | maribavir |
| MEM | Eagle’s minimal essential medium |
| min | minute(s) |
| MoA | mode of action |
| MOI | multiplicity of infection |
| NRA | neutral red assay |
| ORF | open reading frame |
| p.i. | post-infection |
| PBS | phosphate buffered saline |
| PKC | protein kinase C |
| PKI | pharmaceutical kinase inhibitor |
| RNAP | RNA polymerase |
| rpm | rotations per minute |
| SD | standard deviation |
| SEM | standard error of the mean |
| SV | synergy volume(s) |
| TCID | tissue culture infectious dose |
| VGCV | valganciclovir |
| Wb | western blot |
| WT | wild-type |
References
- Wild, M.; Kicuntod, J.; Seyler, L.; Wangen, C.; Bertzbach, L.D.; Conradie, A.M.; Kaufer, B.B.; Wagner, S.; Michel, D.; Eickhoff, J.; et al. Combinatorial Drug Treatments Reveal Promising Anticytomegaloviral Profiles for Clinically Relevant Pharmaceutical Kinase Inhibitors (PKIs). Int. J. Mol. Sci. 2021, 22, 575. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacol. Res. 2019, 139, 471–488. [Google Scholar] [CrossRef] [PubMed]
- Schor, S.; Einav, S. Repurposing of Kinase Inhibitors as Broad-Spectrum Antiviral Drugs. DNA Cell Biol. 2018, 37, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Chamorro, L.; Felip, E.; Ezeonwumelu, I.J.; Margelí, M.; Ballana, E. Cyclin-dependent Kinases as Emerging Targets for Developing Novel Antiviral Therapeutics. Trends Microbiol. 2021, 29, 836–848. [Google Scholar] [CrossRef]
- Steingruber, M.; Marschall, M. The Cytomegalovirus Protein Kinase pUL97:Host Interactions, Regulatory Mechanisms and Antiviral Drug Targeting. Microorganisms 2020, 8, 515. [Google Scholar] [CrossRef]
- Acosta, E.; Bowlin, T.; Brooks, J.; Chiang, L.; Hussein, I.; Kimberlin, D.; Kauvar, L.M.; Leavitt, R.; Prichard, M.; Whitley, R. Advances in the Development of Therapeutics for Cytomegalovirus Infections. J. Infect. Dis. 2020, 221, S32–S44. [Google Scholar] [CrossRef]
- Piret, J.; Boivin, G. Clinical development of letermovir and maribavir: Overview of human cytomegalovirus drug resistance. Antivir. Res. 2019, 163, 91–105. [Google Scholar] [CrossRef]
- Cannon, M.J.; Schmid, D.S.; Hyde, T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 2010, 20, 202–213. [Google Scholar] [CrossRef]
- Mocarski, E.S.; Shenk, T.; Griffiths, P.D.; Pass, R.F. Cytomegaloviruses. In Fields Virology, 6th ed.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1960–2014. [Google Scholar]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef]
- Sever, J.L.; Rakusan, T.A.; Ellaurie, M.; Frenkel, N.; Wyatt, L.S.; Campos, J.M.; O’Donnell, R.M.; Price, M.V. Coinfection with herpesviruses in young children of HIV-infected women. Pediatr. AIDS HIV Infect. 1995, 6, 75–82. [Google Scholar]
- Meesing, A.; Razonable, R.R. Pharmacologic and immunologic management of cytomegalovirus infection after solid organ and hematopoietic stem cell transplantation. Expert Rev. Clin. Pharmacol. 2018, 11, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Neumann, D.M. Persistent HCMV infection of a glioblastoma cell line contributes to the development of resistance to temozolomide. Virus Res. 2020, 276, 197829. [Google Scholar] [CrossRef] [PubMed]
- Revello, M.G.; Gerna, G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin. Microbiol. Rev. 2002, 15, 680–715. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Stamminger, T. Molecular targets for antiviral therapy of cytomegalovirus infections. Future Microbiol. 2009, 4, 731–742. [Google Scholar] [CrossRef]
- Lurain, N.S.; Chou, S. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 2010, 23, 689–712. [Google Scholar] [CrossRef]
- Marty, F.M.; Ljungman, P.; Chemaly, R.F.; Maertens, J.; Dadwal, S.S.; Duarte, R.F.; Haider, S.; Ullmann, A.J.; Katayama, Y.; Brown, J.; et al. Letermovir Prophylaxis for Cytomegalovirus in Hematopoietic-Cell Transplantation. N. Engl. J. Med. 2017, 377, 2433–2444. [Google Scholar] [CrossRef]
- Gerna, G.; Lilleri, D.; Baldanti, F. An overview of letermovir: A cytomegalovirus prophylactic option. Expert Opin. Pharm. 2019, 20, 1429–1438. [Google Scholar] [CrossRef]
- Britt, W.J.; Prichard, M.N. New therapies for human cytomegalovirus infections. Antivir. Res. 2018, 159, 153–174. [Google Scholar] [CrossRef]
- Krishna, B.A.; Wills, M.R.; Sinclair, J.H. Advances in the treatment of cytomegalovirus. Br. Med. Bull. 2019, 131, 5–17. [Google Scholar] [CrossRef]
- Hutterer, C.; Eickhoff, J.; Milbradt, J.; Korn, K.; Zeittrager, I.; Bahsi, H.; Wagner, S.; Zischinsky, G.; Wolf, A.; Degenhart, C.; et al. A novel CDK7 inhibitor of the Pyrazolotriazine class exerts broad-spectrum antiviral activity at nanomolar concentrations. Antimicrob. Agents Chemother 2015, 59, 2062–2071. [Google Scholar] [CrossRef]
- García-Cárceles, J.; Caballero, E.; Gil, C.; Martínez, A. Kinase Inhibitors as Underexplored Antiviral Agents. J. Med. Chem. 2021, 65, 935–954. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2021 update. Pharmacol. Res. 2021, 165, 105463. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, E.; Hahn, F.; Bertzbach, L.D.; Seyler, L.; Wangen, C.; Müller, R.; Tannig, P.; Grau, B.; Baumann, M.; Zent, E.; et al. In vivo proof-of-concept for two experimental antiviral drugs, both directed to cellular targets, using a murine cytomegalovirus model. Antivir. Res. 2019, 161, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, E.; Milbradt, J.; Svrlanska, A.; Strojan, H.; Häge, S.; Kraut, A.; Hesse, A.M.; Amin, B.; Sonnewald, U.; Couté, Y.; et al. Protein kinases responsible for the phosphorylation of the nuclear egress core complex of human cytomegalovirus. J. Gen. Virol. 2017, 98, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Hutterer, C.; Hamilton, S.; Steingruber, M.; Zeitträger, I.; Bahsi, H.; Thuma, N.; Naing, Z.; Örfi, Z.; Örfi, L.; Socher, E.; et al. The chemical class of quinazoline compounds provides a core structure for the design of anticytomegaloviral kinase inhibitors. Antivir. Res. 2016, 134, 130–143. [Google Scholar] [CrossRef]
- Feichtinger, S.; Stamminger, T.; Müller, R.; Graf, L.; Klebl, B.; Eickhoff, J.; Marschall, M. Recruitment of cyclin-dependent kinase 9 to nuclear compartments during cytomegalovirus late replication: Importance of an interaction between viral pUL69 and cyclin T1. J. Gen. Virol. 2011, 92, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Rechter, S.; Scott, G.M.; Eickhoff, J.; Zielke, K.; Auerochs, S.; Müller, R.; Stamminger, T.; Rawlinson, W.D.; Marschall, M. Cyclin-dependent Kinases Phosphorylate the Cytomegalovirus RNA Export Protein pUL69 and Modulate Its Nuclear Localization and Activity. J. Biol. Chem. 2009, 284, 8605–8613. [Google Scholar] [CrossRef] [PubMed]
- Schleiss, M.; Eickhoff, J.; Auerochs, S.; Leis, M.; Abele, S.; Rechter, S.; Choi, Y.; Anderson, J.; Scott, G.; Rawlinson, W.; et al. Protein kinase inhibitors of the quinazoline class exert anti-cytomegaloviral activity in vitro and in vivo. Antivir. Res. 2008, 79, 49–61. [Google Scholar] [CrossRef]
- Herget, T.; Freitag, M.; Morbitzer, M.; Kupfer, R.; Stamminger, T.; Marschall, M. Novel chemical class of pUL97 protein kinase-specific inhibitors with strong anticytomegaloviral activity. Antimicrob. Agents Chemother 2004, 48, 4154–4162. [Google Scholar] [CrossRef]
- Marschall, M.; Stein-Gerlach, M.; Freitag, M.; Kupfer, R.; van den Bogaard, M.; Stamminger, T. Inhibitors of human cytomegalovirus replication drastically reduce the activity of the viral protein kinase pUL97. J. Gen. Virol. 2001, 82, 1439–1450. [Google Scholar] [CrossRef]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Hahn, F.; Hamilton, S.T.; Wangen, C.; Wild, M.; Kicuntod, J.; Brückner, N.; Follett, J.E.L.; Herrmann, L.; Kheimar, A.; Kaufer, B.B.; et al. Development of a PROTAC-Based Targeting Strategy Provides a Mechanistically Unique Mode of Anti-Cytomegalovirus Activity. Int. J. Mol. Sci. 2021, 22, 12858. [Google Scholar] [CrossRef] [PubMed]
- Biron, K.K.; Harvey, R.J.; Chamberlain, S.C.; Good, S.S.; Smith, A.A., 3rd; Davis, M.G.; Talarico, C.L.; Miller, W.H.; Ferris, R.; Dornsife, R.E.; et al. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob. Agents Chemother. 2002, 46, 2365–2372. [Google Scholar] [CrossRef] [PubMed]
- Koszalka, G.W.; Johnson, N.W.; Good, S.S.; Boyd, L.; Chamberlain, S.C.; Townsend, L.B.; Drach, J.C.; Biron, K.K. Preclinical and toxicology studies of 1263W94, a potent and selective inhibitor of human cytomegalovirus replication. Antimicrob. Agents Chemother. 2002, 46, 2373–2380. [Google Scholar] [CrossRef]
- Lalezari, J.P.; Aberg, J.A.; Wang, L.H.; Wire, M.B.; Miner, R.; Snowden, W.; Talarico, C.L.; Shaw, S.; Jacobson, M.A.; Drew, W.L. Phase I dose escalation trial evaluating the pharmacokinetics, anti-human cytomegalovirus (HCMV) activity, and safety of 1263W94 in human immunodeficiency virus-infected men with asymptomatic HCMV shedding. Antimicrob. Agents Chemother. 2002, 46, 2969–2976. [Google Scholar] [CrossRef][Green Version]
- Ma, J.D.; Nafziger, A.N.; Villano, S.A.; Gaedigk, A.; Bertino, J.S. Maribavir Pharmacokinetics and the Effects of Multiple-Dose Maribavir on Cytochrome P450 (CYP) 1A2, CYP 2C9, CYP 2C19, CYP 2D6, CYP 3A, N-Acetyltransferase-2, and Xanthine Oxidase Activities in Healthy Adults. Antimicrob. Agents Chemother. 2006, 50, 1130. [Google Scholar] [CrossRef]
- Marty, F.M.; Boeckh, M. Maribavir and human cytomegalovirus-what happened in the clinical trials and why might the drug have failed? Curr. Opin. Virol. 2011, 1, 555–562. [Google Scholar] [CrossRef]
- Chou, S.; Waldemer, R.H.; Senters, A.E.; Michels, K.S.; Kemble, G.W.; Miner, R.C.; Drew, W.L. Cytomegalovirus UL97 Phosphotransferase Mutations That Affect Susceptibility to Ganciclovir. J. Infect. Dis. 2002, 185, 162–169. [Google Scholar] [CrossRef]
- Chou, S. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev. Med. Virol. 2008, 18, 233–246. [Google Scholar] [CrossRef]
- Sonntag, E.; Hamilton, S.T.; Bahsi, H.; Wagner, S.; Jonjic, S.; Rawlinson, W.D.; Marschall, M.; Milbradt, J. Cytomegalovirus pUL50 is the multi-interacting determinant of the core nuclear egress complex (NEC) that recruits cellular accessory NEC components. J. Gen. Virol. 2016, 97, 1676–1685. [Google Scholar] [CrossRef]
- Goekjian, P.G.; Jirousek, M.R. Protein kinase C in the treatment of disease: Signal transduction pathways, inhibitors, and agents in development. Curr. Med. Chem. 1999, 6, 877–903. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, A.; Wilts, H.; Lenhardt, M.; Hahn, M.; Mertens, T. Indolocarbazoles exhibit strong antiviral activity against human cytomegalovirus and are potent inhibitors of the pUL97 protein kinase. Antivir. Res. 2000, 48, 49–60. [Google Scholar] [CrossRef]
- Marschall, M.; Stein-Gerlach, M.; Freitag, M.; Kupfer, R.; van den Bogaard, M.; Stamminger, T. Direct targeting of human cytomegalovirus protein kinase pUL97 by kinase inhibitors is a novel principle for antiviral therapy. J. Gen. Virol. 2002, 83, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, T.; Reiter, C.; Ibrahim, M.M.; Beutel, J.; Hutterer, C.; Zeitträger, I.; Bahsi, H.; Leidenberger, M.; Friedrich, O.; Kappes, B.; et al. Synthesis of Novel Hybrids of Quinazoline and Artemisinin with High Activities against Plasmodium falciparum, Human Cytomegalovirus, and Leukemia Cells. ACS Omega 2017, 2, 2422–2431. [Google Scholar] [CrossRef] [PubMed]
- Held, F.E.; Guryev, A.A.; Fröhlich, T.; Hampel, F.; Kahnt, A.; Hutterer, C.; Steingruber, M.; Bahsi, H.; von Bojničić-Kninski, C.; Mattes, D.S.; et al. Facile access to potent antiviral quinazoline heterocycles with fluorescence properties via merging metal-free domino reactions. Nat. Commun. 2017, 8, 15071. [Google Scholar] [CrossRef]
- Brehmer, D.; Greff, Z.; Godl, K.; Blencke, S.; Kurtenbach, A.; Weber, M.; Müller, S.; Klebl, B.; Cotten, M.; Kéri, G.; et al. Cellular targets of gefitinib. Cancer Res. 2005, 65, 379–382. [Google Scholar]
- Marschall, M.; Marzi, A.; aus dem Siepen, P.; Jochmann, R.; Kalmer, M.; Auerochs, S.; Lischka, P.; Leis, M.; Stamminger, T. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 2005, 280, 33357–33367. [Google Scholar] [CrossRef]
- Kelso, T.W.; Baumgart, K.; Eickhoff, J.; Albert, T.; Antrecht, C.; Lemcke, S.; Klebl, B.; Meisterernst, M. Cyclin-dependent kinase 7 controls mRNA synthesis by affecting stability of preinitiation complexes, leading to altered gene expression, cell cycle progression, and survival of tumor cells. Mol. Cell Biol. 2014, 34, 3675–3688. [Google Scholar] [CrossRef]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef]
- Martínez-Alonso, D.; Malumbres, M. Mammalian cell cycle cyclins. Semin. Cell Dev. Biol. 2020, 107, 28–35. [Google Scholar] [CrossRef]
- Schütz, M.; Steingruber, M.; Socher, E.; Müller, R.; Wagner, S.; Kögel, M.; Sticht, H.; Marschall, M. Functional Relevance of the Interaction between Human Cyclins and the Cytomegalovirus-Encoded CDK-Like Protein Kinase pUL97. Viruses 2021, 13, 1248. [Google Scholar] [CrossRef] [PubMed]
- Steingruber, M.; Keller, L.; Socher, E.; Ferre, S.; Hesse, A.M.; Couté, Y.; Hahn, F.; Büscher, N.; Plachter, B.; Sticht, H.; et al. Cyclins B1, T1, and H differ in their molecular mode of interaction with cytomegalovirus protein kinase pUL97. J. Biol. Chem. 2019, 294, 6188–6203. [Google Scholar] [CrossRef]
- McCluskey, S.M.; Siedner, M.J.; Marconi, V.C. Management of Virologic Failure and HIV Drug Resistance. Infect. Dis. Clin. North Am. 2019, 33, 707–742. [Google Scholar] [CrossRef] [PubMed]
- Cihlar, T.; Fordyce, M. Current status and prospects of HIV treatment. Curr. Opin. Virol. 2016, 18, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Alvira, D.; Mondragón, L. Drug Delivery Strategies of Chemical CDK Inhibitors. Methods Mol. Biol. 2016, 1336, 141–154. [Google Scholar] [CrossRef]
- Morgan, D.O. The Cell Cycle: Principles of Control; New Science Press Ltd.: London, UK, 2007. [Google Scholar]
- Fu, J.; Zhang, N.; Chou, J.H.; Dong, H.-J.; Lin, S.-F.; Ulrich-Merzenich, G.S.; Chou, T.-C. Drug combination in vivo using combination index method: Taxotere and T607 against colon carcinoma HCT-116 xenograft tumor in nude mice. Synergy 2016, 3, 15–30. [Google Scholar] [CrossRef]
- Marschall, M.; Niemann, I.; Kosulin, K.; Bootz, A.; Wagner, S.; Dobner, T.; Herz, T.; Kramer, B.; Leban, J.; Vitt, D.; et al. Assessment of drug candidates for broad-spectrum antiviral therapy targeting cellular pyrimidine biosynthesis. Antivir. Res. 2013, 100, 640–648. [Google Scholar] [CrossRef]
- Wild, M.; Bertzbach, L.D.; Tannig, P.; Wangen, C.; Müller, R.; Herrmann, L.; Fröhlich, T.; Tsogoeva, S.B.; Kaufer, B.B.; Marschall, M.; et al. The trimeric artesunate derivative TF27 exerts strong anti-cytomegaloviral efficacy: Focus on prophylactic efficacy and oral treatment of immunocompetent mice. Antivir. Res. 2020, 178, 104788. [Google Scholar] [CrossRef]
- Marschall, M.; Freitag, M.; Weiler, S.; Sorg, G.; Stamminger, T. Recombinant green fluorescent protein-expressing human cytomegalovirus as a tool for screening antiviral agents. Antimicrob. Agents Chemother. 2000, 44, 1588–1597. [Google Scholar] [CrossRef]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef]
- Hahn, F.; Hutterer, C.; Henry, C.; Hamilton, S.T.; Strojan, H.; Kraut, A.; Schulte, U.; Schütz, M.; Kohrt, S.; Wangen, C.; et al. Novel cytomegalovirus-inhibitory compounds of the class pyrrolopyridines show a complex pattern of target binding that suggests an unusual mechanism of antiviral activity. Antivir. Res. 2018, 159, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Prichard, M.N.; Shipman, C., Jr. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 1990, 14, 181–205. [Google Scholar] [CrossRef]
- Chou, T.C.; Martin, N. CompuSyn for Drug Combinations: PC Soft-Ware and User’s Guide: A Computer Program for Quantitation of Synergism and Antagonism in Drug Combinations, and the Determination of IC50 and ED50 and LD50 Values; Combo-Syn: Paramus, NJ, USA, 2005. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef]
| Target | Compound | EC50 a | CC50 b | SI c |
|---|---|---|---|---|
| vCDK/ pUL97 | maribavir (MBV) | 0.35 ± 0.42 | >100 | >250 |
| Gö6976 | 1.40 ± 1.99 | >100 | >70 | |
| Ax7396 | 1.93 ± 0.84 | >100 | >50 | |
| Vi7392 | 2.67 ± 0.36 | 57.8 ± 13.9 | 22 | |
| CDK2 | CDK2 Inh II | 6.33 ± 2.80 | >100 | >16 |
| CDK7 | LDC4297 | 0.009 ± 0.002 | >10 | >1000 |
| CDK8 | SEL120 | 0.079 ± 0.001 | 5.90 ± 3.85 | 75 |
| CDK9 | THAL-SNS032 | 0.025 ± 0.002 | 0.14 ± 0.06 | 6 |
| CDK2 | CVT-313 | nd | 5.57 ± 0.05 | nd |
| CDK7 | SY1365 | nd | <0.1 | nd |
| CDK7 | samuraciclib | nd | 0.50 ± 0.21 | nd |
| CDK9 | AZD4573 | nd | 0.35 ± 0.09 | nd |
| CDK1/2/5/9 | dinaciclib | nd | <0.1 | nd |
| CDK1/4/9 | riviciclib | nd | 1.6 ± 0.06 | nd |
| CDK2/9 | CYC065 | nd | <0.1 | nd |
| Compound A | Compound B | Ratio | EC50 a | CC50 b | SI c | |
|---|---|---|---|---|---|---|
| MBV | LDC4297 | 100:1 | 0.06 ± 0.08 | >50 | >800 | |
| Vi7392 | LDC4297 | 100:1 | 0.51 ± 0.44 | >50 | >90 | |
| Gö6976 | LDC4297 | 100:1 | 0.23 ± 0.20 | 19.4 ± 7.7 | 84 | |
| Ax7396 | LDC4297 | 100:1 | 0.58 ± 0.13 | 25.8 ± 2.7 | 44 | |
| SEL120 | LDC4297 | 10:1 | 0.04 ± 0.02 | 0.14 ± 0.09 | 4 | |
| MBV | THAL-SNS032 | 100:1 | 0.26 ± 0.16 | 11.2 ± 0.90 | 43 | |
| SEL120 | THAL-SNS032 | 50:1 | 0.19 ± 0.12 | 0.47 ± 0.04 | 2 | |
| MBV | CDK2 Inh II | 1:1 | 0.01 ± 0.00 | >50 | >5000 | |
| LDC4297 | CDK2 Inh II | 1:100 | 3.3 ± 2.7 | >50 | >15 | |
| SEL120 | CDK2 Inh II | 1:10 | 0.10 ± 0.06 | >50 | >500 | |
| MBV | SEL120 | 10:1 | 0.15 ± 0.07 | 7.7 ± 1.2 | 51 | |
| Compound A | Compound B | Compound C | Ratio | EC50 a | CC50 b | SI c |
| MBV | LDC4297 | SEL120 | 100:1:10 | 0.18 ± 0.01 | 48.7 ± 0.71 | 270 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wild, M.; Hahn, F.; Brückner, N.; Schütz, M.; Wangen, C.; Wagner, S.; Sommerer, M.; Strobl, S.; Marschall, M. Cyclin-Dependent Kinases (CDKs) and the Human Cytomegalovirus-Encoded CDK Ortholog pUL97 Represent Highly Attractive Targets for Synergistic Drug Combinations. Int. J. Mol. Sci. 2022, 23, 2493. https://doi.org/10.3390/ijms23052493
Wild M, Hahn F, Brückner N, Schütz M, Wangen C, Wagner S, Sommerer M, Strobl S, Marschall M. Cyclin-Dependent Kinases (CDKs) and the Human Cytomegalovirus-Encoded CDK Ortholog pUL97 Represent Highly Attractive Targets for Synergistic Drug Combinations. International Journal of Molecular Sciences. 2022; 23(5):2493. https://doi.org/10.3390/ijms23052493
Chicago/Turabian StyleWild, Markus, Friedrich Hahn, Nadine Brückner, Martin Schütz, Christina Wangen, Sabrina Wagner, Mona Sommerer, Stefan Strobl, and Manfred Marschall. 2022. "Cyclin-Dependent Kinases (CDKs) and the Human Cytomegalovirus-Encoded CDK Ortholog pUL97 Represent Highly Attractive Targets for Synergistic Drug Combinations" International Journal of Molecular Sciences 23, no. 5: 2493. https://doi.org/10.3390/ijms23052493
APA StyleWild, M., Hahn, F., Brückner, N., Schütz, M., Wangen, C., Wagner, S., Sommerer, M., Strobl, S., & Marschall, M. (2022). Cyclin-Dependent Kinases (CDKs) and the Human Cytomegalovirus-Encoded CDK Ortholog pUL97 Represent Highly Attractive Targets for Synergistic Drug Combinations. International Journal of Molecular Sciences, 23(5), 2493. https://doi.org/10.3390/ijms23052493






