A Genome-Wide Screen Reveals That Endocytic Genes Are Important for Pma1p Asymmetry during Cell Division in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Results
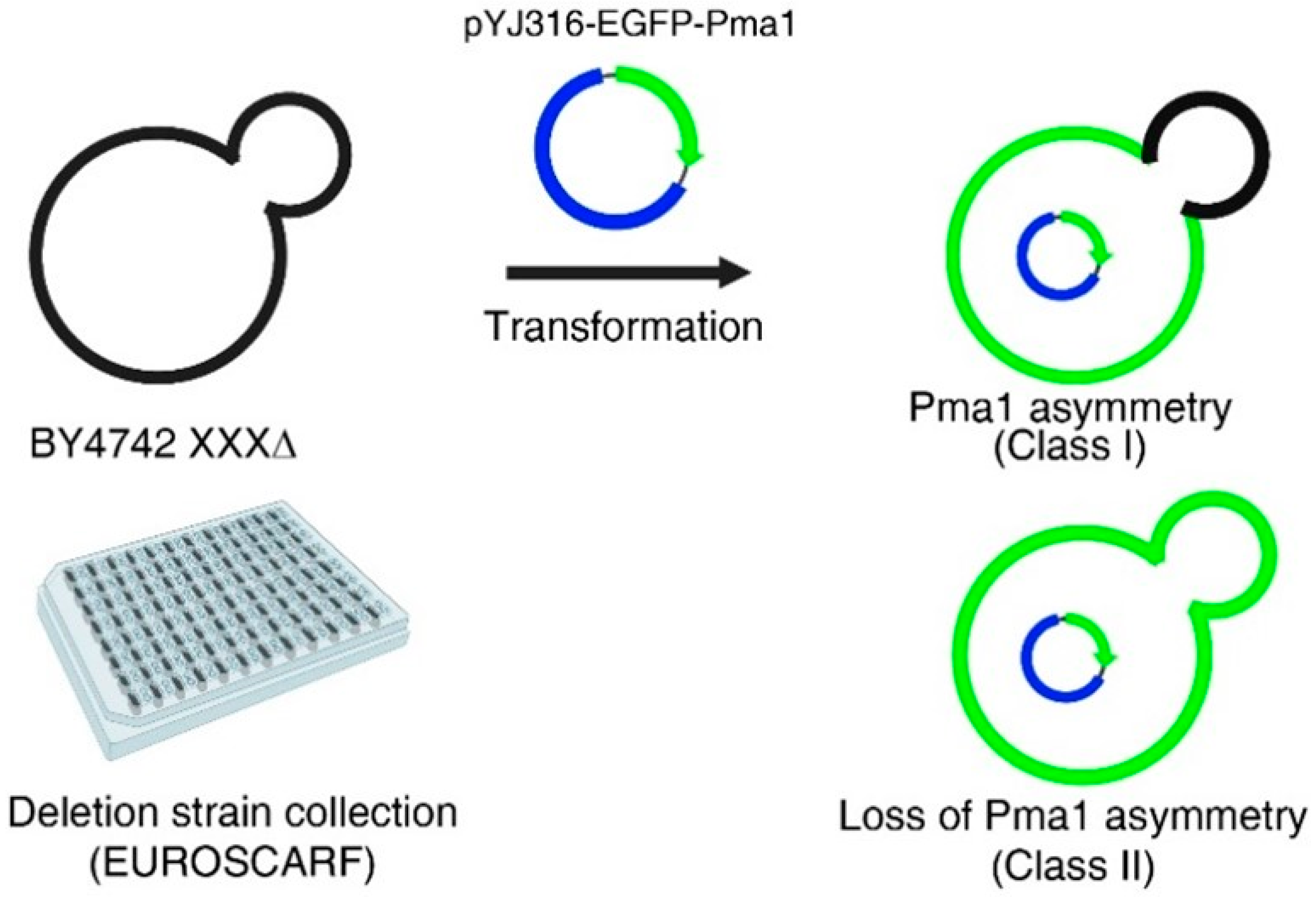
2.1. A Genome-Wide Screen to Identify Genes That Regulate the Asymmetric Distribution of Pma1p between Mother and Daughter Cells during Cell Division
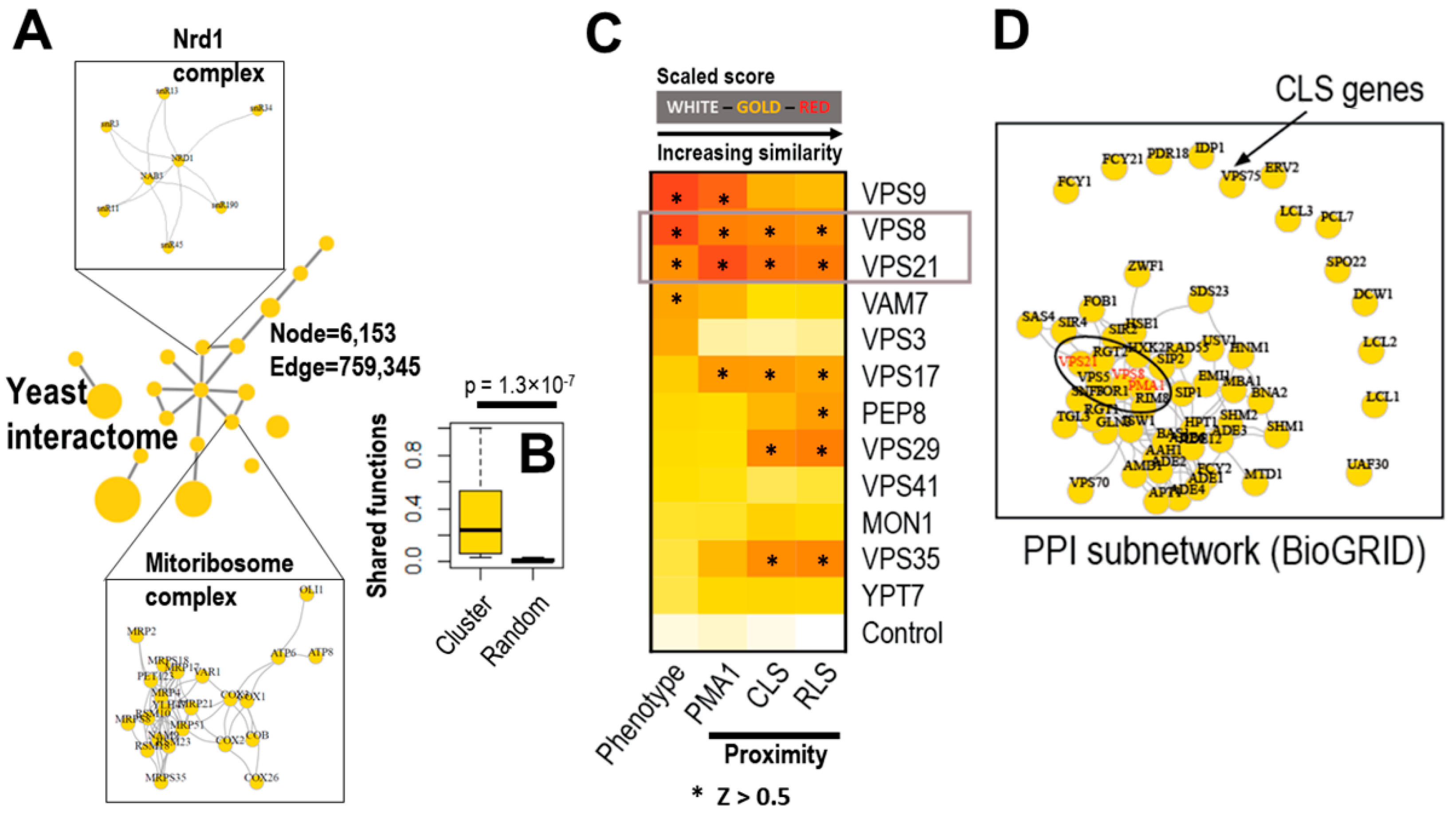
2.2. Endocytic Genes Seem to Be Critical for the Asymmetric Distribution of Plasma Membrane LARPs but Not of Cytoplasmic LARPs
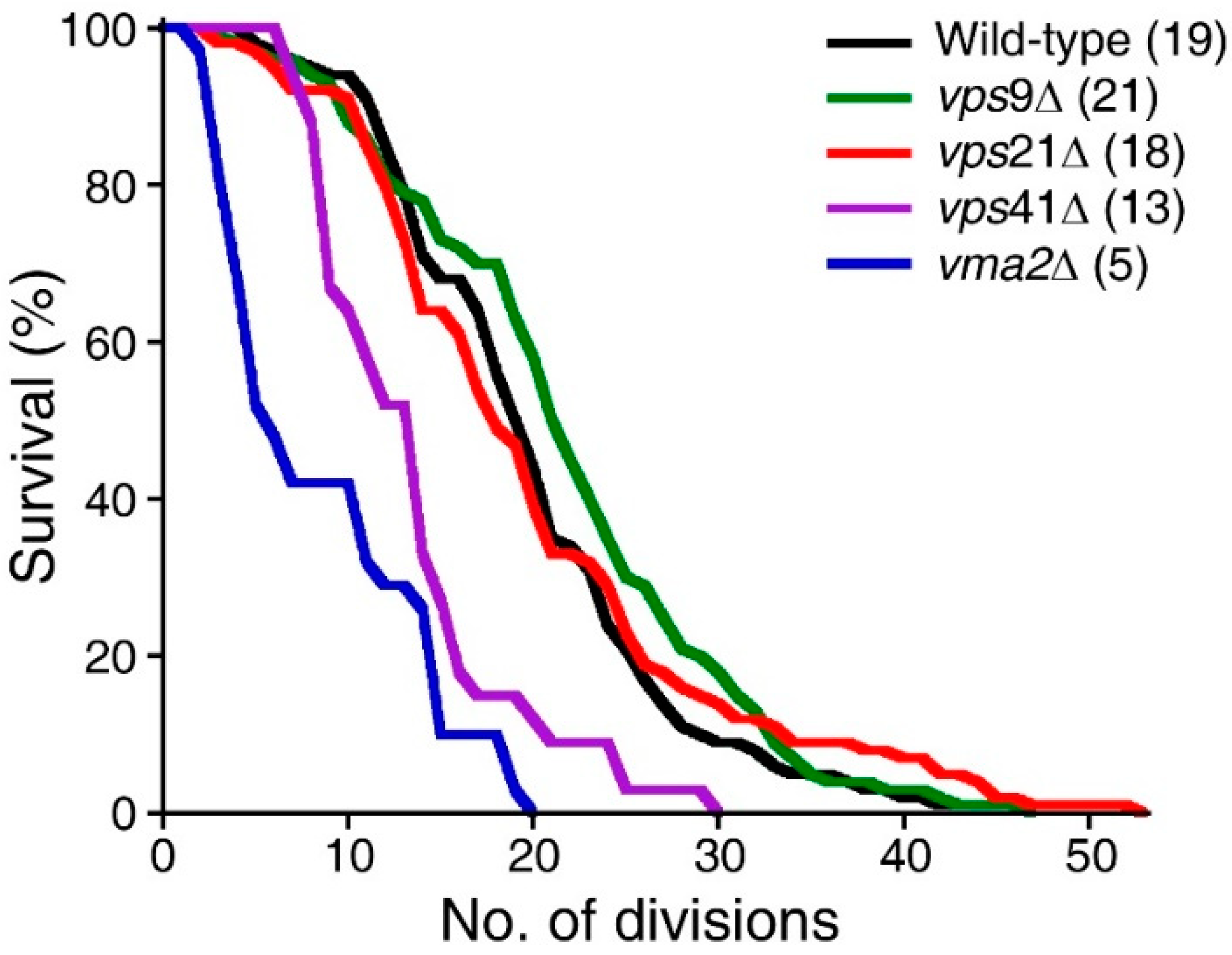
2.3. Disruption of the Asymmetric Distribution of Pma1p during Cell Division Correlates Little with RLS
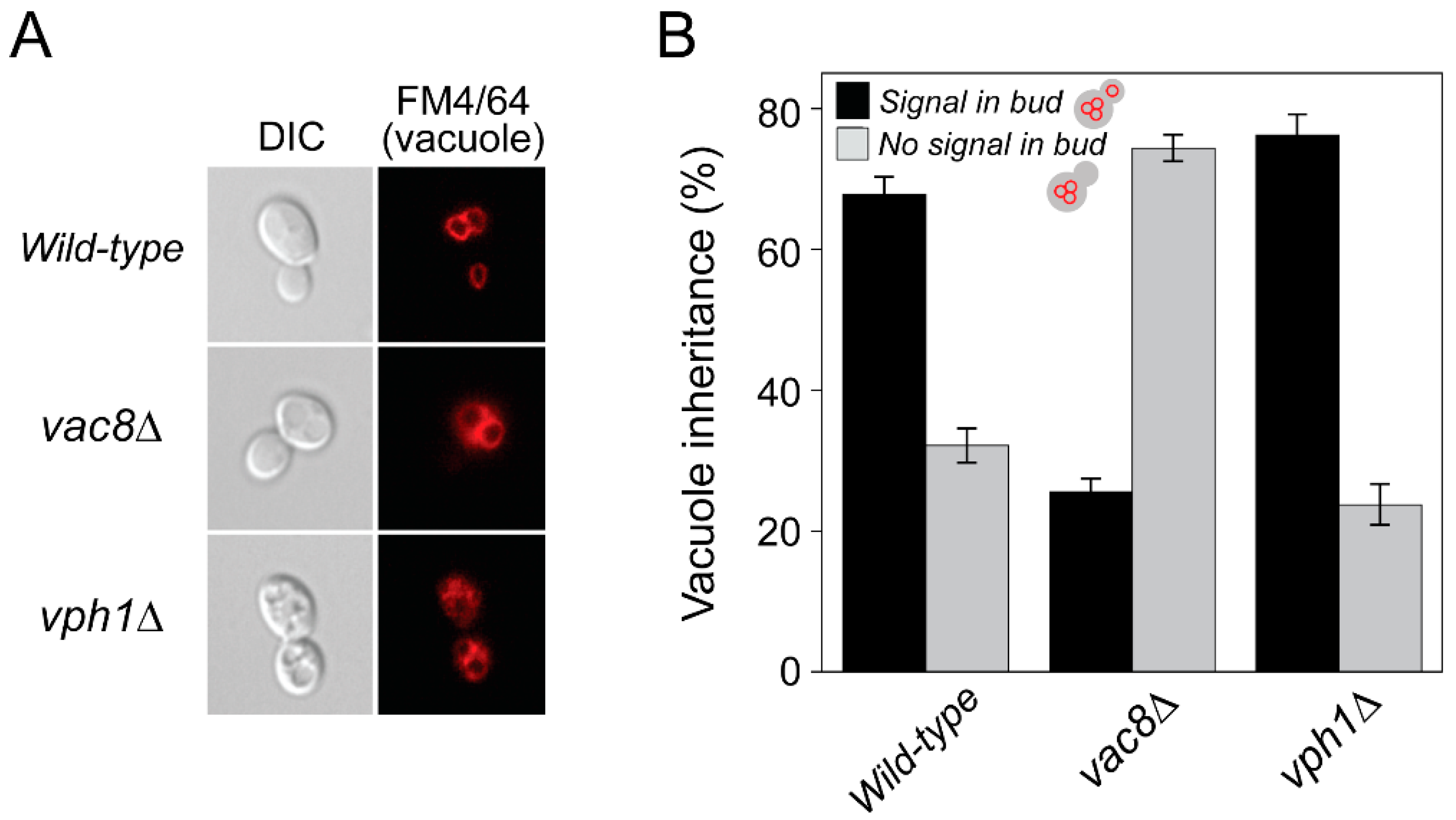
2.4. Damaged Vacuoles Are Successfully Transported to Daughter Cells during Cell Division
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Plasmids
4.2. Yeast Transformation
4.3. Microscopy
4.4. Measurement of RLS Using a Tetrad Dissection Microscope Equipped with a Micromanipulator
4.5. Vacuole Staining with FM4/64
4.6. Protein Interaction Network Construction and Proximity Analysis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [Green Version]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Steinkraus, K.A.; Kaeberlein, M.; Kennedy, B.K. Replicative aging in yeast: The means to the end. Annu. Rev. Cell Dev. Biol. 2008, 24, 29–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFaline-Figueroa, J.R.; Vevea, J.; Swayne, T.C.; Zhou, C.; Liu, C.; Leung, G.; Boldogh, I.R.; Pon, L.A. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 2011, 10, 885–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spokoini, R.; Moldavski, O.; Nahmias, Y.; England, J.L.; Schuldiner, M.; Kaganovich, D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012, 2, 738–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Slaughter, B.D.; Unruh, J.R.; Guo, F.; Yu, Z.; Mickey, K.; Narkar, A.; Ross, R.T.; McClain, M.; Li, R. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell 2014, 159, 530–542. [Google Scholar] [CrossRef] [Green Version]
- Henderson, K.A.; Hughes, A.L.; Gottschling, D.E. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. eLife 2014, 3, e03504. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Gottschling, D.E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 2012, 492, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Klecker, T.; Westermann, B. Asymmetric inheritance of mitochondria in yeast. Biol. Chem. 2020, 401, 779–791. [Google Scholar] [CrossRef]
- Ferreira, T.; Mason, A.B.; Slayman, C.W. The yeast Pma1 proton pump: A model for understanding the biogenesis of plasma membrane proteins. J. Biol. Chem. 2001, 276, 29613–29616. [Google Scholar] [CrossRef] [Green Version]
- Thayer, N.H.; Leverich, C.K.; Fitzgibbon, M.P.; Nelson, Z.W.; Henderson, K.A.; Gafken, P.R.; Hsu, J.J.; Gottschling, D.E. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc. Natl. Acad. Sci. USA 2014, 111, 14019–14026. [Google Scholar] [CrossRef] [Green Version]
- Balderhaar, H.J.; Lachmann, J.; Yavavli, E.; Brocker, C.; Lurick, A.; Ungermann, C. The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes. Proc. Natl. Acad. Sci. USA 2013, 110, 3823–3828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balderhaar, H.J.; Ungermann, C. CORVET and HOPS tethering complexes—Coordinators of endosome and lysosome fusion. J. Cell Sci. 2013, 126 Pt 6, 1307–1316. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, M.; Arlt, H.; Epp, N.; Lachmann, J.; Griffith, J.; Perz, A.; Reggiori, F.; Ungermann, C. Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J. Biol. Chem. 2013, 288, 5166–5175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epp, N.; Ungermann, C. The N-terminal domains of Vps3 and Vps8 are critical for localization and function of the CORVET tethering complex on endosomes. PLoS ONE 2013, 8, e67307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.; Johnston, W.; Kovtun, O.; Mureev, S.; Brocker, C.; Ungermann, C.; Alexandrov, K. Subunit organisation of in vitro reconstituted HOPS and CORVET multisubunit membrane tethering complexes. PLoS ONE 2013, 8, e81534. [Google Scholar] [CrossRef] [PubMed]
- Markgraf, D.F.; Ahnert, F.; Arlt, H.; Mari, M.; Peplowska, K.; Epp, N.; Griffith, J.; Reggiori, F.; Ungermann, C. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol. Biol. Cell 2009, 20, 5276–5289. [Google Scholar] [CrossRef] [Green Version]
- Solinger, J.A.; Spang, A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013, 280, 2743–2757. [Google Scholar] [CrossRef]
- Noree, C.; Sato, B.K.; Broyer, R.M.; Wilhelm, J.E. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 2010, 190, 541–551. [Google Scholar] [CrossRef] [Green Version]
- Higuchi, R.; Vevea, J.D.; Swayne, T.C.; Chojnowski, R.; Hill, V.; Boldogh, I.R.; Pon, L.A. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr. Biol. 2013, 23, 2417–2422. [Google Scholar] [CrossRef] [Green Version]
- Pernice, W.M.; Vevea, J.D.; Pon, L.A. A role for Mfb1p in region-specific anchorage of high-functioning mitochondria and lifespan in Saccharomyces cerevisiae. Nat. Commun. 2016, 7, 10595. [Google Scholar] [CrossRef] [Green Version]
- Vida, T.A.; Emr, S.D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995, 128, 779–792. [Google Scholar] [CrossRef]
- Wang, Y.X.; Catlett, N.L.; Weisman, L.S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 1998, 140, 1063–1074. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.X.; Zhao, H.; Harding, T.M.; de Mesquita, D.S.G.; Woldringh, C.L.; Klionsky, D.J.; Munn, A.L.; Weisman, L.S. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol. Biol. Cell 1996, 7, 1375–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manolson, M.F.; Proteau, D.; Preston, R.A.; Stenbit, A.; Roberts, B.T.; Hoyt, M.A.; Preuss, D.; Mulholland, J.; Botstein, D.; Jones, E.W. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 1992, 267, 14294–14303. [Google Scholar] [CrossRef]
- Bagnat, M.; Chang, A.; Simons, K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 2001, 12, 4129–4138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabrizio, P.; Hoon, S.; Shamalnasab, M.; Galbani, A.; Wei, M.; Giaever, G.; Nislow, C.; Longo, V.D. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010, 6, e1001024. [Google Scholar] [CrossRef] [Green Version]
- Matecic, M.; Smith, D.L.; Pan, X.; Maqani, N.; Bekiranov, S.; Boeke, J.D.; Smith, J.S. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010, 6, e1000921. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.D.; Tsuchiya, M.; Fox, L.A.; Dang, N.; Hu, D.; Kerr, E.O.; Johnston, E.D.; Tchao, B.N.; Pak, D.N.; Welton, K.L.; et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008, 18, 564–570. [Google Scholar] [CrossRef] [Green Version]
- Tang, F.; Watkins, J.W.; Bermudez, M.; Gray, R.; Gaban, A.; Portie, K.; Grace, S.; Kleve, M.; Craciun, G. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy 2008, 4, 874–886. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Kim, D.U.; Hayles, J.; Kim, D.; Wood, V.; Park, H.O.; Won, M.; Yoo, H.S.; Duhig, T.; Nam, M.; Palmer, G.; et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010, 28, 617–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Echevarria, E.; Gonzalez-Polo, N.; Zorrilla, S.; Martinez-Lumbreras, S.; Santiveri, C.M.; Campos-Olivas, R.; Sanchez, M.; Calvo, O.; Gonzalez, B.; Perez-Canadillas, J.M. The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition. Nucleic Acids Res. 2017, 45, 10293–10305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCormick, M.A.; Delaney, J.R.; Tsuchiya, M.; Tsuchiyama, S.; Shemorry, A.; Sim, S.; Chou, A.C.; Ahmed, U.; Carr, D.; Murakami, C.J.; et al. A Comprehensive Analysis of Replicative Lifespan in 4698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metab. 2015, 22, 895–906. [Google Scholar] [CrossRef] [Green Version]
- Starai, V.J.; Jun, Y.; Wickner, W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc. Natl. Acad. Sci. USA 2007, 104, 13551–13558. [Google Scholar] [CrossRef] [Green Version]


| Strain | Percentage of Cells with the Class II Phenotype | Gene Name | Known Functions |
|---|---|---|---|
| Wild-type BY4742 | 30 | ||
| BY4742 vps9Δ | 89 | VPS9 | A guanine nucleotide-exchange factor (GEF) that functions in vacuolar protein transport by stimulating Vps21p |
| BY4742 vps8Δ | 88 | VPS8 | A component of the CORVET complex involved in endosomal vesicle tethering and fusion in the endosome-to-vacuole protein targeting pathway |
| BY4742 vps21Δ | 76 | VPS21 | An endosomal Rab family GTPase required for endocytic transport and sorting of vacuolar hydrolases and endosomal localization of the CORVET complex |
| BY4742 vam7Δ | 72 | VAM7 | A vacuolar SNARE protein that functions with Vam3p and Vti1p in vacuolar protein trafficking |
| BY4742 vps3Δ | 71 | VPS3 | A component of the CORVET membrane tethering complex |
| BY4742 vps17Δ | 65 | VPS17 | A subunit of the retromer complex essential for endosome-to-Golgi retrograde transport |
| BY4742 vps26Δ | 62 | VPS26 | A vacuolar component of the retromer complex |
| BY4742 vps29Δ | 58 | VPS29 | A subunit of the retromer complex essential for endosome-to-Golgi retrograde transport |
| BY4742 vps41Δ | 56 | VPS41 | A subunit of the HOPS endocytic tethering complex involved in vacuolar membrane fusion |
| BY4742 mon1Δ | 53 | MON1 | A subunit of the heterodimeric Mon1p-Ccz1p GEF complex that stimulates activation of Ypt7p |
| BY4742 vps35Δ | 51 | VPS35 | An endosomal subunit of the retromer complex required for retrograde transport |
| BY4742 ypt7Δ | 50 | YPT7 | A Rab family GTPase required for vacuole fusion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, S.-Y.; Jang, E.; Ko, N.; Kim, M.; Kim, S.Y.; Moon, Y.; Nam, J.-S.; Lee, S.; Jun, Y. A Genome-Wide Screen Reveals That Endocytic Genes Are Important for Pma1p Asymmetry during Cell Division in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 2364. https://doi.org/10.3390/ijms23042364
Yoon S-Y, Jang E, Ko N, Kim M, Kim SY, Moon Y, Nam J-S, Lee S, Jun Y. A Genome-Wide Screen Reveals That Endocytic Genes Are Important for Pma1p Asymmetry during Cell Division in Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2022; 23(4):2364. https://doi.org/10.3390/ijms23042364
Chicago/Turabian StyleYoon, So-Young, Eunhong Jang, Naho Ko, Minseok Kim, Su Yoon Kim, Yeojin Moon, Jeong-Seok Nam, Sunjae Lee, and Youngsoo Jun. 2022. "A Genome-Wide Screen Reveals That Endocytic Genes Are Important for Pma1p Asymmetry during Cell Division in Saccharomyces cerevisiae" International Journal of Molecular Sciences 23, no. 4: 2364. https://doi.org/10.3390/ijms23042364
APA StyleYoon, S.-Y., Jang, E., Ko, N., Kim, M., Kim, S. Y., Moon, Y., Nam, J.-S., Lee, S., & Jun, Y. (2022). A Genome-Wide Screen Reveals That Endocytic Genes Are Important for Pma1p Asymmetry during Cell Division in Saccharomyces cerevisiae. International Journal of Molecular Sciences, 23(4), 2364. https://doi.org/10.3390/ijms23042364








