A Transcriptomic Analysis of T98G Human Glioblastoma Cells after Exposure to Cadmium-Selenium Quantum Dots Mainly Reveals Alterations in Neuroinflammation Processes and Hypothalamus Regulation
Abstract
1. Introduction
2. Results
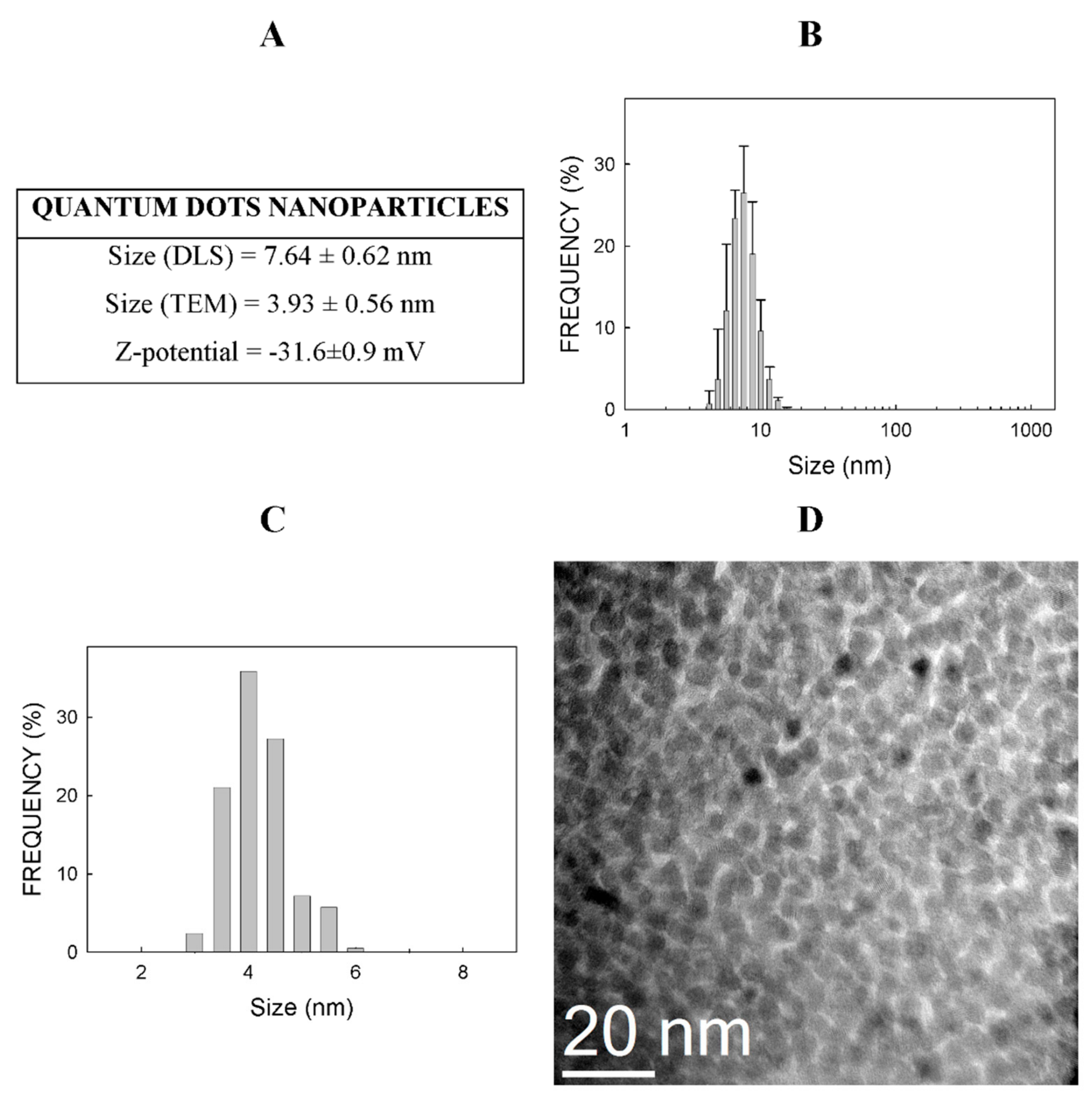
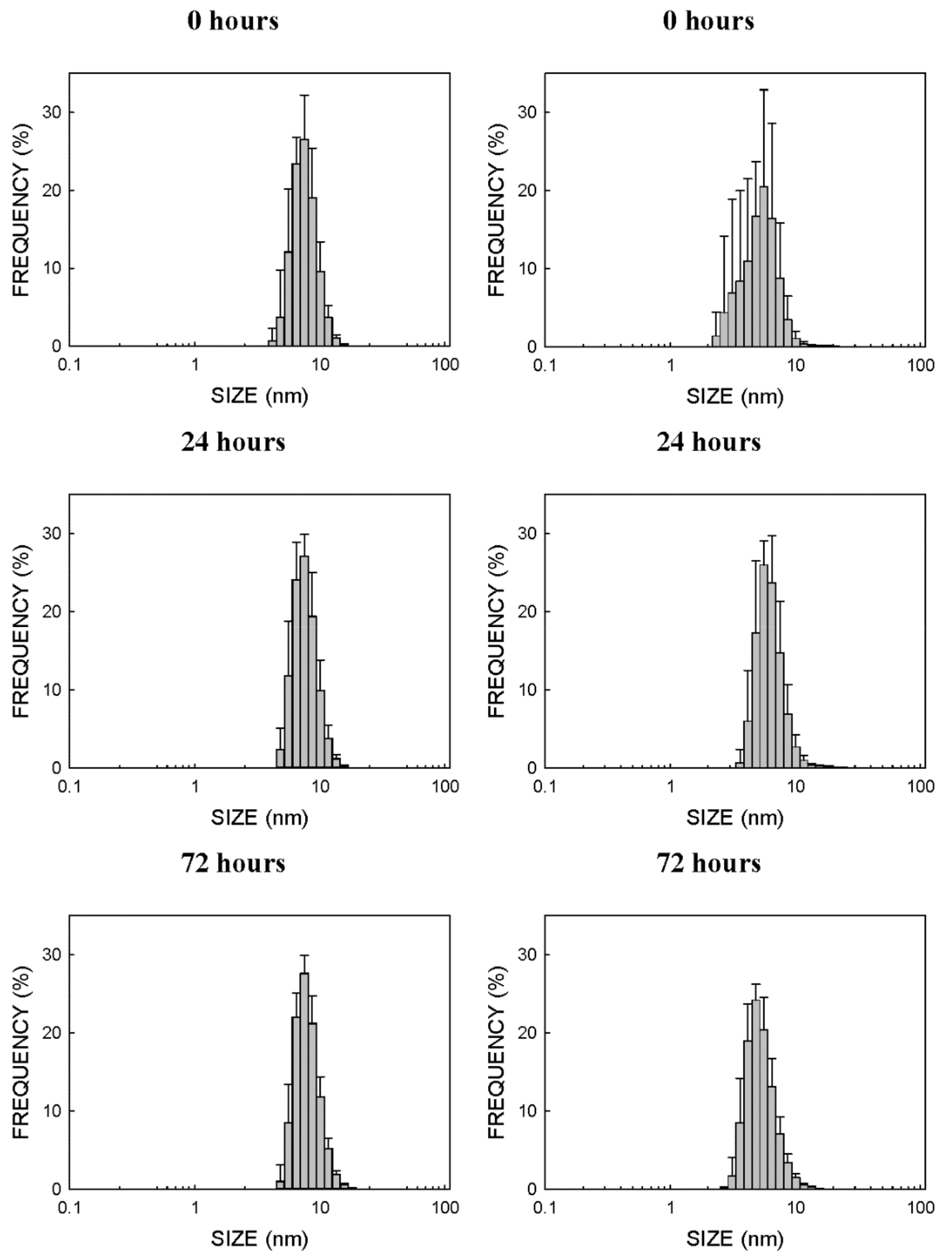
2.1. Physico-Chemical Properties of CdSe-QDs
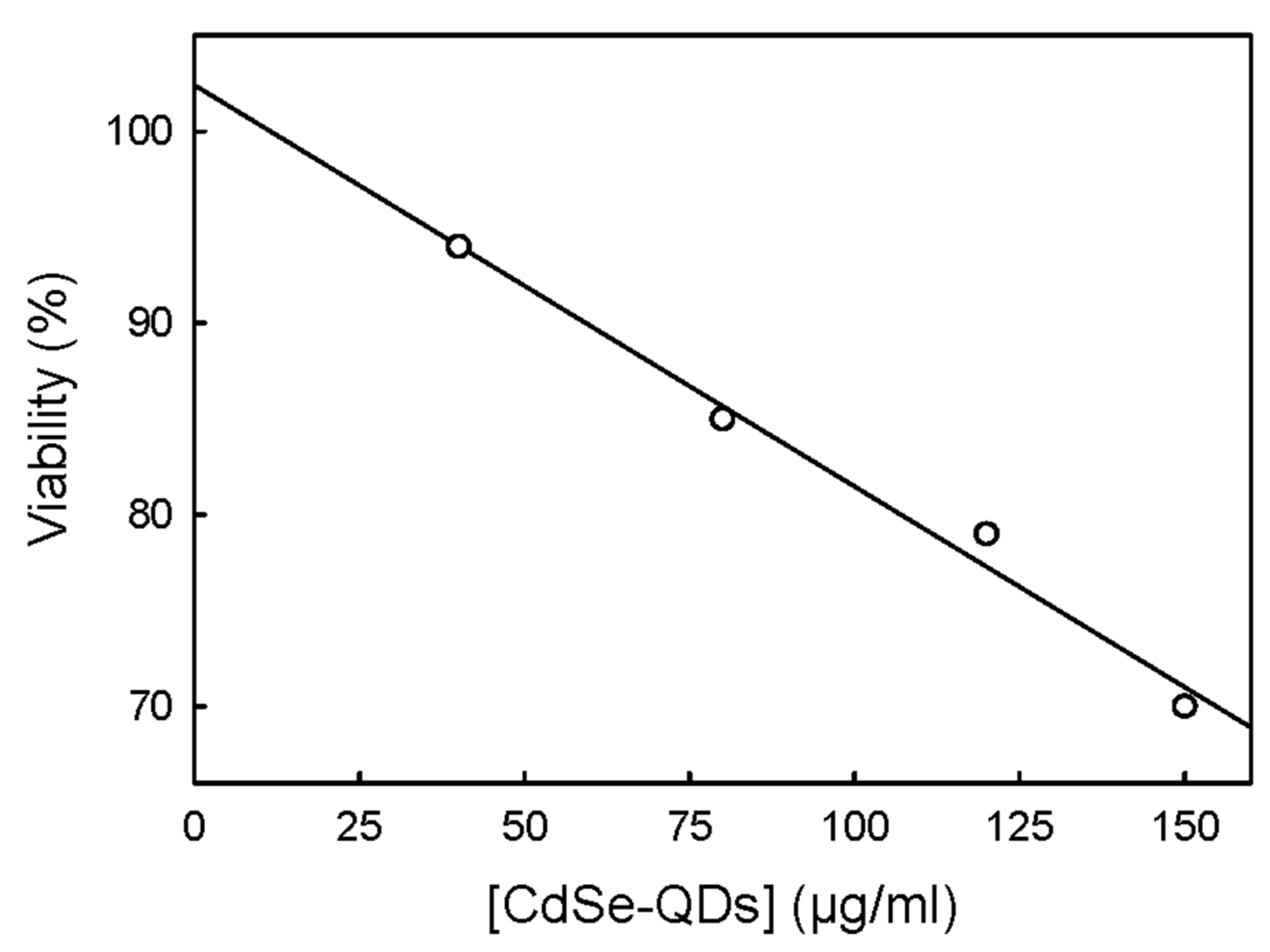
2.2. Effects of CdSe-QDs on T98G Cell Viability
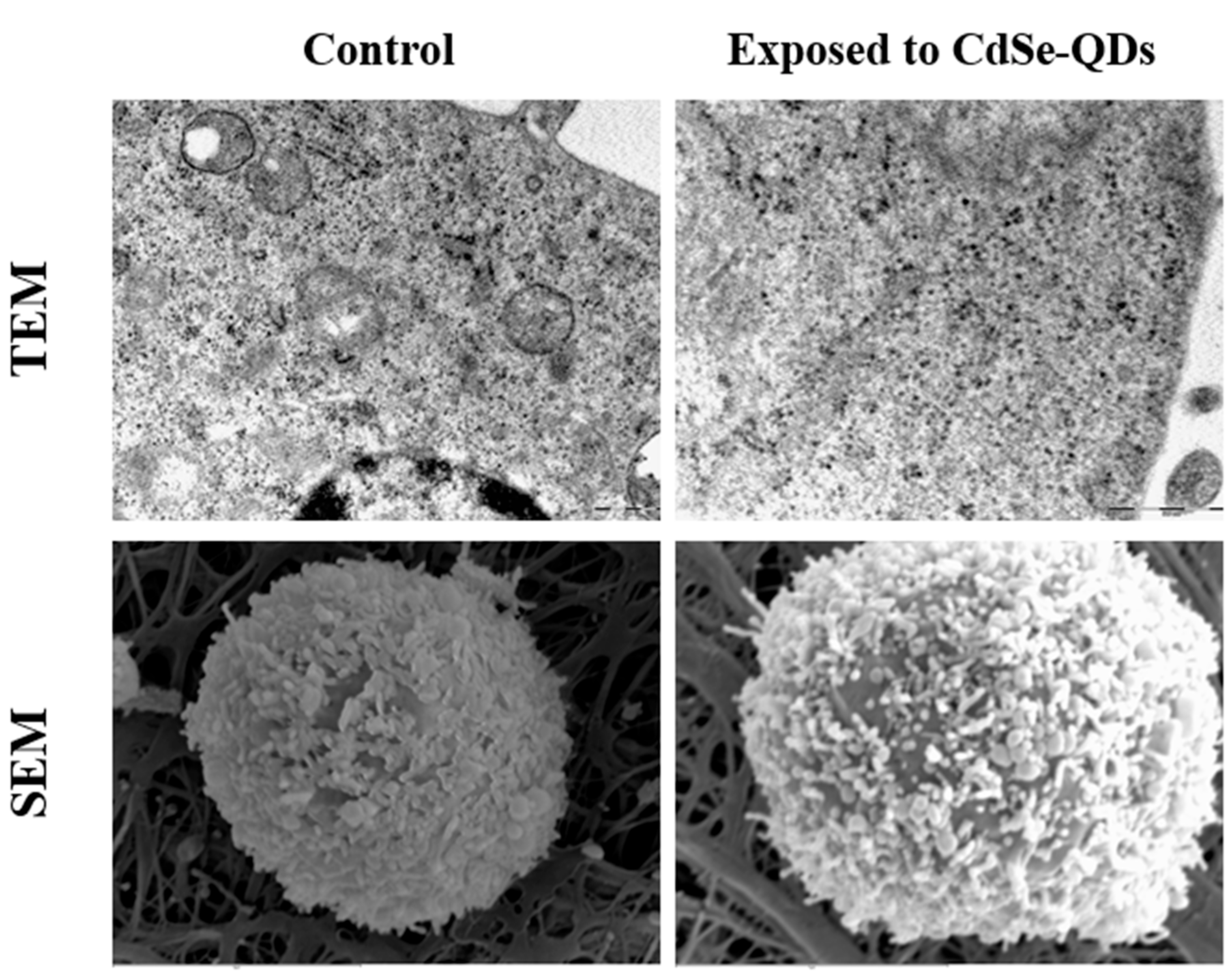
2.3. Microscopy
2.4. Massively Parallel Sequencing of the RNA of the Cells Exposed to CdSe-QDs
2.4.1. RNA Quality
2.4.2. Alignment of Reads to the Reference Genome
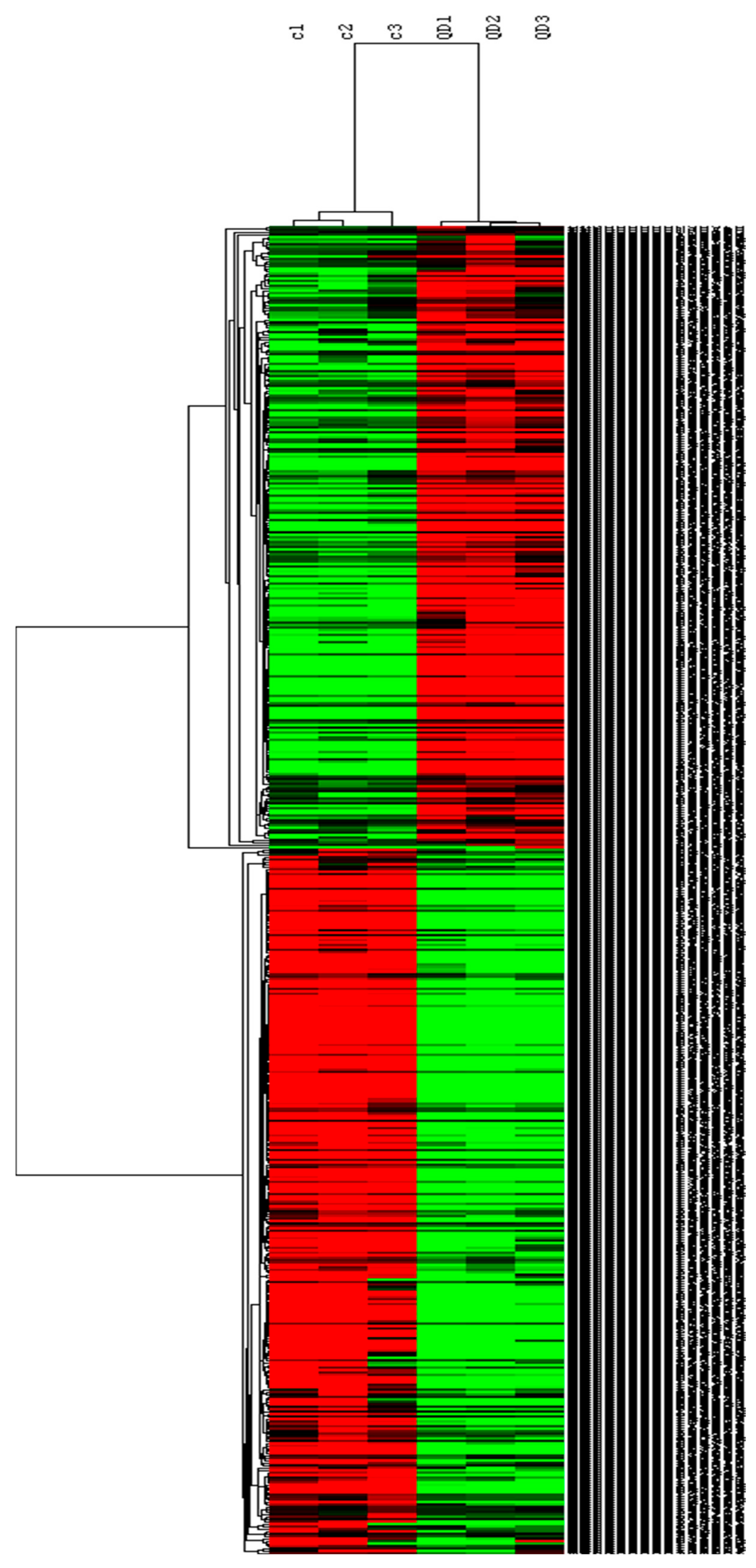
2.4.3. Differentially Expressed Genes
2.4.4. Annotation of Differentially Expressed Genes
2.4.5. Altered Molecular Pathways
3. Discussion
3.1. Physico-Chemical Properties of Nanoparticles
3.2. Cytotoxicity
3.3. Cellular Uptake
3.4. Altered Molecular Pathways and Biological Processes
3.4.1. Gonadotropin-Releasing Hormone Receptor Pathway
3.4.2. Inflammation and Immune System Alterations
3.5. Risk
4. Materials and Methods
4.1. Nanoparticles
4.2. Physico-Chemical Characterisation of QD-NPs
4.3. Cellular Cultures
4.4. Cellular Exposure to CdSe-QDs
4.5. Cellular Viability Tests
4.6. Electronic Microscopy
4.7. Massively Parallel RNA Sequencing
4.8. Bioinformatic Analysis
4.9. Ontological Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Union. Commission Recommendation of 18 October 2011 on the Definition of Nanomaterial. 2011. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2011:275:0038:0040:ES:PDF (accessed on 27 January 2022).
- Mo, D.; Hu, L.; Zeng, G.; Chen, G.; Wan, J.; Yu, Z.; Huang, Z.; He, K.; Zhang, C.; Cheng, M. Cadmium-containing quantum dots: Properties, applications, and toxicity. Appl. Microbiol. Biotechnol. 2017, 101, 2713–2733. [Google Scholar] [CrossRef] [PubMed]
- Matea, C.T.; Mocan, T.; Tabaran, F.; Pop, T.; Mosteanu, O.; Puia, C.; Iancu, C.; Mocan, L. Quantum dots in imaging, drug delivery and sensor applications. Int. J. Nanomed. 2017, 12, 5421–5431. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Shim, Y.; Myong Song, J. Quantum dot as probe for disease diagnosis and monitoring. Biotechnol. J. 2016, 11, 31–42. [Google Scholar] [CrossRef] [PubMed]
- European Union Observatory for Nanoparticles. Available online: https://euon.echa.europa.eu/search-for-nanomaterials (accessed on 27 January 2022).
- Chang, X.; Li, J.; Niu, S.; Xue, Y.; Tang, M. Neurotoxicity of metal-containing nanoparticles and implications in glial cells. J. Appl. Toxicol. 2021, 41, 65–81. [Google Scholar] [CrossRef]
- Fuster, E.; Candela, H.; Estévez, J.; Arias, A.J.; Vilanova, E.; Sogorb, M.A. Effects of silver nanoparticles on T98G human glioblastoma cells. Toxicol. Appl. Pharmacol. 2020, 404, 115178. [Google Scholar] [CrossRef]
- Fuster, E.; Candela, H.; Estévez, J.; Vilanova, E.; Sogorb, M.A. Titanium Dioxide, but Not Zinc Oxide, Nanoparticles Cause Severe Transcriptomic Alterations in T98G Human Glioblastoma Cells. Int. J. Mol. Sci. 2021, 22, 2084. [Google Scholar] [CrossRef]
- Panossian, A.; Seo, E.J.; Efferth, T. Effects of anti-inflammatory and adaptogenic herbal extracts on gene expression of eicosanoids signaling pathways in isolated brain cells. Phytomedicine 2019, 60, 152881. [Google Scholar] [CrossRef]
- Uniprot Database Accession Number P30414: NK-Tumor Recognition Protein. Available online: https://www.uniprot.org/uniprot/P30414 (accessed on 27 January 2022).
- Uniprot Database Accession Number P25103: Substance-P Receptor. Available online: https://www.uniprot.org/uniprot/P25103 (accessed on 27 January 2022).
- Uniprot Database Accession Number P18146: Early Growth Response Protein 1. Available online: https://www.uniprot.org/uniprot/P18146 (accessed on 27 January 2022).
- Uniprot Database Accession Number P01100: Proto-Oncogene c-Fos. Available online: https://www.uniprot.org/uniprot/P01100 (accessed on 27 January 2022).
- National Academy of Sciences. Committee on Toxicity Testing and Assessment of Environmental Agents, National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy. 2007. Available online: http://nap.edu/11970 (accessed on 27 January 2022).
- Wu, T.; Liang, X.; He, K.; Wei, T.; Wang, Y.; Zou, L.; Lu, J.; Yao, Y.; Liu, N.; Zhang, T.; et al. Transcriptome analysis of different sizes of 3-mercaptopropionic acid-modified cadmium telluride quantum dot-induced toxic effects reveals immune response in rat hippocampus. J. Appl. Toxicol. 2018, 38, 1177–1194. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, X.; Wu, Q.; Li, Y.; Wang, D. Translocation and neurotoxicity of CdTe quantum dots in RMEs motor neurons in nematode Caenorhabditis elegans. J. Hazard Mater. 2015, 283, 480–489. [Google Scholar] [CrossRef]
- Yang, Z.; Zou, W.; Pan, Y.; Yong, K.T.; Li, L.; Wang, X.; Liu, D.; Chen, T.; Xue, D.; Lin, G. PEGylated CuInS2/ZnS quantum dots inhibit neurite outgrowth by downregulating the NGF/p75NTR/MAPK pathway. Ecotoxicol. Environ. Saf. 2021, 207, 111378. [Google Scholar] [CrossRef]
- Huang, X.; Tang, M. Research advance on cell imaging and cytotoxicity of different types of quantum Dots. J. Appl. Toxicol. 2021, 41, 342–361. [Google Scholar] [CrossRef]
- He, K.; Liang, X.; Wei, T.; Liu, N.; Wang, Y.; Zou, L.; Bai, C.; Yao, Y.; Wu, T.; Kong, L.; et al. A metabolomics study: CdTe/ZnS quantum dots induce polarization in mice microglia. Chemosphere 2020, 246, 125629. [Google Scholar] [CrossRef] [PubMed]
- Turovsky, E.A.; Mal’tseva, V.N.; Sarimov, R.M.; Simakin, A.V.; Gudkov, S.V.; Plotnikov, E.Y. Features of the cytoprotective effect of selenium nanoparticles on primary cortical neurons and astrocytes during oxygen-glucose deprivation and reoxygenation. Sci. Rep. 2022, 12, 1710. [Google Scholar] [CrossRef] [PubMed]
- Sharif, A.; Baroncini, M.; Prevot, V. Role of glia in the regulation of gonadotropin-releasing hormone neuronal activity and secretion. Neuroendocrinology 2013, 98, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Smedlund, K.B.; Hill, J.W. The role of non-neuronal cells in hypogonadotropic hypogonadism. Mol. Cell. Endocrinol. 2020, 518, 110996. [Google Scholar] [CrossRef]
- Garcia-Segura, L.M.; Lorenz, B.; DonCarlos, L.L. The role of glia in the hypothalamus: Implications for gonadal steroid feedback and reproductive neuroendocrine output. Reproduction 2008, 135, 419–429. [Google Scholar] [CrossRef][Green Version]
- Rosin, J.M.; Kurrasch, D.M. Emerging roles for hypothalamic microglia as regulators of physiological homeostasis. Front. Neuroendocrinol. 2019, 54, 100748. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, S.; Chen, H.; He, M.; Deng, Y.; Cao, Z.; Pi, H.; Chen, C.; Li, M.; Ma, Q.; et al. CdSe/ZnS quantum dots induce hepatocyte pyroptosis and liver inflammation via NLRP3 inflammasome activation. Biomaterials 2016, 90, 27–39. [Google Scholar] [CrossRef]
- He, K.; Liang, X.; Wei, T.; Liu, N.; Wang, Y.; Zou, L.; Lu, J.; Yao, Y.; Kong, L.; Zhang, T.; et al. DNA damage in BV-2 cells: An important supplement to the neurotoxicity of CdTe quantum dots. J. Appl. Toxicol. 2019, 39, 525–539. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, T.; Chen, Y.; Tang, M. Research advances on potential neurotoxicity of quantum dots. J. Appl. Toxicol. 2016, 36, 345–351. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, M. Review of in vitro toxicological research of quantum dot and potentially involved mechanisms. Sci. Total Environ. 2018, 625, 940–962. [Google Scholar] [CrossRef] [PubMed]
- Terron, A.; Bal-Price, A.; Paini, A.; Monnet-Tschudi, F.; Bennekou, S.H.; EFSA WG EPI1 Members; Leist, M.; Schildknecht, S. An adverse outcome pathway for parkinsonian motor deficits associated with mitochondrial complex I inhibition. Arch. Toxicol. 2018, 92, 41–82. [Google Scholar] [CrossRef] [PubMed]
- Tosic, J.; Stanojevic, Z.; Vidicevic, S.; Isakovic, A.; Ciric, D.; Martinovic, T.; Kravic-Stevovic, T.; Bumbasirevic, V.; Paunovic, V.; Jovanovic, S.; et al. Graphene quantum dots inhibit T cell-mediated neuroinflammation in rats. Neuropharmacology 2019, 146, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lie, Q.; Liu, Y.; Jia, Z.; Gong, Y.; Yuan, X.; Liu, J. Multifunctional Selenium Quantum Dots for the Treatment of Alzheimer’s Disease by Reducing Aβ-Neurotoxicity and Oxidative Stress and Alleviate Neuroinflammation. ACS Appl. Mater. Interfaces 2021, 13, 30261–30273. [Google Scholar] [CrossRef] [PubMed]
- Stein, G.H. T98G: An anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro. J. Cell. Physiol. 1979, 99, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java Tree view-extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–34248. [Google Scholar] [CrossRef]
| Concentration (µg/mL) | Test | % of Viability (n) |
|---|---|---|
| 40 | MTT | 89 ± 8 (9) |
| 30 | MTT | 91 ± 1 (3) |
| 40 | Neutral red | 96 ± 1 (7) |
| Control Samples | Treated Samples | |||||
|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | Replicate 1 | Replicate 2 | Replicate 3 | |
| Initial | 31,016,624 | 30,419,346 | 32,022,288 | 34,786,612 | 29,827,860 | 40,211,724 |
| Filtered | 26,681,128 | 26,192,232 | 26,451,976 | 29,101,028 | 24,773,250 | 36,251,964 |
| Aligned (%) | 26,342,795 (98.7) | 25,793,274 (98.5) | 26,083,336 (98.6) | 28,717,867 (98.7) | 24,455,042 (98.7) | 35,782,526 (98.7) |
| Duplicated (%) | 28.1 | 28.1 | 26.9 | 28.7 | 26.6 | 31.6 |
| Biological Process Sub-Ontology (GO Term) | Genes in Reference List | Genes among DEG | Expected among DEG | EF | p Value | FDR |
|---|---|---|---|---|---|---|
| Temperature homeostasis (GO:0001659) | 14 | 4 | 0.07 | 56 | 1.78 × 10−6 | 8.35 × 10−3 |
| Inflammatory response (GO:0006954) | 208 | 8 | 1.07 | 7.5 | 1.27 × 10−5 | 2.24 × 10−2 |
| Cytokine-mediated signaling pathway (GO:0019221) | 390 | 12 | 2.00 | 6.0 | 6.66 × 10−7 | 9.38 × 10−3 |
| Positive regulation of developmental process (GO:0051094) | 746 | 15 | 3.83 | 3.9 | 4.18 × 10−6 | 1.18 × 10−2 |
| Regulation of cell differentiation (GO:0045595) | 966 | 18 | 4.96 | 3.63 | 1.01 × 10−6 | 7.13 × 10−3 |
| Regulation of cell death (GO:0010941) | 1091 | 17 | 5.61 | 3.0 | 2.36 × 10−5 | 3.32 × 10−2 |
| Reactome Pathway | Genes in Reference List | Genes among DEG | Expected among DEG | EF | p Value | FDR |
|---|---|---|---|---|---|---|
| Interleukin-10 signaling | 16 | 3 | 0.08 | 36 | 1.16 × 10−4 | 4.92 × 10−2 |
| Interleukin-4 and Interleukin-13 signaling | 62 | 7 | 0.32 | 22 | 5.27 × 10−8 | 1.12 × 10−4 |
| Class A/1 (Rhodopsin-like receptors) | 50 | 5 | 0.26 | 19 | 8.36 × 10−6 | 8.87 × 10−3 |
| Pathway | Genes Involved | Log2 Fold Change |
|---|---|---|
| 5-Hydroxytryptamine degradation | Aldehyde dehydrogenase family 1 member A3 | 1.12 |
| Angiogenesis | Proto-oncogene c-fos | −2.16 |
| Tissue factor F3 ortholog | −1.05 | |
| Angiotensin II-stimulated signaling through G proteins and beta-arrestin | Early growth response protein 1 | −2.44 |
| Apoptosis signaling pathway | Proto-oncogene c-Fos | −2.16 |
| Baculoviral IAP repeat-containing protein 3 | −1.19 | |
| B cell activation | Proto-oncogene c-Fos | −2.16 |
| B-cell receptor CD22 | 1.41 | |
| Blood coagulation | Tissue factor F3 | −1.04 |
| CCKR signaling map | Early growth response protein 1 | −2.44 |
| Proto-oncogene c-Fos | −2.16 | |
| Interleukin-8 | −1.67 | |
| Substance-P receptor | 2.54 | |
| Baculoviral IAP repeat-containing protein 3 | −1.19 | |
| Coenzyme A biosynthesis | Pantothenate kinase 1 | 1.04 |
| Dopamine receptor-mediated signaling pathway | D(2) dopamine receptor | 1.07 |
| EGF receptor signaling pathway | Protein sprouty homolog 4 | −1.98 |
| FGF signaling pathway | Protein sprouty homolog 4 | −1.98 |
| Gonadotropin-releasing hormone receptor pathway | Early growth response protein 1 | −2.44 |
| Proto-oncogene c-Fos | −2.16 | |
| D(2) dopamine receptor | 1.07 | |
| DNA-binding protein inhibitor ID-3 | −1.07 | |
| Transcription factor jun-B | −1.16 | |
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha-mediated pathway | D(2) dopamine receptor | 1.07 |
| Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha-mediated pathway | D(2) dopamine receptor | 1.07 |
| B2 bradykinin receptor | 1.06 | |
| Huntington disease | Proto-oncogene c-Fos | −2.16 |
| Inflammation mediated by chemokine and cytokine signaling pathway | Interleukin-8 | −1.67 |
| Transcription factor jun-B | −1.16 | |
| Integrin alpha-M | −1.80 | |
| Interleukin-6 | −1.84 | |
| Interleukin-1 beta | 1.57 | |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | Proto-oncogene c-Fos | −2.16 |
| Integrin signaling pathway | Integrin alpha-M | −1.80 |
| Interleukin signaling pathway | Proto-oncogene c-Fos | −2.16 |
| Interleukin-8 | −1.67 | |
| Interleukin-6 | −1.84 | |
| Nicotine pharmacodynamics pathway | D(2) dopamine receptor | 1.07 |
| PDGF signaling pathway | Proto-oncogene c-Fos | −2.16 |
| T cell activation | Proto-oncogene c-Fos | −2.16 |
| TGF-beta signaling pathway | Transcription factor jun-B | −1.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuster, E.; Candela, H.; Estévez, J.; Vilanova, E.; Sogorb, M.A. A Transcriptomic Analysis of T98G Human Glioblastoma Cells after Exposure to Cadmium-Selenium Quantum Dots Mainly Reveals Alterations in Neuroinflammation Processes and Hypothalamus Regulation. Int. J. Mol. Sci. 2022, 23, 2267. https://doi.org/10.3390/ijms23042267
Fuster E, Candela H, Estévez J, Vilanova E, Sogorb MA. A Transcriptomic Analysis of T98G Human Glioblastoma Cells after Exposure to Cadmium-Selenium Quantum Dots Mainly Reveals Alterations in Neuroinflammation Processes and Hypothalamus Regulation. International Journal of Molecular Sciences. 2022; 23(4):2267. https://doi.org/10.3390/ijms23042267
Chicago/Turabian StyleFuster, Encarnación, Héctor Candela, Jorge Estévez, Eugenio Vilanova, and Miguel A. Sogorb. 2022. "A Transcriptomic Analysis of T98G Human Glioblastoma Cells after Exposure to Cadmium-Selenium Quantum Dots Mainly Reveals Alterations in Neuroinflammation Processes and Hypothalamus Regulation" International Journal of Molecular Sciences 23, no. 4: 2267. https://doi.org/10.3390/ijms23042267
APA StyleFuster, E., Candela, H., Estévez, J., Vilanova, E., & Sogorb, M. A. (2022). A Transcriptomic Analysis of T98G Human Glioblastoma Cells after Exposure to Cadmium-Selenium Quantum Dots Mainly Reveals Alterations in Neuroinflammation Processes and Hypothalamus Regulation. International Journal of Molecular Sciences, 23(4), 2267. https://doi.org/10.3390/ijms23042267







