Abstract
Pituitary adenylate cyclase-activating polypeptide (PACAP) is a broadly expressed neuropeptide which has diverse effects in both the peripheral and central nervous systems. While its neuroprotective effects have been shown in a variety of disease models, both animal and human data support the role of PACAP in migraine generation. Both PACAP and its truncated derivative PACAP(6-38) increased calcium influx in rat trigeminal ganglia (TG) primary sensory neurons in most experimental settings. PACAP(6-38), however, has been described as an antagonist for PACAP type I (known as PAC1), and Vasoactive Intestinal Polypeptide Receptor 2 (also known as VPAC2) receptors. Here, we aimed to compare the signaling pathways induced by the two peptides using transcriptomic analysis. Rat trigeminal ganglion cell cultures were incubated with 1 µM PACAP-38 or PACAP(6-38). Six hours later RNA was isolated, next-generation RNA sequencing was performed and transcriptomic changes were analyzed to identify differentially expressed genes. Functional analysis was performed for gene annotation using the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome databases. We found 200 common differentially expressed (DE) genes for these two neuropeptides. Both PACAP-38 and PACAP(6-38) treatments caused significant downregulation of NADH: ubiquinone oxidoreductase subunit B6 and upregulation of transient receptor potential cation channel, subfamily M, member 8. The common signaling pathways induced by both peptides indicate that they act on the same target, suggesting that PACAP activates trigeminal primary sensory neurons via a mechanism independent of the identified and cloned PAC1/VPAC2 receptor, either via another target structure or a different splice variant of PAC1/VPAC2 receptors. Identification of the target could help to understand key mechanisms of migraine.
1. Introduction
Pituitary adenylate cyclase-activating polypeptide (PACAP), a neuropeptide belonging to the vasoactive intestinal polypeptide (VIP)-secretin family, is broadly expressed throughout the body. It exists in 27- and 38-amino acid-containing forms (PACAP-27 and PACAP-38), the latter one is the predominant form in mammals. PACAP was shown to have diverse actions in both the central nervous system (CNS) and the periphery. Its physiological roles include the regulation of circadian rhythm, energy homeostasis, as well as the activity of the hypothalamic-pituitary-adrenal axis. PACAP also has a major role in neurodevelopment, neuroprotection and regeneration in a variety of CNS injuries and neurodegenerative processes [1,2]. PACAP exerts most of its effects via the specific PAC1 receptor, as well as the VPAC1 and 2 receptors [3,4,5] shared with VIP having comparable affinities. In addition, Mas-related G-protein coupled receptor (Mrgpr) activation by PACAP-38 or its truncated derivatives, PACAP(6-38) or PACAP(6-27), was described on mast cells [6,7]. Mrgpr receptors belong to a diverse family of receptors, many of which are also expressed on sensory neurons [8]. Still, Mrgpr activation by PACAP has not been explored up to date.
Conflicting results were reported for the effects of PACAP on nociception in rodent models. While electrophysiological recordings showed that PACAP had a direct activating and sensitizing effect on nociceptive neurons, both pronociceptive and analgesic effects were reported in pain models, in vivo. Intrathecally administered PACAP induced nocifensive behavior in mice [9,10], but attenuated formalin-induced responses in rats [11,12]. Peripherally administered PACAP reduced hyperalgesia and pain-related behaviors in acute/subacute models of inflammatory and visceral pain [13]. In PACAP gene-deleted mice, the overall effect of PACAP was concluded to be pronociceptive in a variety of chronic pain models by contributing to central nociceptive sensitization [14,15,16,17].
It is of particular relevance that PACAP was implicated in the generation of migraine headaches. PACAP plasma levels were found to be elevated in migraine patients [18] and PACAP infusion could trigger headache in healthy volunteers and also induced a delayed, migraine-like headache in migraine patients [19,20]. Neurogenic inflammation induced by dural sensory nerve stimulation and consequent release of proinflammatory neuropeptides such as calcitonin-gene related peptide (CGRP), tachykinins and PACAP, is an important component of migraine [21] It consists of meningeal vasodilatation, plasma protein extravasation (oedema formation) and activation of inflammatory cells including mast cells [22], which in turn also release inflammatory mediators i.e., cytokines and peptides, such as PACAP [23]. These mediators further trigger the sensory nerve terminals leading to the aggravation of local neurogenic inflammation and pain [24,25].
Both PACAP and its receptors are expressed in the primary and secondary sensory neurons in the trigeminal ganglia (TG) [26,27] and trigeminal nucleus caudalis (TNC), respectively [4,28]. Animal experiments also confirmed that PACAP induced meningeal vasodilation and neuronal activation in the trigeminovascular system [29,30].
The receptorial mechanisms of PACAP in the trigeminovascular system are not understood. Since the headache-inducing effect of PACAP was not shared with VIP, the contribution of VPAC receptors could be discarded [31]. Additional data also demonstrate that PACAP can induce CGRP release independently of VPAC2 and PAC1 receptors, as well [32]. Since various Mrgprs are also expressed on primary sensory neurons [33,34], it can also be suggested that Mrgpr activation might contribute to the pronociceptive effects of PACAP.
Even though PACAP(6-38) can antagonize the effect of PACAP on heterologously expressed PAC1 receptors and various neuronal cell lines, our previous results clearly showed that PACAP(6-38) treatment did not inhibit PACAP-38, but produced identical effects by itself in rat primary sensory neurons. Both PACAP-38 and PACAP(6-38) could inhibit neuropeptide release from sensory nerve terminals of isolated trachea [35] and induce Ca2+-influx in primary cultures of trigeminal ganglion cells [36].
In order to elucidate the receptorial and signaling mechanisms of PACAP-induced trigeminal ganglia primary sensory neuronal activation related to migraine, here we analyzed and compared the transcriptome changes in cell cultures treated with PACAP-38 or PACAP(6-38).
2. Results
We determined the common differentially-expressed (DE) genes identified 6 h after PACAP-38 and PACAP(6-38) treatment. Administration of the two peptides has a high impact related to migraine-like cellular processes. All data are demonstrated in the Supplementary section.
2.1. Expression of PAC1, VPAC2 and Mrgpr Receptor Transcripts in TG Cultures
Transcripts of the known receptors of PACAP, PAC1 (Adcyap1r1) and VPAC2 receptors, as well as several Mrgpr receptors were detected in most samples as presented in Figure 1. These receptors were not differentially expressed in the PACAP-38- and PACAP(6-38)-treated groups compared to the control. VPAC1 was not expressed in either group.
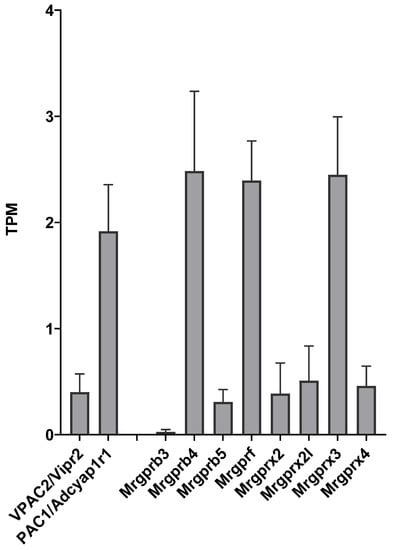
Figure 1.
Transcripts Per Million (TPM) values for receptors with known or potential affinity for PACAP in untreated control trigeminal ganglion (TG) cell cultures. Data represent mean ± SD (n = 3).
2.2. Differentially-Expressed (DE) Genes in Both PACAP-38- and PACAP(6-38)-Treated Trigeminal Ganglion Cultures
Figure 2 shows the numerical analysis of differentially expressed genes. Sample collection 6 h after the treatment yielded 200 common differentially expressed genes for PACAP-38 and PACAP6-38. For PACAP-38, 70 other DE genes, for PACAP6-38 132 other DE genes were found at the 6 h samplings. All DE genes are listed in Table A1.
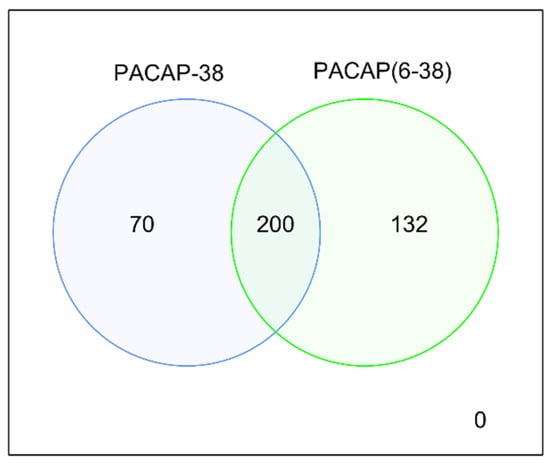
Figure 2.
Numerical representation of differentially expressed genes for PACAP-38 and PACAP6-38. The expression of genes was compared to the respective untreated control groups.
Figure 3 shows common DE genes potentially involved in neurological pathophysiology for PACAP-38 and PACAP(6-38) treatments compared to the untreated control cell culture. Noteworthy findings in the DE list are Cenpb, Gnal, Hsp90aa1, Hmga1, Tomm70, Gnai1 and Tomm34 which were upregulated regarding FC value in the case of both PACAP-38- and PACAP(6-38)-treated TG cell culture (Figure 2). It is highly notable in both neuropeptide-treated cell cultures that Ndufb6 (NADH:ubiquinone oxidoreductase subunit B6) was extensively downregulated compared to the control group (value of FC was −50.7 and −80.9 for PACAP-38- and PACAP(6-38)-treated cells, respectively) and Trpm8 was upregulated in both cases. Fbl (Fibrillarin); Fhl2 (four and a half LIM domains 2), Slc25a5 (solute carrier family 25 member 5), Tomm6 (translocase of outer mitochondrial membrane 6) were also highly downregulated in both. Listed abbreviations can be found in Appendix A.

Figure 3.
Heatmap of fold change (FC) values for differentially expressed (DE) genes shared in trigeminal ganglion (TG) cell cultures treated with PACAP-38 or PACAP(6-38) compared to the control group (1 µM, 6 h). p < 0.05 was considered as significantly different. (See the list of gene name abbreviations in Appendix A). DE genes associated with neurological disorders were selected from total hit according to databases (https://rgd.mcw.edu/; https://www.genecards.org/; https://www.ncbi.nlm.nih.gov last accessed date: 10 April 2021).
2.3. Pathway Analysis with Common DE Genes
Figure 4 shows the results for GO analyses of downregulated genes. This search highlighted mitochondrial dysfunction, regarding p-values for each term or number of significant DE genes for both administration types (Figure 4A,B).
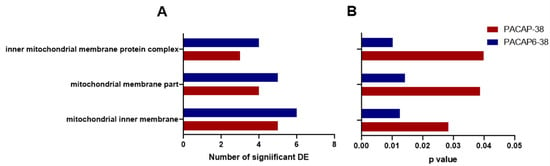
Figure 4.
Highlighted GO analysis results which were significant in both PACAP-38- (red) and PACAP(6-38)- (blue) treated trigeminal ganglion cells (1 µM, 6 h). Panel A represents the number of differentially expressed (DE) genes related to each GO term, while panel B shows p values for the identical terms (p < 0.05).
Reactome analysis was used to determine the intracellular pathways the DE genes were involved in. Figure 5 shows pathways for the common DE genes gained from the Reactome database. CREB1 phosphorylation through the activation of adenylate cyclase, protein kinase A (PKA) activation in glucagon signaling, glucagon signaling in metabolic regulation, PKA activation, PKA-mediated phosphorylation of CREB, Post NMDA receptor activation events were found in the case of upregulated genes. Ca-dependent events were upregulated, while Complex I biogenesis was downregulated in response to both PACAP-38- and PACAP(6-38)-treatment, referring to potential mitochondrial dysfunction mechanism. The results of GO (Figure 4) and Reactome analysis also revealed inhibiting effects of both peptides on mitochondrial functions supporting this concept.
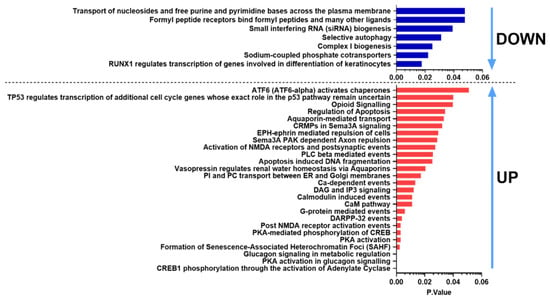
Figure 5.
Significantly affected Reactome pathways found for both PACAP-38 and PACAP(6-38)-treated cultures ranked according to the p-value. Blue bars indicate downregulated, while red bars show upregulated pathways.
In the KEGG analysis, one of the common pathway results was the calcium signaling pathway. Figure 6 highlights the common DE genes in this signaling pathway. In both cases GnaI, Prkacb were upregulated, and F2R, Slc25a5 were downregulated. This result also highlighted the possible negative effect on the mitochondria. For TG culture treated with PACAP-38, Figure 6 showed Plcb3 as upregulated, in contrast to TG culture administered with PACAP(6-38), while Gnaq was significantly upregulated in the PACAP(6-38) but not PACAP-38 group. Expression of some genes also presented with a distinct pattern: Calm2, MCU were upregulated in the PACAP(6-38) and Camk2g in the PACAP-38 treated cells. These results showed the effect of PACAP-38/PACAP(6-38) on the calcium signaling pathway. The KEGG Ca2+ pathway was also implicated significantly when ranked list enrichment was performed, which takes into account not only DE genes but all genes whose transcripts were detected. The Ca2+ pathway containing all genes can be found in Supplementary Figure S1.
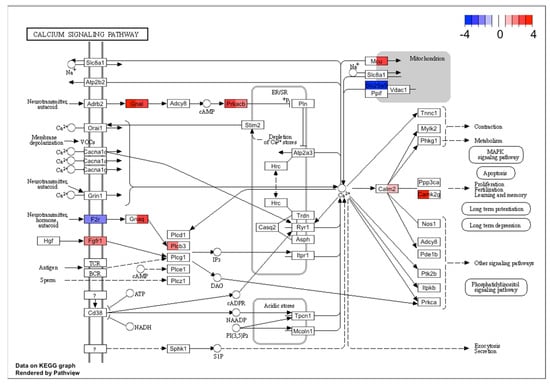
Figure 6.
The KEGG Ca2+ signaling pathway was significantly altered in both PACAP-38 and PACAP(6-38)-treated trigeminal ganglion cells. Rectangles in color indicate genes that were significantly changed after treatment (left sides of rectangles represent PACAP-38, while right sides stand for PACAP(6-38)). The red and blue color scale show normalized fold change values for each differentially expressed gene. The KEGG Ca2+ signaling pathway also significantly implicated by ranked list enrichment performed on all genes can be found in Supplementary Figure S1.
3. Discussion
This is the first description of transcriptomic changes of rat trigeminal ganglion cells in response to PACAP-38 related to mitochondrial dysfunction and neuroinflammatory mechanisms potentially mediated by calcium signaling. Moreover, these alterations were similar after treatment with PACAP(6-38), known to be PAC1 receptor antagonist at the cloned receptor, which indicates that trigeminal ganglion cell activation is independent of the PAC1 receptor. In our previous study, both PACAP-38 and PACAP(6-38) were found to induce increased intracellular Ca2+ levels in the same trigeminal ganglion cell culture [36]. Interestingly, similar agonistic effects for the two peptides were found on sensory nerve terminals of the rat trachea and human cytotrophoblast cells [35,37], mouse macrophages [38], chicken chondroblasts [39] and rat mast cells [7]. The stimulating effect of PACAP(6-38) was a surprising finding, since it is a well-established antagonist on the identified and cloned receptors of PACAP-38, PAC1 and VPAC1/2 receptors, heterologously expressed in CHO, Cos7 cells and Xenopus oocytes [7,36,40]. Our previous and present results raises the possibility that PACAP-induced trigeminovascular activation involved in migraine [21] is not mediated by these known receptors. This virtually contradictory, stimulating effect of PACAP(6-38) might be explained by an action on a receptor other than PAC1 [22]. The MrgB3 receptor was proposed as a potential target of both peptides to induce rat meningeal mast cell activation [7], but the involvement of a different PAC1 splice variant with modified binding site cannot be excluded.
Among the known receptors of PACAP-38, transcripts of PAC1 and VPAC2 receptors were detected in trigeminal ganglion cell cultures which is in line with previously reported data [27]. We have also detected several subtypes of the Mrgpr family which could be putative targets mediating the shared effects of PACAP-38 and PACAP(6-38). Mrgprs constitute a large, diverse family of G-protein coupled receptors originally described on primary sensory neurons [8], but expressed by other cell types including mast cells [6,7,41]. The endogenous ligands of these receptors are still not well defined, but various Mrgprs can be activated by amino acids or peptides, e.g., MrgprD by β-alanine or MrgprX1 by proenkephalin and proopiomelanocortin cleavage products [34]. In sensory neurons Mrgprs have been implicated in the transduction of pain and itch, as well [42]. However, interestingly, a small subset of MrgprB4-positive C-fibers were also identified as transmitting gentle stroking, but not noxious mechanical stimuli [43]. In mast cells, Mrgprs are responsible for the non-IgE-mediated activation by basic secretagogues [41]. In particular, degranulation induced by PACAP, PACAP(6-27) or PACAP(6-38) was shown to be mediated by MrgprX2 in human and MrgprB3 receptors in rat mast cells [6,7]. In our TG culture, we have detected the presence of MrgprB3 and transcripts of seven other Mrgprs. According to our results, we cannot exclude totally the role of PAC1 receptor, because we did not have a group for inhibiting PAC1 receptor or deleting PAC1 receptor in cells. It can be suggested, that either MrgprB3 or one of the other Mrgpr subtypes were activated by PACAP-38 and PACAP(6-38), but further experiments are needed to confirm this mechanism.
In order to identify the PACAP-induced intracellular signaling pathways in primary sensory neurons functional enrichment analysis of commonly altered DE genes by PACAP-38 and PACAP(6-38) pointed to significantly altered calcium signaling and PKA activation. Since PAC1 receptors can activate adenylate cyclase and phospholipase C signaling pathways as well [4], these data does not conclusively exclude the role of PAC1 receptors in the response to PACAP. However, as mentioned above, it is plausible that a common target is mediating the effects of the two peptides. Both calcium and PKA of these signaling pathways can be associated with increased nociceptor sensitivity [44,45] which supports the pronociceptive effects of PACAP. PKA activation can increase the sensitivity of transient receptor potential vanilloid 1 (TRPV1) channels, a key transducer of noxious heat and chemical stimuli [46,47] and voltage-gated sodium channels [48,49], which result in lowered activations thresholds and increased suprathreshold responses of nociceptive neurons. Intracellular calcium binding to calmodulin can activate calmodulin-dependent kinase (CaMKII) which was shown to be involved in changes of synaptic plasticity behind chronic pain conditions [45]. Increased intracellular calcium levels can also affect mitochondrial function. Functional enrichment analysis results point to mitochondrial alteration associated with a mitochondrial electron transport chain dysfunction. In particular, the B6 subunit of NADH: ubiquinone oxidoreductase (Complex I) was strongly downregulated by both treatments. This is an intriguing finding which can also link the effect of PACAP to migraine as metabolic changes and mitochondrial dysfunction such as decreased activity of Complex I, III, IV and citrate synthase have been detected in migraine patients [50,51]. Our recent study investigating the transcriptome of peripheral blood mononuclear cells of migraine patients also revealed that the mitochondrial electron transport chain was significantly affected even in headache-free periods and during headache when compared to healthy control samples [52]. It is plausible therefore that the migraine headache-inducing effect of PACAP is generated with the involvement of mitochondria. Regarding the importance of mitochondria in nociceptive sensory neurons, there is accumulating evidence demonstrating mitochondrial dysfunction in several chronic pain conditions and pain is a common complaint in patients with mitochondrial diseases [53,54,55]. Mitochondria together with the endoplasmic reticulum are key regulators of intracellular calcium homeostasis which set neuronal excitability. Moreover, mitochondrial dysfunction can lead to increased generation of reactive oxygen species which can also contribute to nociceptor sensitization by acting on multiple targets [56]. There is direct evidence that experimental inhibition of mitochondrial complex III in airway C fibers resulted in increased excitability by activation of TRP channels and protein kinase C [57,58]. Moreover, reactive oxygen species and the release of mtDNA can induce an inflammatory reaction promoting the sensitization of nociceptors.
Other interesting pain-related genes in the DE list include upregulation of the TRPM8 ion channel, which is a menthol- and cold-sensitive ion channel expressed in both dorsal root and trigeminal ganglion cells [59,60]. TRPM8 is expressed by both nociceptive and non-nociceptive sensory neurons, co-expressed with the TRPV1 ion channels in the first case [61,62,63]. Under physiological conditions TRPM8 is responsible for detecting innocuous and noxious cold temperatures [64,65,66], but it can also reduce the activation of nociceptors by other stimuli, which explains the alleviation of pain by cooling or menthol. However, there are also data showing the involvement of TRPM8 in cold allodynia in chronic inflammatory and neuropathic pain models [67]. Remarkably, there is also a possible role of TRPM8 in migraine as several large genome-wide association studies identified single nucleotide polymorphisms within and near the TRPM8 gene which confer a reduced risk of migraine [68,69,70]. Cutaneous application of menthol reduced the headache in migraine patients [71]. Likewise, another TRPM8 receptor agonist icilin reversed allodynia in an animal model of dural sensitization [72]. The relevance of the upregulation of Trpm8 after PACAP-38- or PACAP(6-38)-treatment is not yet clear as there are no literature data linking TRPM8 ion channel function with PACAP. There is, however, an interesting potential connection between TRPM8 channels and mitochondrial dysfunction. The presence of TRPM8 was shown in the endoplasmic reticulum (ER) of several cell types, including human keratinocytes [73], bronchial epithelial cells [74], prostate cancer cells [75] as well as mouse vascular smooth muscle cells [76]. In these studies, TRPM8 channels in the ER were shown to participate in the regulation of calcium homeostasis between intracellular compartments and influence mitochondrial function. While the presence of TRPM8 in the ER has not yet been reported in sensory neurons, other TRP channels, such as TRPV1 are expressed in both the plasma and ER membranes of dorsal root ganglion cells [77,78] therefore it is plausible that the ER TRPM8 can affect mitochondrial Ca2+ levels in trigeminal neurons, as well.
Figure 7 summarizes our findings in a schematic graph. Both neuropeptides act on G-protein coupled receptors (GPCR) which could either be a different target structure such as Mrgpr receptors or a splice variant of PAC1/VPAC2 receptors. GPCR trigger Guanine nucleotide-binding protein G(i) subunit alpha-1 (GnaI). GnaI activates protein kinase cAMP-Activated Catalytic Subunit Beta via increased cAMP levels. Phosphorylation by PKA may induce TRPM8 channel inhibition [79] and this inhibitory activity might potentiallly cause overexpression of TRPM8, potentiating calcium overload in mitochondria resulting Complex I suppression.
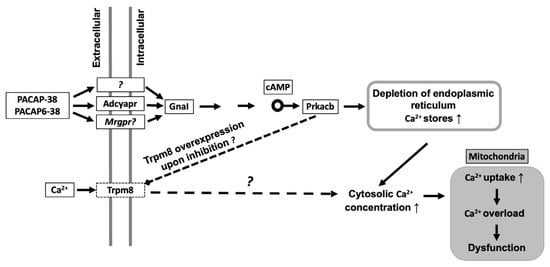
Figure 7.
Schematic representation of the hypothetical mechanisms initiated by PACAP in trigeminal ganglion cells. PACAP-38/PACAP6-38 activate G- protein coupled receptors (GPCR). Activation of Guanine nucleotide-binding protein G(i) subunit alpha-1 (GnaI) leads to protein kinase A (PKA; Protein Kinase cAMP-Activated Catalytic Subunit Beta—Prkacb) activation via increased cAMP levels. Phosphorylation by PKA may induce TRPM8 channel inhibition which could cause overexpression of TRPM8 ion channel. Calcium overload could lead to suppressed Complex I biogenesis.
In conclusion, transcriptomic changes induced by PACAP-38 and PACAP6-38 in cultured TG cells indicated cellular processes which can be associated with mechanisms occurring in migraine patients, in particular nociceptive sensitization and mitochondrial dysfunction. A limitation of our approach is the lack of functional confirmation of the affected cellular pathways which needs future research. Results, reported in this article are supported by previous human and animal experimental data on the mechanisms of migraine headache. Although, we could not identify the target of PACAP-38 for sensory neuronal activation, the common transcriptome alterations with PACAP(6-38) can potentially suggest a PAC1 independent shared pathway. These results open novel perspectives for antimigraine drug research connected to mitochondrial function.
4. Materials and Methods
4.1. Primary Cultures of TG Neurons
Primary cell cultures of TG neurons were made from 1–4-day-old Wistar rat pups as described elsewhere [80]. TG cells were dissected in ice-cold phosphate-buffered solution (PBS), incubated for 35 min at 37 °C in PBS containing collagenase (Type XI, 1 mg/mL) and then in PBS with deoxyribonuclease I (1000 units/mL) for 8 min. The ganglia were washed with Ca2+ and Mg2+-free PBS and dissociated by trituration. TG cells were plated on poly-D-lysin-coated glass coverslips and grown in a nutrient-supplemented medium for the experiment. The cell culture medium contained 180 mL Dulbecco’s- Modified Eagle Medium (D-MEM), 20 mL horse serum, 20 mL fetal bovine albumin, 2 mL insulin-transferrinselenium- S, 3.2 mL putrescin dihydrochloride (100 µg/mL), 20 µL triiodo-thyronine (0.2 mg/mL), 1.24 mL progesterone (0.5 mg/mL), 100 µL penicillin, 100 µL streptomycin and nerve growth factor (NGF, 200 ng/mL). The coverslips were maintained at 37 °C in an atmosphere containing 5% CO2. The cell cultures were incubated with 1 µM PACAP-38 or PACAP6-38. Untreated cultures served as controls. After 6 h after PACAP-38 or PACAP6-38 administration, samples were collected for RNA isolation. Conditions were repeated in triplicates.
4.2. RNA Isolation and Quality Control
Total RNA isolation and purification was performed as previously described [81] applying the phenol-based TRI Reagent procedure (Molecular Research Center, Cincinnati, OH, USA), up to the step of acquiring the RNA containing aqueous layer. The aqueous phase was mixed with an equal volume of absolute ethanol and was loaded into Zymo-Spin™ IICR Column. Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA, USA) was used according to the manufacturer’s protocol including the optional on-column DNase digestion. Qubit 3.0 (Invitrogen, Carlsbad, CA, USA) was used for RNA concentration measurement. The RNA quality verification was carried out with TapeStation 4200 using RNA ScreenTape (Agilent Technologies, Santa Clara, CA, USA). Sequencing libraries were prepared from high quality (RIN > 8) RNA samples.
4.3. Illumina Library Preparation and Sequencing
The library for Illumina sequencing was prepared using QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen, Vienna, Austria). 400 ng of total RNA was used as input for first strand cDNA generation using oligodT primer followed by RNA removing. Thereafter, the second strand synthesis is initiated by random priming and the products were purified with magnetic beads. Finally, the libraries were amplified and barcoded using PCR. All libraries were assessed on the TapeStation 4200 (Agilent Technologies, Santa Clara, CA, USA) to examine if adapter dimers formed during PCR. The QuantSeq libraries were sequenced using the Illumina NextSeq550 platform to produce 75 bp single end reads.
4.4. Bioinformatics
The sequencing reads were aligned against the Rattus norvegicus reference genome (Rnor 6.0 Ensembl release) with STAR v2.5.3a [82]. After alignment, the reads were associated with known protein-coding genes and the number of reads aligned within each gene was counted using HTSeq library v0.11.1 [83]. Gene count data were normalized using the trimmed mean of M values (TMM) normalization method of the edgeR R/Bioconductor package (v3.28, R v3.6.0, Bioconductor v3.9) [84]. Data were further log transformed using the voom approach for statistical evaluation [85] in the limma package [86]. Fold change (FC) values between the compared groups resulting from linear modeling process and modified t-test p-values were produced by the limma package. When determining differentially expressed (DE) genes, filtering thresholds were set to FC 2 and p-value 0.05 when the PACAP-38 treatment was compared to the untreated control group, and to FC 1.5 and p-value 0.001 for the PACAP(6-38) versus untreated control comparison (p-values are provided after correction for multiple comparisons by the Benjamini-Hochberg method. Normalized counts were represented as transcripts per million (TPM) values. Functional analysis (annotations of genes) was performed using the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome databases. Detection of functional enrichment was performed in the differentially expressed gene list (DE list enrichment: Fisher’s exact test for GO, hypergeometric test for KEGG and Reactome) and towards the top of the list when all genes have been ranked according to the evidence for being differentially expressed (ranked list enrichment: non-parametric Kolmogorov-Smirnov test for GO and KEGG, hypergeometric test for Reactome) applying the topGO v2.37.0, ReactomePA v1.30.0, gage v2.36.0 packages. The pathview package v1.26.0 [86] was used to visualize mapping data to KEGG pathways. All data for each gene at different time can be found in Supplementary section.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms23042120/s1.
Author Contributions
Conceptualization, K.B., É.S. and Z.H.; data curation, K.T.-L. and J.K.; formal analysis, K.T.-L. and J.K.; funding acquisition, A.G., K.B. and Z.H.; investigation, K.T.-L., T.A., P.U., J.K.; methodology, J.K., T.A., Peter Urbán, K.B. and É.S.; supervision, J.K., A.G., K.B., É.S. and Z.H.; visualization, K.T.-L.; writing—original draft, K.T.-L. and K.B.; writing—review and editing, K.T.-L., J.K., T.A., K.B., P.U., A.G., É.S. and Z.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Brain Research Program 2017-1.2.1-NKP-2017-00002 (NAP-2; Chronic Pain Research Group), Gazdaságfejlesztési és Innovációs Operatív Program (Economy Development and Innovation Operative Programme) (GINOP)-2.3.2-15-2016-00050 (Peptidergic Signaling in Health and Disease; PEPSYS), GINOP 2.3.2-15-2016-0034, Emberi Erőforrás Operatív Program (Human Resource Operative Programme) (EFOP) 3.6.2-16-2017-00008 (2017-2019), EFOP-3.6.1-16-2016-00004 and National Research Development and Innovation Office grant OTKA FK132587. A.Gy. and J.K. were supported by the grants GINOP-2.3.4-15-2020-00010, GINOP-2.3.1-20-2020-00001 and Educating Experts of the Future: Developing Bioinformatics and Biostatistics competencies of European Biomedical Students (BECOMING, 2019-1-HU01-KA203-061251). Bioinformatics infrastructure was supported by ELIXIR Hungary (http://elixir-hungary.org/). Project no. TKP2020-IKA-08 has been implemented with the support provided from the National Research, Development and Innovation Fund of Hungary, financed under the 2020-4.1.1-TKP2020 funding scheme. This work was also financed by the Thematic Excellence Program 2021 Health Sub-program of the Ministry for Innovation and Technology in Hungary, within the framework of the EGA-16 project of University of Pécs. The research was performed in collaboration with the Genomics and Bioinformatics Core Facility at the Szentágothai Research Centre of the University of Pécs.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All RNA-Seq data sets generated as part of this study will be publicly available at the European Nucleotide Archive (https://www.ebi.ac.uk/ena, accessed on 1 Ocotber 2021), under accession number PRJEB47291.
Acknowledgments
The authors wish to thank Cecília Disztl for expert technical assistance and Genomics and Bioinformatics Core Facility at the Szentágothai Research Centre of the University of Pécs.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Appendix A

Table A1.
List of DE Gene Names.
Table A1.
List of DE Gene Names.
| Abbreviation | Description |
| Abca3 | ATP binding cassette subfamily A member 3 |
| Adam23 | ADAM metallopeptidase domain 23 |
| Akap7 | A-kinase anchoring protein 7 |
| Ankh | ANKH inorganic pyrophosphate transport regulator |
| Antxr2 | ANTXR cell adhesion molecule 2 |
| Arhgap21 | Rho GTPase activating protein 21 |
| Arhgap23 | Rho GTPase activating protein 23 |
| Arhgap42 | Rho GTPase activating protein 42 |
| Art3 | ADP-ribosyltransferase 3 |
| Atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| Atp2c1 | ATPase secretory pathway Ca2+ transporting 1 |
| Bmp2k | BMP-2 inducible kinase |
| Cask | calcium/calmodulin dependent serine protein kinase |
| Ccl2 | C-C motif chemokine ligand 2 |
| Cenpb | centromere protein B |
| Clic4 | chloride intracellular channel 4 |
| Cluh | clustered mitochondria homolog |
| Col4a2 | collagen type IV alpha 2 chain |
| Ctsl | cathepsin L |
| Dgkd | diacylglycerol kinase, delta |
| Dnajc13 | DnaJ heat shock protein family (Hsp40) member C13 |
| Dync1h1 | dynein cytoplasmic 1 heavy chain 1 |
| Dync1li1 | dynein cytoplasmic 1 light intermediate chain 1 |
| Epb41l2 | erythrocyte membrane protein band 4.1-like 2 |
| Esyt2 | extended synaptotagmin 2 |
| F2r | coagulation factor II (thrombin) receptor |
| Fbl | fibrillarin |
| Fgfr1 | Fibroblast growth factor receptor 1 |
| Fhl2 | four and a half LIM domains 2 |
| Fpgs | folylpolyglutamate synthase |
| Gas7 | growth arrest specific 7 |
| Gnai1 | G protein subunit alpha i1 |
| Gnal | G protein subunit alpha L |
| Hbb | hemoglobin subunit beta |
| Hmga1 | high mobility group AT-hook 1 |
| Hsp90aa1 | heat shock protein 90 alpha family class A member 1 |
| Hspa9 | heat shock protein family A (Hsp70) member 9 |
| Lonp1 | lon peptidase 1, mitochondrial |
| Map2k4 | mitogen activated protein kinase kinase 4 |
| Map3k20 | mitogen-activated protein kinase kinase kinase 20 |
| Mapk8ip3 | mitogen-activated protein kinase 8 interacting protein 3 |
| Myef2 | myelin expression factor 2 |
| Nav1 | neuron navigator 1 |
| Ncs1 | neuronal calcium sensor 1 |
| Ndfip2 | Nedd4 family interacting protein 2 |
| Ndufab1 | NADH:ubiquinone oxidoreductase subunit AB1 |
| Ndufb6 | NADH:ubiquinone oxidoreductase subunit B6 |
| Opa1 | OPA1, mitochondrial dynamin-like GTPase |
| Osmr | oncostatin M receptor |
| Parp14 | poly (ADP-ribose) polymerase family, member 14 |
| Pcdhgc3 | protocadherin gamma subfamily C, 3 |
| Pitpnb | phosphatidylinositol transfer protein, beta |
| Plin2 | perilipin 2 |
| Plxna4 | plexin A4 |
| Ppp1cb | protein phosphatase 1 catalytic subunit beta |
| Praf2 | PRA1 domain family, member 2 |
| Prkacb | protein kinase cAMP-activated catalytic subunit beta |
| Prkar1a | protein kinase cAMP-dependent type I regulatory subunit alpha |
| Prkar2a | protein kinase cAMP-dependent type II regulatory subunit alpha |
| Prrc2b | proline-rich coiled-coil 2B |
| Psen1 | presenilin 1 |
| Ptprg | protein tyrosine phosphatase, receptor type, G |
| Rcn1 | reticulocalbin 1 |
| Slc20a2 | solute carrier family 20 member 2 |
| Slc25a5 | solute carrier family 25 member 5 |
| Slc30a9 | solute carrier family 30 member 9 |
| Slc3a2 | solute carrier family 3 member 2 |
| Snca | synuclein alpha |
| Tiam1 | TIAM Rac1 associated GEF 1 |
| Tmem128 | transmembrane protein 128 |
| Tmem131 | transmembrane protein 131 |
| Tollip | toll interacting protein |
| Tomm34 | translocase of outer mitochondrial membrane 34 |
| Tomm6 | translocase of outer mitochondrial membrane 6 |
| Tomm70 | translocase of outer mitochondrial membrane 70 |
| Tpm4 | tropomyosin 4 |
| Trio | trio Rho guanine nucleotide exchange factor |
| Trpm8 | transient receptor potential cation channel, subfamily M, member 8 |
| Txlna | taxilin alpha |
| Txn1 | thioredoxin 1 |
| Txndc17 | thioredoxin domain containing 17 |
References
- Somogyvári-Vigh, A.; Reglődi, D. Pituitary Adenylate Cyclase Activating Polypeptide: A Potential Neuroprotective Peptide. Curr. Pharm. Des. 2004, 10, 2861–2889. [Google Scholar] [CrossRef] [PubMed]
- Reglődi, D.; Kiss, P.; Lubics, A.; Tamás, A. Review on the Protective Effects of PACAP in Models of Neurodegenerative Diseases in Vitro and in Vivo. Curr. Pharm. Des. 2011, 17, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Dickson, L.; Finlayson, K. VPAC and PAC Receptors: From Ligands to Function. Pharmacol. Ther. 2009, 121, 294–316. [Google Scholar] [CrossRef] [PubMed]
- Vaudry, D.; Falluel-Morel, A.; Bourgault, S.; Basille, M.; Burel, D.; Wurtz, O.; Fournier, A.; Chow, B.K.C.; Hashimoto, H.; Galas, L.; et al. Pituitary Adenylate Cyclase-Activating Polypeptide and Its Receptors: 20 Years after the Discovery. Pharm. Rev. 2009, 61, 283–357. [Google Scholar] [CrossRef]
- Harmar, A.J.; Fahrenkrug, J.; Gozes, I.; Laburthe, M.; May, V.; Pisegna, J.R.; Vaudry, D.; Vaudry, H.; Waschek, J.A.; Said, S.I. Pharmacology and Functions of Receptors for Vasoactive Intestinal Peptide and Pituitary Adenylate Cyclase-Activating Polypeptide: IUPHAR Review 1. Br. J. Pharmacol. 2012, 166, 4–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatemoto, K.; Nozaki, Y.; Tsuda, R.; Konno, S.; Tomura, K.; Furuno, M.; Ogasawara, H.; Edamura, K.; Takagi, H.; Iwamura, H.; et al. Immunoglobulin E-Independent Activation of Mast Cell Is Mediated by Mrg Receptors. Biochem. Biophys. Res. Commun. 2006, 349, 1322–1328. [Google Scholar] [CrossRef]
- Pedersen, S.H.; la Cour, S.H.; Calloe, K.; Hauser, F.; Olesen, J.; Klaerke, D.A.; Jansen-Olesen, I. PACAP-38 and PACAP(6-38) Degranulate Rat Meningeal Mast Cells via the Orphan MrgB3-Receptor. Front. Cell. Neurosci. 2019, 13, 114. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Han, S.; Zylka, M.J.; Simon, M.I.; Anderson, D.J. A Diverse Family of GPCRs Expressed in Specific Subsets of Nociceptive Sensory Neurons. Cell 2001, 106, 619–632. [Google Scholar] [CrossRef] [Green Version]
- Narita, M.; Dun, S.L.; Dun, N.J.; Tseng, L.F. Hyperalgesia Induced by Pituitary Adenylate Cyclase-Activating Polypeptide in the Mouse Spinal Cord. Eur. J. Pharmacol. 1996, 311, 121–126. [Google Scholar] [CrossRef]
- Shimizu, T.; Katahira, M.; Sugawara, H.; Inoue, K.; Miyata, A. Diverse Effects of Intrathecal Pituitary Adenylate Cyclase-Activating Polypeptide on Nociceptive Transmission in Mice Spinal Cord. Regul. Pept. 2004, 123, 117–122. [Google Scholar] [CrossRef]
- Yamamoto, T.; Tatsuno, I. Antinociceptive Effect of Intrathecally Administered Pituitary Adenylate Cyclase Activating Polypeptide (PACAP) on the Rat Formalin Test. Neurosci. Lett. 1995, 184, 32–35. [Google Scholar] [CrossRef]
- Zhang, Y.; Malmberg, A.B.; Sjölund, B.; Yaksh, T.L. The Effect of Pituitary Adenylate Cyclase Activating Peptide (PACAP) on the Nociceptive Formalin Test. Neurosci. Lett. 1996, 207, 187–190. [Google Scholar] [CrossRef]
- Sándor, K.; Bölcskei, K.; McDougall, J.J.; Schuelert, N.; Reglődi, D.; Elekes, K.; Pethő, G.; Pintér, E.; Szolcsányi, J.; Helyes, Z. Divergent Peripheral Effects of Pituitary Adenylate Cyclase-Activating Polypeptide-38 on Nociception in Rats and Mice. Pain 2009, 141, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, T.; Shintani, N.; Matsumura, S.; Okuda-Ashitaka, E.; Hashimoto, H.; Muratani, T.; Minami, T.; Baba, A.; Ito, S. Pituitary Adenylate Cyclase-Activating Polypeptide Is Required for the Development of Spinal Sensitization and Induction of Neuropathic Pain. J. Neurosci. 2004, 24, 7283–7291. [Google Scholar] [CrossRef]
- Sándor, K.; Kormos, V.; Botz, B.; Imreh, A.; Bölcskei, K.; Gaszner, B.; Markovics, A.; Szolcsányi, J.; Shintani, N.; Hashimoto, H.; et al. Impaired Nocifensive Behaviours and Mechanical Hyperalgesia, but Enhanced Thermal Allodynia in Pituitary Adenylate Cyclase-Activating Polypeptide Deficient Mice. Neuropeptides 2010, 44, 363–371. [Google Scholar] [CrossRef]
- Botz, B.; Imreh, A.; Sándor, K.; Elekes, K.; Szolcsányi, J.; Reglődi, D.; Quinn, J.P.; Stewart, J.; Zimmer, A.; Hashimoto, H.; et al. Role of Pituitary Adenylate-Cyclase Activating Polypeptide and Tac1 Gene Derived Tachykinins in Sensory, Motor and Vascular Functions under Normal and Neuropathic Conditions. Peptides 2013, 43, 105–112. [Google Scholar] [CrossRef]
- Botz, B.; Bölcskei, K.; Kereskai, L.; Kovács, M.; Németh, T.; Szigeti, K.; Horváth, I.; Máthé, D.; Kovács, N.; Hashimoto, H.; et al. Differential Regulatory Role of Pituitary Adenylate Cyclase-Activating Polypeptide in the Serum-Transfer Arthritis Model: Complex Regulatory Role of PACAP in Arthritis. Arthritis Rheumatol. 2014, 66, 2739–2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuka, B.; Helyes, Z.; Markovics, A.; Bagoly, T.; Szolcsányi, J.; Szabó, N.; Tóth, E.; Kincses, Z.T.; Vécsei, L.; Tajti, J. Alterations in PACAP-38-like Immunoreactivity in the Plasma during Ictal and Interictal Periods of Migraine Patients. Cephalalgia 2013, 33, 1085–1095. [Google Scholar] [CrossRef] [Green Version]
- Schytz, H.W.; Birk, S.; Wienecke, T.; Kruuse, C.; Olesen, J.; Ashina, M. PACAP38 Induces Migraine-like Attacks in Patients with Migraine without Aura. Brain 2009, 132, 16–25. [Google Scholar] [CrossRef] [Green Version]
- Amin, F.M.; Asghar, M.S.; Guo, S.; Hougaard, A.; Hansen, A.E.; Schytz, H.W.; van der Geest, R.J.; de Koning, P.J.; Larsson, H.B.; Olesen, J.; et al. Headache and Prolonged Dilatation of the Middle Meningeal Artery by PACAP38 in Healthy Volunteers. Cephalalgia 2012, 32, 140–149. [Google Scholar] [CrossRef]
- Lukacs, M.; Tajti, J.; Fulop, F.; Toldi, J.; Edvinsson, L.; Vecsei, L. Migraine, Neurogenic Inflammation, Drug Development—Pharmacochemical Aspects. Curr. Med. Chem. 2017, 24, 3649–3665. [Google Scholar] [CrossRef] [PubMed]
- Baun, M.; Pedersen, M.H.F.; Olesen, J.; Jansen-Olesen, I. Dural Mast Cell Degranulation Is a Putative Mechanism for Headache Induced by PACAP-38. Cephalalgia 2012, 32, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Okragly, A.J.; Morin, S.M.; DeRosa, D.; Martin, A.P.; Johnson, K.W.; Johnson, M.P.; Benschop, R.J. Human Mast Cells Release the Migraine-Inducing Factor Pituitary Adenylate Cyclase-Activating Polypeptide (PACAP). Cephalalgia 2018, 38, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Tajti, J.; Tuka, B.; Botz, B.; Helyes, Z.; Vecsei, L. Role of Pituitary Adenylate Cyclase-Activating Polypeptide in Nociception and Migraine. CNS Neurol. Disord. Drug Targets 2015, 14, 540–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyuncu Irmak, D.; Kilinc, E.; Tore, F. Shared Fate of Meningeal Mast Cells and Sensory Neurons in Migraine. Front. Cell. Neurosci. 2019, 13, 136. [Google Scholar] [CrossRef] [Green Version]
- Moller, K.; Zhang, Y.-Z.; Håkanson, R.; Luts, A.; Sjölund, B.; Uddman, R.; Sundler, F. Pituitary Adenylate Cyclase Activating Peptide Is a Sensory Neuropeptide: Immunocytochemical and Immunochemical Evidence. Neuroscience 1993, 57, 725–732. [Google Scholar] [CrossRef]
- Chaudhary, P.; Baumann, T.K. Expression of VPAC2 Receptor and PAC1 Receptor Splice Variants in the Trigeminal Ganglion of the Adult Rat. Brain Res. Mol. Brain Res. 2002, 104, 137–142. [Google Scholar] [CrossRef]
- Uddman, R.; Tajti, J.; Hou, M.; Sundler, F.; Edvinsson, L. Neuropeptide Expression in the Human Trigeminal Nucleus Caudalis and in the Cervical Spinal Cord C1 and C2. Cephalalgia 2002, 22, 112–116. [Google Scholar] [CrossRef] [Green Version]
- Markovics, A.; Kormos, V.; Gaszner, B.; Lashgarara, A.; Szoke, E.; Sandor, K.; Szabadfi, K.; Tuka, B.; Tajti, J.; Szolcsanyi, J.; et al. Pituitary Adenylate Cyclase-Activating Polypeptide Plays a Key Role in Nitroglycerol-Induced Trigeminovascular Activation in Mice. Neurobiol. Dis. 2012, 45, 633–644. [Google Scholar] [CrossRef]
- Tuka, B.; Helyes, Z.; Markovics, A.; Bagoly, T.; Németh, J.; Márk, L.; Brubel, R.; Reglődi, D.; Párdutz, Á.; Szolcsányi, J.; et al. Peripheral and Central Alterations of Pituitary Adenylate Cyclase Activating Polypeptide-like Immunoreactivity in the Rat in Response to Activation of the Trigeminovascular System. Peptides 2012, 33, 307–316. [Google Scholar] [CrossRef]
- Rahmann, A.; Wienecke, T.; Hansen, J.M.; Fahrenkrug, J.; Olesen, J.; Ashina, M. Vasoactive Intestinal Peptide Causes Marked Cephalic Vasodilation, but Does Not Induce Migraine. Cephalalgia 2008, 28, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Jansen-Olesen, I.; Baun, M.; Amrutkar, D.V.; Ramachandran, R.; Christophersen, D.V.; Olesen, J. PACAP-38 but Not VIP Induces Release of CGRP from Trigeminal Nucleus Caudalis via a Receptor Distinct from the PAC1 Receptor. Neuropeptides 2014, 48, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Solinski, H.J.; Gudermann, T.; Breit, A. Pharmacology and Signaling of MAS-Related G Protein-Coupled Receptors. Pharm. Rev. 2014, 66, 570–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bader, M.; Alenina, N.; Andrade-Navarro, M.A.; Santos, R.A. Mas and Its Related G Protein–Coupled Receptors, Mrgprs. Pharm. Rev. 2014, 66, 1080–1105. [Google Scholar] [CrossRef] [PubMed]
- Németh, J.; Reglődi, D.; Pozsgai, G.; Szabó, Á.; Elekes, K.; Pintér, E.; Szolcsányi, J.; Helyes, Z. Effect of Pituitary Adenylate Cyclase Activating Polypeptide-38 on Sensory Neuropeptide Release and Neurogenic Inflammation in Rats and Mice. Neuroscience 2006, 143, 223–230. [Google Scholar] [CrossRef]
- Sághy, É.; Payrits, M.; Helyes, Z.; Reglődi, D.; Bánki, E.; Tóth, G.; Couvineau, A.; Szőke, É. Stimulatory Effect of Pituitary Adenylate Cyclase-Activating Polypeptide 6-38, M65 and Vasoactive Intestinal Polypeptide 6–28 on Trigeminal Sensory Neurons. Neuroscience 2015, 308, 144–156. [Google Scholar] [CrossRef]
- Walker, C.S.; Sundrum, T.; Hay, D.L. PACAP Receptor Pharmacology and Agonist Bias: Analysis in Primary Neurons and Glia from the Trigeminal Ganglia and Transfected Cells: PACAP Pharmacology in the Trigeminal Ganglia. Br. J. Pharmacol. 2014, 171, 1521–1533. [Google Scholar] [CrossRef] [Green Version]
- Reglődi, D.; Börzsei, R.; Bagoly, T.; Boronkai, A.; Rácz, B.; Tamás, A.; Kiss, P.; Horváth, G.; Brubel, R.; Németh, J.; et al. Agonistic Behavior of PACAP6-38 on Sensory Nerve Terminals and Cytotrophoblast Cells. J. Mol. Neurosci. 2008, 36, 270–278. [Google Scholar] [CrossRef]
- Ichinose, M.; Asai, M.; Imai, K.; Sawada, M. Enhancement of Phagocytosis in Mouse Macrophages by Pituitary Adenylate Cyclase Activating Polypeptide (PACAP) and Related Peptides. Immunopharmacology 1995, 30, 217–224. [Google Scholar] [CrossRef]
- Juhász, T.; Matta, C.; Katona, É.; Somogyi, C.; Takács, R.; Gergely, P.; Csernoch, L.; Panyi, G.; Tóth, G.; Reglődi, D.; et al. Pituitary Adenylate Cyclase Activating Polypeptide (PACAP) Signalling Exerts Chondrogenesis Promoting and Protecting Effects: Implication of Calcineurin as a Downstream Target. PLoS ONE 2014, 9, e91541. [Google Scholar] [CrossRef] [Green Version]
- McNeil, B.D.; Pundir, P.; Meeker, S.; Han, L.; Undem, B.J.; Kulka, M.; Dong, X. Identification of a Mast-Cell-Specific Receptor Crucial for Pseudo-Allergic Drug Reactions. Nature 2015, 519, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, D.P. The Role of Mrgprs in Pain. Neurosci. Lett. 2021, 744, 135544. [Google Scholar] [CrossRef] [PubMed]
- Vrontou, S.; Wong, A.M.; Rau, K.K.; Koerber, H.R.; Anderson, D.J. Genetic Identification of C Fibres That Detect Massage-like Stroking of Hairy Skin in Vivo. Nature 2013, 493, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Hucho, T.; Levine, J.D. Signaling Pathways in Sensitization: Toward a Nociceptor Cell Biology. Neuron 2007, 55, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.-Q.; Liu, D.-Q.; Chen, S.-P.; Sun, J.; Zhou, X.-R.; Luo, F.; Tian, Y.-K.; Ye, D.-W. Cellular and Molecular Mechanisms of Calcium/Calmodulin-Dependent Protein Kinase II in Chronic Pain. J. Pharm. Exp. Ther. 2017, 363, 176–183. [Google Scholar] [CrossRef] [Green Version]
- Bhave, G.; Zhu, W.; Wang, H.; Brasier, D.J.; Oxford, G.S.; Gereau, R.W. CAMP-Dependent Protein Kinase Regulates Desensitization of the Capsaicin Receptor (VR1) by Direct Phosphorylation. Neuron 2002, 35, 721–731. [Google Scholar] [CrossRef] [Green Version]
- Varga, A.; Bölcskei, K.; Szöke, é.; Almási, R.; Czéh, G.; Szolcsányi, J.; Pethö, G. Relative Roles of Protein Kinase A and Protein Kinase C in Modulation of Transient Receptor Potential Vanilloid Type 1 Receptor Responsiveness in Rat Sensory Neurons in Vitro and Peripheral Nociceptors in Vivo. Neuroscience 2006, 140, 645–657. [Google Scholar] [CrossRef]
- Gold, M.S.; Levine, J.D.; Correa, A.M. Modulation of TTX-R INa by PKC and PKA and Their Role in PGE2-Induced Sensitization of Rat Sensory Neurons In Vitro. J. Neurosci. 1998, 18, 10345–10355. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, E.M.; Okuse, K.; Wood, J.N.; Dolphin, A.C.; Moss, S.J. CAMP-Dependent Phosphorylation of the Tetrodotoxin-Resistant Voltage-Dependent Sodium Channel SNS. J. Physiol. 1999, 516, 433–446. [Google Scholar] [CrossRef]
- Fuentes, E.; Araya-Maturana, R.; Urra, F.A. Regulation of Mitochondrial Function as a Promising Target in Platelet Activation-Related Diseases. Free Radic. Biol. Med. 2019, 136, 172–182. [Google Scholar] [CrossRef]
- Gross, E.C.; Lisicki, M.; Fischer, D.; Sándor, P.S.; Schoenen, J. The Metabolic Face of Migraine—From Pathophysiology to Treatment. Nat. Rev. Neurol. 2019, 15, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Aczél, T.; Körtési, T.; Kun, J.; Urbán, P.; Bauer, W.; Herczeg, R.; Farkas, R.; Kovács, K.; Vásárhelyi, B.; Karvaly, G.B.; et al. Identification of Disease- and Headache-Specific Mediators and Pathways in Migraine Using Blood Transcriptomic and Metabolomic Analysis. J. Headache Pain 2021, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Meeus, M.; Nijs, J.; Hermans, L.; Goubert, D.; Calders, P. The Role of Mitochondrial Dysfunctions Due to Oxidative and Nitrosative Stress in the Chronic Pain or Chronic Fatigue Syndromes and Fibromyalgia Patients: Peripheral and Central Mechanisms as Therapeutic Targets? Expert Opin. Ther. Targets 2013, 17, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- van den Ameele, J.; Fuge, J.; Pitceathly, R.D.S.; Berry, S.; McIntyre, Z.; Hanna, M.G.; Lee, M.; Chinnery, P.F. Chronic Pain Is Common in Mitochondrial Disease. Neuromuscul. Disord. 2020, 30, 413–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousuf, M.S.; Maguire, A.D.; Simmen, T.; Kerr, B.J. Endoplasmic Reticulum–Mitochondria Interplay in Chronic Pain: The Calcium Connection. Mol. Pain 2020, 16, 1744806920946889. [Google Scholar] [CrossRef] [PubMed]
- Salvemini, D.; Little, J.W.; Doyle, T.; Neumann, W.L. Roles of Reactive Oxygen and Nitrogen Species in Pain. Free Radic. Biol. Med. 2011, 51, 951. [Google Scholar] [CrossRef] [Green Version]
- Nesuashvili, L.; Hadley, S.H.; Bahia, P.K.; Taylor-Clark, T.E. Sensory Nerve Terminal Mitochondrial Dysfunction Activates Airway Sensory Nerves via Transient Receptor Potential (TRP) Channels. Mol. Pharmacol. 2013, 83, 1007–1019. [Google Scholar] [CrossRef] [Green Version]
- Hadley, S.H.; Bahia, P.K.; Taylor-Clark, T.E. Sensory Nerve Terminal Mitochondrial Dysfunction Induces Hyperexcitability in Airway Nociceptors via Protein Kinase C. Mol. Pharm. 2014, 85, 839–848. [Google Scholar] [CrossRef] [Green Version]
- McKemy, D.D.; Neuhausser, W.M.; Julius, D. Identification of a Cold Receptor Reveals a General Role for TRP Channels in Thermosensation. Nature 2002, 416, 52–58. [Google Scholar] [CrossRef]
- Peier, A.M.; Moqrich, A.; Hergarden, A.C.; Reeve, A.J.; Andersson, D.A.; Story, G.M.; Earley, T.J.; Dragoni, I.; McIntyre, P.; Bevan, S.; et al. A TRP Channel That Senses Cold Stimuli and Menthol. Cell 2002, 108, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Okazawa, M.; Inoue, W.; Hori, A.; Hosokawa, H.; Matsumura, K.; Kobayashi, S. Noxious Heat Receptors Present in Cold-Sensory Cells in Rats. Neurosci. Lett. 2004, 359, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Abe, J.; Hosokawa, H.; Okazawa, M.; Kandachi, M.; Sawada, Y.; Yamanaka, K.; Matsumura, K.; Kobayashi, S. TRPM8 Protein Localization in Trigeminal Ganglion and Taste Papillae. Brain Res. Mol. Brain Res. 2005, 136, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Sarria, I.; Gu, J. Menthol Response and Adaptation in Nociceptive-like and Nonnociceptive-like Neurons: Role of Protein Kinases. Mol. Pain 2010, 6, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bautista, D.M.; Siemens, J.; Glazer, J.M.; Tsuruda, P.R.; Basbaum, A.I.; Stucky, C.L.; Jordt, S.-E.; Julius, D. The Menthol Receptor TRPM8 Is the Principal Detector of Environmental Cold. Nature 2007, 448, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Dhaka, A.; Murray, A.N.; Mathur, J.; Earley, T.J.; Petrus, M.J.; Patapoutian, A. TRPM8 Is Required for Cold Sensation in Mice. Neuron 2007, 54, 371–378. [Google Scholar] [CrossRef] [Green Version]
- Knowlton, W.M.; Bifolck-Fisher, A.; Bautista, D.M.; McKemy, D.D. TRPM8, but Not TRPA1, Is Required for Neural and Behavioral Responses to Acute Noxious Cold Temperatures and Cold-Mimetics in Vivo. Pain 2010, 150, 340–350. [Google Scholar] [CrossRef] [Green Version]
- Colburn, R.W.; Lubin, M.L.; Stone, D.J.; Wang, Y.; Lawrence, D.; D’Andrea, M.R.; Brandt, M.R.; Liu, Y.; Flores, C.M.; Qin, N. Attenuated Cold Sensitivity in TRPM8 Null Mice. Neuron 2007, 54, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Chasman, D.I.; Schürks, M.; Anttila, V.; de Vries, B.; Schminke, U.; Launer, L.J.; Terwindt, G.M.; van den Maagdenberg, A.M.J.M.; Fendrich, K.; Völzke, H.; et al. Genome-Wide Association Study Reveals Three Susceptibility Loci for Common Migraine in the General Population. Nat. Genet. 2011, 43, 695–698. [Google Scholar] [CrossRef] [Green Version]
- Freilinger, T.; Anttila, V.; de Vries, B.; Malik, R.; Kallela, M.; Terwindt, G.M.; Pozo-Rosich, P.; Winsvold, B.; Nyholt, D.R.; van Oosterhout, W.P.J.; et al. Genome-Wide Association Analysis Identifies Susceptibility Loci for Migraine without Aura. Nat. Genet. 2012, 44, 777–782. [Google Scholar] [CrossRef]
- Gormley, P.; Anttila, V.; Winsvold, B.S.; Palta, P.; Esko, T.; Pers, T.H.; Farh, K.-H.; Cuenca-Leon, E.; Muona, M.; Furlotte, N.A.; et al. Meta-Analysis of 375,000 Individuals Identifies 38 Susceptibility Loci for Migraine. Nat. Genet. 2016, 48, 856–866. [Google Scholar] [CrossRef] [Green Version]
- Borhani Haghighi, A.; Motazedian, S.; Rezaii, R.; Mohammadi, F.; Salarian, L.; Pourmokhtari, M.; Khodaei, S.; Vossoughi, M.; Miri, R. Cutaneous Application of Menthol 10% Solution as an Abortive Treatment of Migraine without Aura: A Randomised, Double-Blind, Placebo-Controlled, Crossed-over Study. Int. J. Clin. Pract. 2010, 64, 451–456. [Google Scholar] [CrossRef]
- Kayama, Y.; Shibata, M.; Takizawa, T.; Ibata, K.; Shimizu, T.; Ebine, T.; Toriumi, H.; Yuzaki, M.; Suzuki, N. Functional Interactions between Transient Receptor Potential M8 and Transient Receptor Potential V1 in the Trigeminal System: Relevance to Migraine Pathophysiology. Cephalalgia 2018, 38, 833–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidaux, G.; Borowiec, A.; Gordienko, D.; Beck, B.; Shapovalov, G.G.; Lemonnier, L.; Flourakis, M.; Vandenberghe, M.; Slomianny, C.; Dewailly, E.; et al. Epidermal TRPM8 Channel Isoform Controls the Balance between Keratinocyte Proliferation and Differentiation in a Cold-Dependent Manner. Proc. Natl. Acad. Sci. USA 2015, 112, E3345–E3354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabnis, A.S.; Shadid, M.; Yost, G.S.; Reilly, C.A. Human Lung Epithelial Cells Express a Functional Cold-Sensing TRPM8 Variant. Am. J. Respir. Cell Mol. Biol. 2008, 39, 466. [Google Scholar] [CrossRef] [Green Version]
- Thebault, S.; Lemonnier, L.; Bidaux, G.; Flourakis, M.; Bavencoffe, A.; Gordienko, D.; Roudbaraki, M.; Delcourt, P.; Panchin, Y.; Shuba, Y.; et al. Novel Role of Cold/Menthol-Sensitive Transient Receptor Potential Melastatine Family Member 8 (TRPM8) in the Activation of Store-Operated Channels in LNCaP Human Prostate Cancer Epithelial Cells. J. Biol. Chem. 2005, 280, 39423–39435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, S.; Wang, B.; Lin, S.; Zhang, H.; Li, Y.; Wei, X.; Cui, Y.; Wei, X.; Lu, Z.; Gao, P.; et al. Activation of Transient Receptor Potential Melastatin Subtype 8 Attenuates Cold-Induced Hypertension Through Ameliorating Vascular Mitochondrial Dysfunction. JAHA 2017, 6, e005495. [Google Scholar] [CrossRef]
- Kárai, L.J.; Russell, J.T.; Iadarola, M.J.; Oláh, Z. Vanilloid Receptor 1 Regulates Multiple Calcium Compartments and Contributes to Ca2+-Induced Ca2+ Release in Sensory Neurons. J. Biol. Chem. 2004, 279, 16377–16387. [Google Scholar] [CrossRef] [Green Version]
- Gallego-Sandín, S.; Rodríguez-García, A.; Alonso, M.T.; García-Sancho, J. The Endoplasmic Reticulum of Dorsal Root Ganglion Neurons Contains Functional TRPV1 Channels. J. Biol. Chem. 2009, 284, 32591. [Google Scholar] [CrossRef] [Green Version]
- Szőke, É.; Balla, Z.; Csernoch, L.; Czéh, G.; Szolcsányi, J. Interacting Effects of Capsaicin and Anandamide on Intracellular Calcium in Sensory Neurones. NeuroReport 2000, 11, 1949–1952. [Google Scholar] [CrossRef]
- Aczél, T.; Kun, J.; Szőke, É.; Rauch, T.; Junttila, S.; Gyenesei, A.; Bölcskei, K.; Helyes, Z. Transcriptional Alterations in the Trigeminal Ganglia, Nucleus and Peripheral Blood Mononuclear Cells in a Rat Orofacial Pain Model. Front. Mol. Neurosci. 2018, 11, 219. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).