Network-Assisted Systems Biology Analysis of the Mitochondrial Proteome in a Pre-Clinical Model of Ischemia, Revascularization and Post-Conditioning
Abstract
1. Introduction
2. Results
2.1. Ischemia, Revascularization, and Post-Conditioning Impacts on the Swine Mitochondrial Proteome
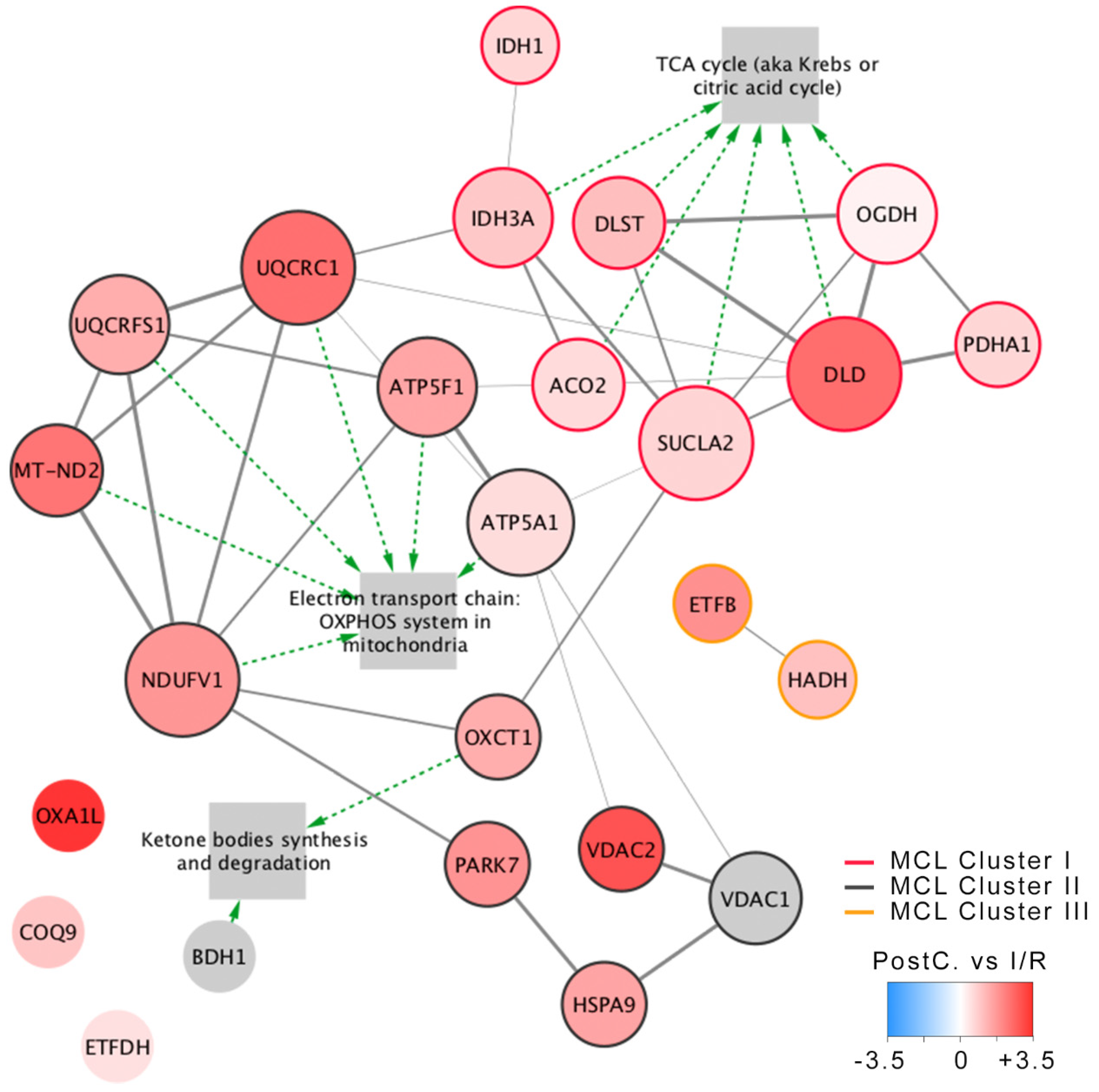
2.2. Tricarboxylic Acid Cycle
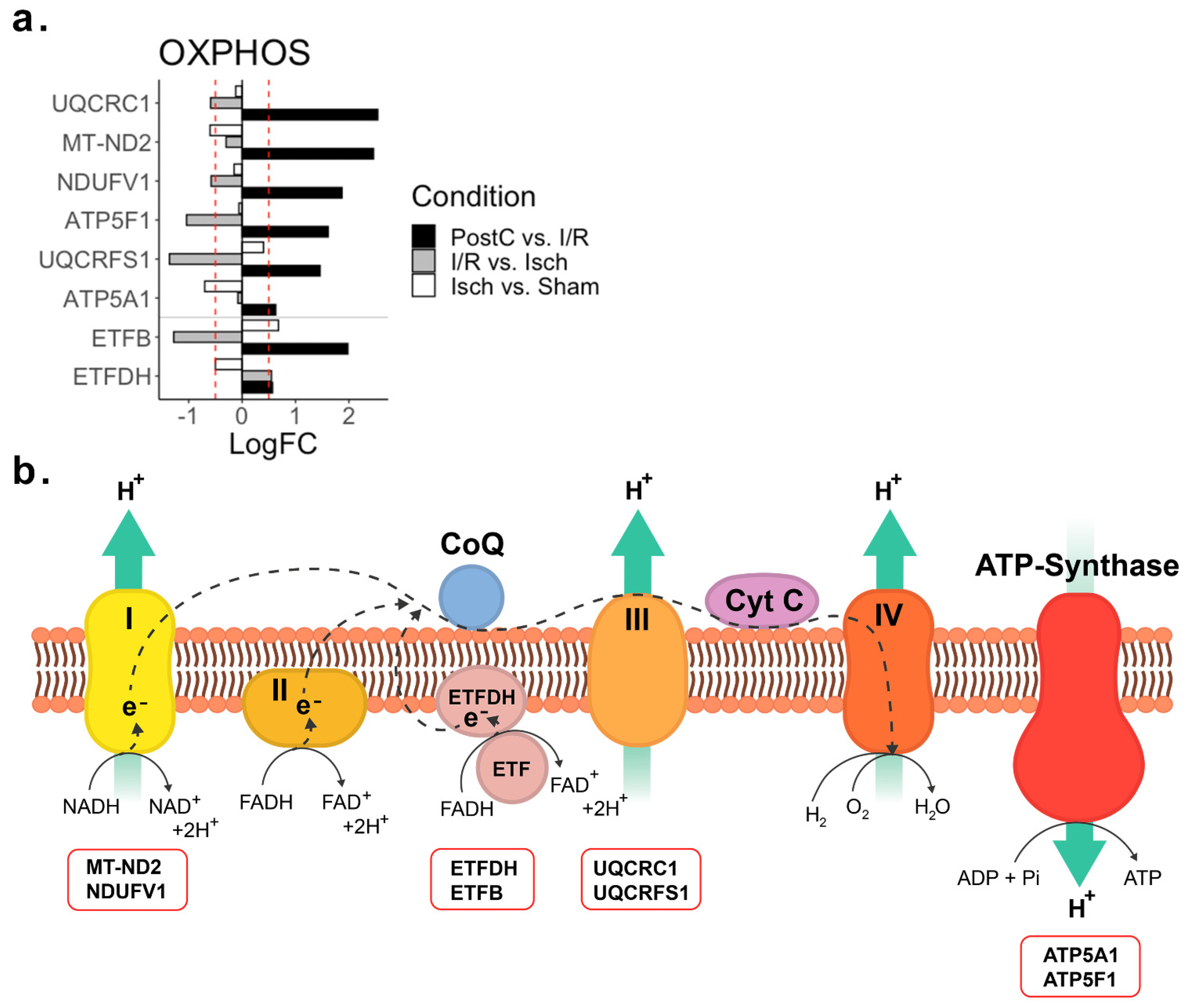
2.3. Oxidative Phosphorylation
2.4. Non Metabolic Proteins
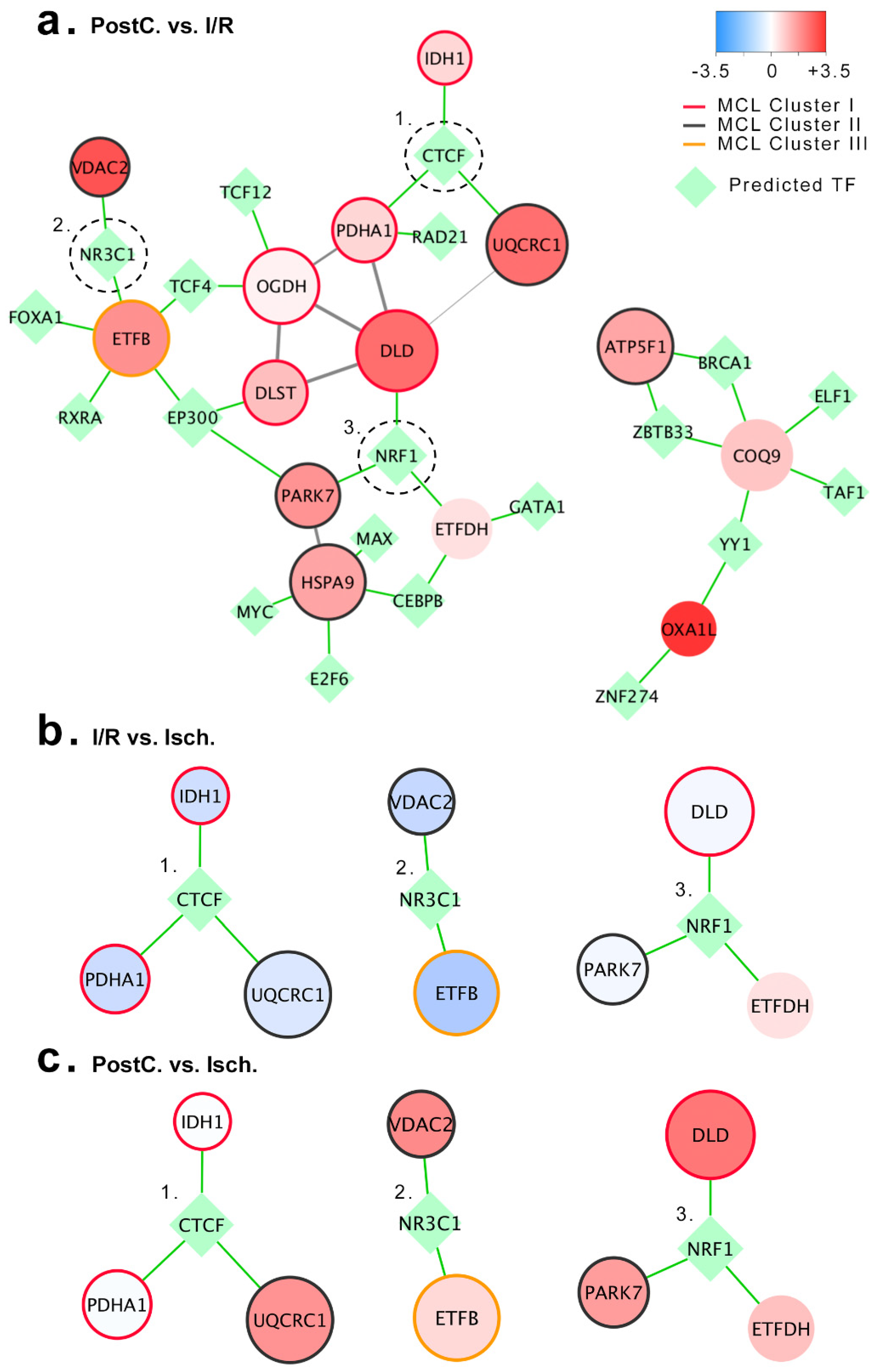
2.5. Prediction of Ischemic Post-Conditioning Mitochondrial Regulatory Network
3. Discussion
3.1. Mitochondrial Response to Ischemia in the Pig Heart
3.2. Mitochondrial Response to Revascularization in the Pig Heart
3.3. Mitochondrial Response to Post-Conditioning in the Pig Heart
4. Materials and Methods
4.1. Experimental Model
4.2. Sample Collection and Protein Extraction
4.3. Proteomic Analysis
4.4. In Silico Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nabel, E.G.; Braunwald, E. A Tale of Coronary Artery Disease and Myocardial Infarction. N. Engl. J. Med. 2012, 366, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Smilowitz, N.R.; Feit, F. The History of Primary Angioplasty and Stenting for Acute Myocardial Infarction. Curr. Cardiol. Rep. 2016, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Bahit, M.C.; Kochar, A.; Granger, C.B. Post-Myocardial Infarction Heart Failure. JACC Heart Fail. 2018, 6, 179–186. [Google Scholar] [CrossRef]
- Cahill, T.J.; Kharbanda, R.K. Heart failure after myocardial infarction in the era of primary percutaneous coronary intervention: Mechanisms, incidence and identification of patients at risk. World J. Cardiol. 2017, 9, 407. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, E.J.; Blaha, M.J.; Chiuve, S.E.; Cushman, M.; Das, S.R.; Deo, R.; de Ferranti, S.D.; Floyd, J.; Fornage, M.; Gillespie, C.; et al. Heart Disease and Stroke Statistics—2017 Update: A Report From the American Heart Association. Circulation 2017, 135, e146–e603. [Google Scholar] [CrossRef]
- Moran, A.E.; Forouzanfar, M.H.; Roth, G.A.; Mensah, G.A.; Ezzati, M.; Flaxman, A.; Murray, C.J.L.; Naghavi, M. The global burden of ischemic heart disease in 1990 and 2010: The Global Burden of Disease 2010 study. Circulation 2014, 129, 1493–1501. [Google Scholar] [CrossRef]
- Reimer, K.A.; Jennings, R.B. The “wavefront phenomenon” of myocardial ischemic cell death. II. Transmural progression of necrosis within the framework of ischemic bed size (myocardium at risk) and collateral flow. Lab. Investig. 1979, 40, 633–644. [Google Scholar]
- Frangogiannis, N.G. The Mechanistic Basis of Infarct Healing. Antioxid. Redox Signal. 2006, 8, 1907–1939. [Google Scholar] [CrossRef]
- Kelle, S.; Roes, S.D.; Klein, C.; Kokocinski, T.; de Roos, A.; Fleck, E.; Bax, J.J.; Nagel, E. Prognostic Value of Myocardial Infarct Size and Contractile Reserve Using Magnetic Resonance Imaging. J. Am. Coll. Cardiol. 2009, 54, 1770–1777. [Google Scholar] [CrossRef]
- Sobel, B.E.; Bresnahan, G.F.; Shell, W.E.; Yoder, R.D. Estimation of infarct size in man and its relation to prognosis. Circulation 1972, 46, 640–648. [Google Scholar] [CrossRef]
- Yellon, D.M.; Hausenloy, D.J. Myocardial Reperfusion Injury. N. Engl. J. Med. 2007, 357, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Dorado, D.; Ruiz-Meana, M.; Piper, H.M. Lethal reperfusion injury in acute myocardial infarction: Facts and unresolved issues. Cardiovasc. Res. 2009, 83, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Hausenloy, D.J.; Erik Bøtker, H.; Condorelli, G.; Ferdinandy, P.; Garcia-Dorado, D.; Heusch, G.; Lecour, S.; van Laake, L.W.; Madonna, R.; Ruiz-Meana, M.; et al. Translating cardioprotection for patient benefit: Position paper from the Working Group of Cellular Biology of the Heart of the European Society of Cardiology. Cardiovasc. Res. 2013, 98, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Heusch, G. Critical Issues for the Translation of Cardioprotection. Circ. Res. 2017, 120, 1477–1486. [Google Scholar] [CrossRef]
- Murry, C.E.; Jennings, R.B.; Reimer, K.A. Preconditioning with ischemia: A delay of lethal cell injury in ischemic myocardium. Circulation 1986, 74, 1124–1136. [Google Scholar] [CrossRef]
- Zhao, Z.-Q.; Corvera, J.S.; Halkos, M.E.; Kerendi, F.; Wang, N.-P.; Guyton, R.A.; Vinten-Johansen, J. Inhibition of myocardial injury by ischemic postconditioning during reperfusion: Comparison with ischemic preconditioning. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, H579–H588. [Google Scholar] [CrossRef]
- Staat, P.; Rioufol, G.; Piot, C.; Cottin, Y.; Cung, T.T.; L’Huillier, I.; Aupetit, J.-F.; Bonnefoy, E.; Finet, G.; André-Fouët, X.; et al. Postconditioning the Human Heart. Circulation 2005, 112, 2143–2148. [Google Scholar] [CrossRef]
- Vinten-Johansen, J.; Yellon, D.M.; Opie, L.H. Postconditioning: A Simple, Clinically Applicable Procedure to Improve Revascularization in Acute Myocardial Infarction. Circulation 2005, 112, 2085–2088. [Google Scholar] [CrossRef]
- Bøtker, H.E.; Lassen, T.R.; Jespersen, N.R. Clinical translation of myocardial conditioning. Am. J. Physiol. Circ. Physiol. 2018, 314, H1225–H1252. [Google Scholar] [CrossRef]
- Gao, J.; Luo, J.; Liu, F.; Zheng, Y.; Chen, B.; Chen, Q.; Yang, Y. Short-and long-term effects of ischemic postconditioning in STEMI patients: A meta-analysis. Lipids Health Dis. 2015, 14, 147. [Google Scholar] [CrossRef]
- Mir, T.; Uddin, M.; Changal, K.H.; Perveiz, E.; Kaur, J.; Sattar, Y.; Ullah, W.; Sheikh, M. Long-term outcomes of ischemic post-conditioning primary PCI and conventional primary PCI in acute STEMI: A meta-analysis of randomized trials. Expert Rev. Cardiovasc. Ther. 2021, 19, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Boengler, K.; Schulz, R.; Heusch, G. Loss of cardioprotection with ageing. Cardiovasc. Res. 2009, 83, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Ludman, A.J.; Yellon, D.M.; Hausenloy, D.J. Cardiac preconditioning for ischaemia: Lost in translation. Dis. Model. Mech. 2010, 3, 35–38. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Whittington, H.J.; Harding, I.; Stephenson, C.I.M.; Bell, R.; Hausenloy, D.J.; Mocanu, M.M.; Yellon, D.M. Cardioprotection in the aging, diabetic heart: The loss of protective Aktsignalling. Cardiovasc. Res. 2013, 99, 694–704. [Google Scholar] [CrossRef]
- Cohen, M.V.; Downey, J.M. Signalling pathways and mechanisms of protection in pre- and postconditioning: Historical perspective and lessons for the future. Br. J. Pharmacol. 2015, 172, 1913–1932. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Silverman, E.K.; Schmidt, H.H.H.W.; Anastasiadou, E.; Altucci, L.; Angelini, M.; Badimon, L.; Balligand, J.; Benincasa, G.; Capasso, G.; Conte, F.; et al. Molecular networks in Network Medicine: Development and applications. WIREs Syst. Biol. Med. 2020, 12, e1489. [Google Scholar] [CrossRef]
- Charitou, T.; Bryan, K.; Lynn, D.J. Using biological networks to integrate, visualize and analyze genomics data. Genet. Sel. Evol. 2016, 48, 27. [Google Scholar] [CrossRef]
- Di Lisa, F.; Canton, M.; Carpi, A.; Kaludercic, N.; Menabò, R.; Menazza, S.; Semenzato, M. Mitochondrial injury and protection in ischemic pre- and postconditioning. Antioxid. Redox Signal. 2011, 14, 881–891. [Google Scholar] [CrossRef]
- Ramachandra, C.J.A.; Hernandez-Resendiz, S.; Crespo-Avilan, G.E.; Lin, Y.-H.; Hausenloy, D.J. Mitochondria in acute myocardial infarction and cardioprotection. EBioMedicine 2020, 57, 102884. [Google Scholar] [CrossRef]
- Di Lisa, F.; Canton, M.; Menabò, R.; Kaludercic, N.; Bernardi, P. Mitochondria and cardioprotection. Heart Fail. Rev. 2007, 12, 249–260. [Google Scholar] [CrossRef]
- Heusch, G. Molecular Basis of Cardioprotection. Circ. Res. 2015, 116, 674–699. [Google Scholar] [CrossRef]
- Vilahur, G.; Cubedo, J.; Casani, L.; Padro, T.; Sabate-Tenas, M.; Badimon, J.J.; Badimon, L. Reperfusion-triggered stress protein response in the myocardium is blocked by post-conditioning. Systems biology pathway analysis highlights the key role of the canonical aryl-hydrocarbon receptor pathway. Eur. Heart J. 2013, 34, 2082–2093. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Van Dongen, S. Graph Clustering by Flow Simulation; University of Utrecht: Utrecht, The Netherlands, 2000. [Google Scholar]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Miller, A.R.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting communities. Nucleic Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef]
- Gerstein, M.B.; Kundaje, A.; Hariharan, M.; Landt, S.G.; Yan, K.-K.; Cheng, C.; Mu, X.J.; Khurana, E.; Rozowsky, J.; Alexander, R.; et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012, 489, 91–100. [Google Scholar] [CrossRef]
- Hochachka, P.; Owen, T.; Allen, J.; Whittow, G. Multiple end products of anaerobiosis in diving vertebrates. Comp. Biochem. Physiol. B. Comp. Biochem. 1975, 50, 17–22. [Google Scholar] [CrossRef]
- Chinopoulos, C. Which way does the citric acid cycle turn during hypoxia? The critical role of α-ketoglutarate dehydrogenase complex. J. Neurosci. Res. 2013, 91, 1030–1043. [Google Scholar] [CrossRef]
- Czibik, G.; Steeples, V.; Yavari, A.; Ashrafian, H. Citric Acid Cycle Intermediates in Cardioprotection. Circ. Cardiovasc. Genet. 2014, 7, 711–719. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Pell, V.R.; Gaude, E.; Aksentijević, D.; Sundier, S.Y.; Robb, E.L.; Logan, A.; Nadtochiy, S.M.; Ord, E.N.J.; Smith, A.C.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Pell, V.R.; James, A.M.; Work, L.M.; Saeb-Parsy, K.; Frezza, C.; Krieg, T.; Murphy, M.P. A Unifying Mechanism for Mitochondrial Superoxide Production during Ischemia-Reperfusion Injury. Cell Metab. 2016, 23, 254–263. [Google Scholar] [CrossRef]
- Zweier, J.L.; Talukder, M.A.H. The role of oxidants and free radicals in reperfusion injury. Cardiovasc. Res. 2006, 70, 181–190. [Google Scholar] [CrossRef]
- Becker, L. New concepts in reactive oxygen species and cardiovascular reperfusion physiology. Cardiovasc. Res. 2004, 61, 461–470. [Google Scholar] [CrossRef]
- Treberg, J.R.; Brand, M.D. A model of the proton translocation mechanism of complex I. J. Biol. Chem. 2011, 286, 17579–17584. [Google Scholar] [CrossRef]
- Korge, P.; Calmettes, G.; John, S.A.; Weiss, J.N. Reactive oxygen species production induced by pore opening in cardiac mitochondria: The role of complex III. J. Biol. Chem. 2017, 292, 9882–9895. [Google Scholar] [CrossRef]
- Bleier, L.; Dröse, S. Superoxide generation by complex III: From mechanistic rationales to functional consequences. Biochim. Biophys. Acta BBA Bioenerg. 2013, 1827, 1320–1331. [Google Scholar] [CrossRef]
- Chen, Q.; Vazquez, E.J.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Production of Reactive Oxygen Species by Mitochondria: Central Role of Complex III. J. Biol. Chem. 2003, 278, 36027–36031. [Google Scholar] [CrossRef]
- Petrosillo, G.; Francesca, M.R.; Di Venosa, N.; Paradies, A.G. Decreased complex III activity in mitochondria isolated from rat heart subjected to ischemia and reperfusion: Role of reactive oxygen species and cardiolipin. FASEB J. 2003, 17, 714–716. [Google Scholar] [CrossRef]
- Wong, R.; Aponte, A.M.; Steenbergen, C.; Murphy, E. Cardioprotection leads to novel changes in the mitochondrial proteome. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H75–H91. [Google Scholar] [CrossRef][Green Version]
- Cao, S.; Liu, Y.; Wang, H.; Mao, X.; Chen, J.; Liu, J.; Xia, Z.; Zhang, L.; Liu, X.; Yu, T. Ischemic postconditioning influences electron transport chain protein turnover in Langendorff-perfused rat hearts. PeerJ 2016, 4, e1706. [Google Scholar] [CrossRef]
- Yao, G.-Y.; Zhu, Q.; Xia, J.; Chen, F.-J.; Huang, M.; Liu, J.; Zhou, T.-T.; Wei, J.-F.; Cui, G.-Y.; Zheng, K.-Y.; et al. Ischemic postconditioning confers cerebroprotection by stabilizing VDACs after brain ischemia. Cell Death Dis. 2018, 9, 1033. [Google Scholar] [CrossRef]
- Das, S.; Steenbergen, C.; Murphy, E. Does the Voltage Dependent Anion Channel Modulate Cardiac Ischemia-Reperfusion Injury? Biochim. Biophys. Acta BBA Biomembr. 2012, 1818, 1451–1456. [Google Scholar] [CrossRef]
- De Lazzari, F.; Prag, H.A.; Gruszczyk, A.V.; Whitworth, A.J.; Bisaglia, M. DJ-1: A promising therapeutic candidate for ischemia-reperfusion injury. Redox Biol. 2021, 41, 101884. [Google Scholar] [CrossRef]
- Zhou, T.T.; Wang, X.Y.; Huang, J.; Deng, Y.Z.; Qiu, L.J.; Liu, H.Y.; Xu, X.W.; Ma, Z.X.; Tang, L.; Chen, H.P. Mitochondrial Translocation of DJ-1 Is Mediated by Grp75: Implication in Cardioprotection of Resveratrol Against Hypoxia/Reoxygenation-Induced Oxidative Stress. J. Cardiovasc. Pharmacol. 2020, 75, 305–313. [Google Scholar] [CrossRef]
- Ibanez, B.; Prat-González, S.; Speidl, W.S.; Vilahur, G.; Pinero, A.; Cimmino, G.; García, M.J.; Fuster, V.; Sanz, J.; Badimon, J.J. Early metoprolol administration before coronary reperfusion results in increased myocardial salvage: Analysis of ischemic myocardium at risk using cardiac magnetic resonance. Circulation 2007, 115, 2909–2916. [Google Scholar] [CrossRef]
- Skyschally, A.; Van Caster, P.; Iliodromitis, E.; Schulz, R.; Kremastinos, D.; Heusch, G. Ischemic postconditioning: Experimental models and protocol algorithms. Basic Res. Cardiol. 2009, 104, 469–483. [Google Scholar] [CrossRef]
- Skyschally, A.; Van Caster, P.; Boengler, K.; Gres, P.; Musiolik, J.; Schilawa, D.; Schulz, R.; Heusch, G. Ischemic postconditioning in pigs: No causal role for RISK activation. Circ. Res. 2009, 104, 15–18. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Kutmon, M.; Kelder, T.; Mandaviya, P.; Evelo, C.T.A.; Coort, S.L. CyTargetLinker: A Cytoscape App to Integrate Regulatory Interactions in Network Analysis. PLoS ONE 2013, 8, e82160. [Google Scholar] [CrossRef]
- Banjanovic, E.S.; Osborne, J.W. Confidence Intervals for Effect Sizes: Applying Bootstrap Confidence Intervals for Effect Sizes: Applying Bootstrap Resampling Resampling. Pract. Assess. Res. Eval. 2016, 21, 5. [Google Scholar] [CrossRef]
- Zuurbier, C.J.; Bertrand, L.; Beauloye, C.R.; Andreadou, I.; Ruiz-Meana, M.; Jespersen, N.R.; Kula-Alwar, D.; Prag, H.A.; Eric Botker, H.; Dambrova, M.; et al. Cardiac metabolism as a driver and therapeutic target of myocardial infarction. J. Cell. Mol. Med. 2020, 24, 5937–5954. [Google Scholar] [CrossRef]


| GO_Term | Description | Strength | FDR |
|---|---|---|---|
| GO:0006091 | Generation of precursor metabolites and energy | 1.59 | 2.88 × 10−25 |
| GO:0045333 | Cellular respiration | 1.9 | 1.24 × 10−23 |
| GO:0055114 | Oxidation-reduction process | 1.2 | 6.17 × 10−17 |
| GO:0009060 | Aerobic respiration | 2 | 2.41 × 10−14 |
| GO:0006099 | Tricarboxylic acid cycle | 2.27 | 6.75 × 10−13 |
| GO:0044281 | Small molecule metabolic process | 0.92 | 4.85 × 10−11 |
| GO:0022904 | Respiratory electron transport chain | 1.77 | 2.40 × 10−9 |
| GO:0006119 | Oxidative phosphorylation | 1.72 | 4.49 × 10−9 |
| GO:0046034 | ATP metabolic process | 1.54 | 5.51 × 10−9 |
| GO:0019752 | Carboxylic acid metabolic process | 1.08 | 1.14 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallinat, A.; Vilahur, G.; Padró, T.; Badimon, L. Network-Assisted Systems Biology Analysis of the Mitochondrial Proteome in a Pre-Clinical Model of Ischemia, Revascularization and Post-Conditioning. Int. J. Mol. Sci. 2022, 23, 2087. https://doi.org/10.3390/ijms23042087
Gallinat A, Vilahur G, Padró T, Badimon L. Network-Assisted Systems Biology Analysis of the Mitochondrial Proteome in a Pre-Clinical Model of Ischemia, Revascularization and Post-Conditioning. International Journal of Molecular Sciences. 2022; 23(4):2087. https://doi.org/10.3390/ijms23042087
Chicago/Turabian StyleGallinat, Alex, Gemma Vilahur, Teresa Padró, and Lina Badimon. 2022. "Network-Assisted Systems Biology Analysis of the Mitochondrial Proteome in a Pre-Clinical Model of Ischemia, Revascularization and Post-Conditioning" International Journal of Molecular Sciences 23, no. 4: 2087. https://doi.org/10.3390/ijms23042087
APA StyleGallinat, A., Vilahur, G., Padró, T., & Badimon, L. (2022). Network-Assisted Systems Biology Analysis of the Mitochondrial Proteome in a Pre-Clinical Model of Ischemia, Revascularization and Post-Conditioning. International Journal of Molecular Sciences, 23(4), 2087. https://doi.org/10.3390/ijms23042087









