The Biosynthesis and Transport of Ophiobolins in Aspergillus ustus 094102
Abstract
:1. Introduction
2. Results
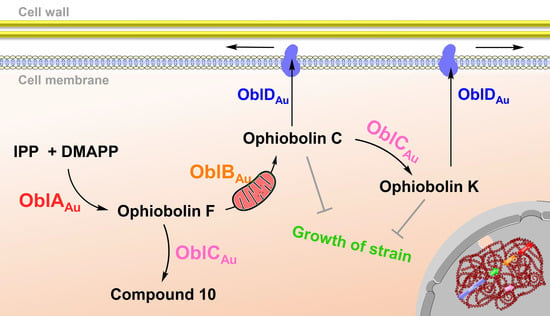
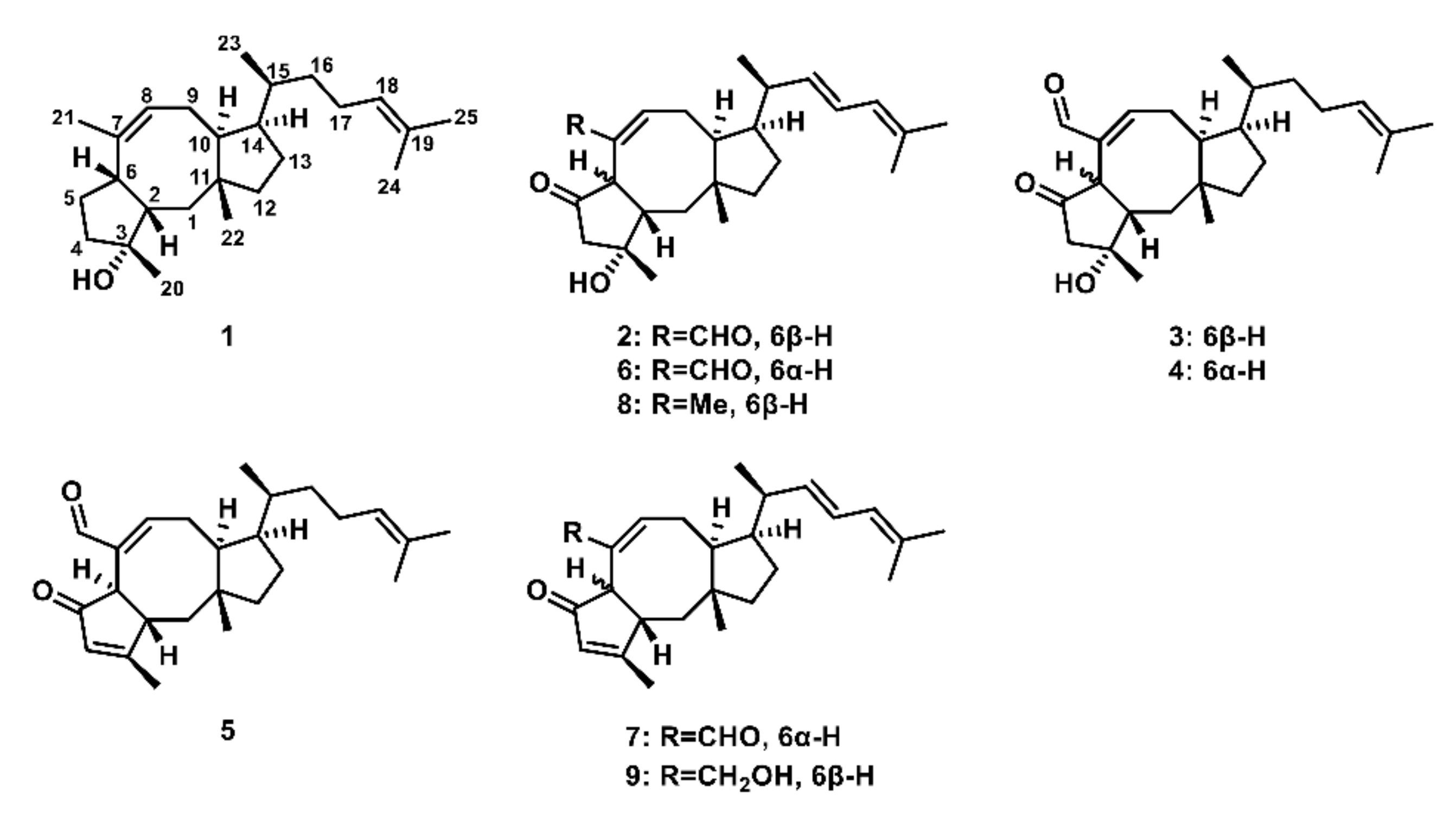
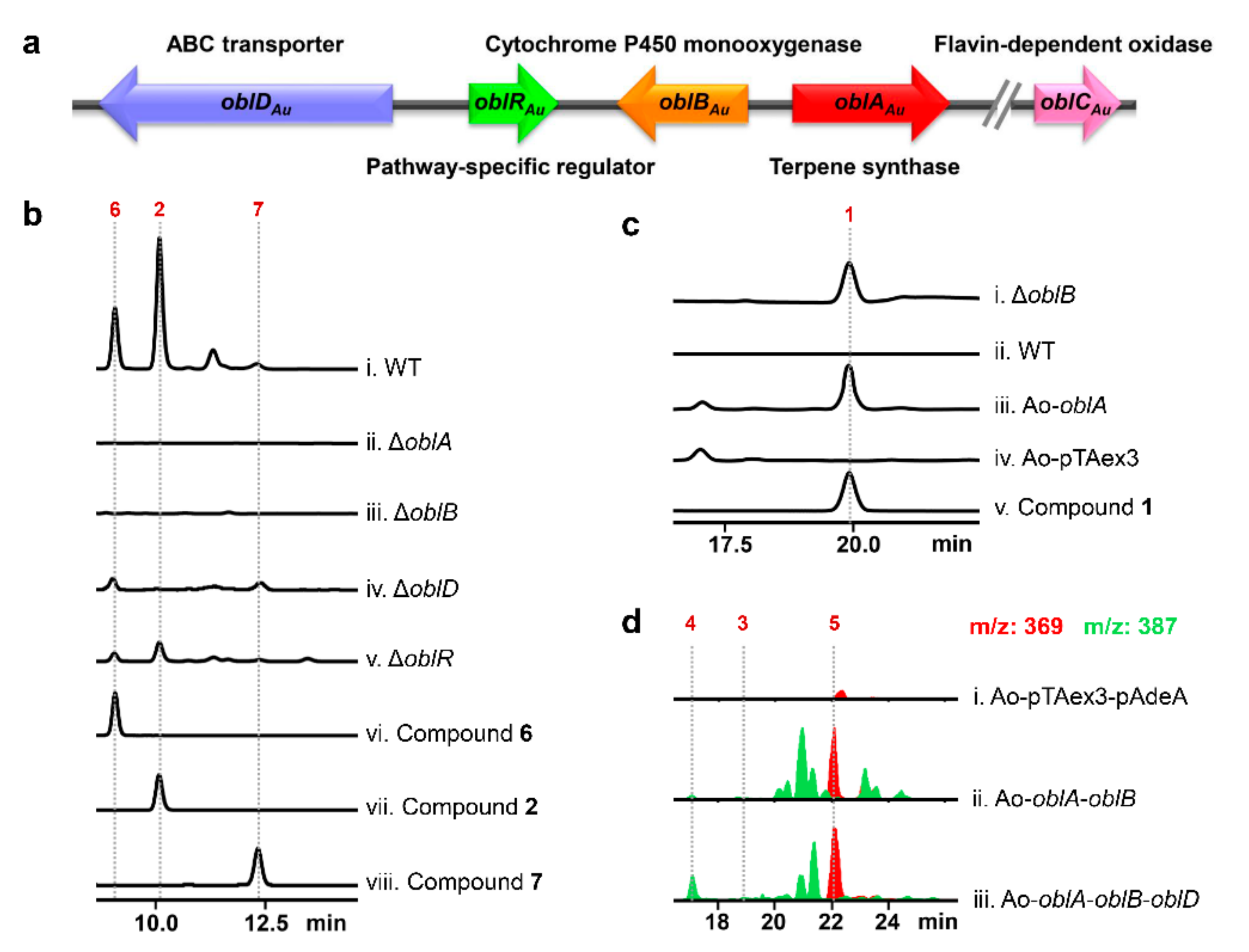
2.1. Confirmation of Ophiobolin Biosynthesis Genes in oblAu of A. ustus 094102
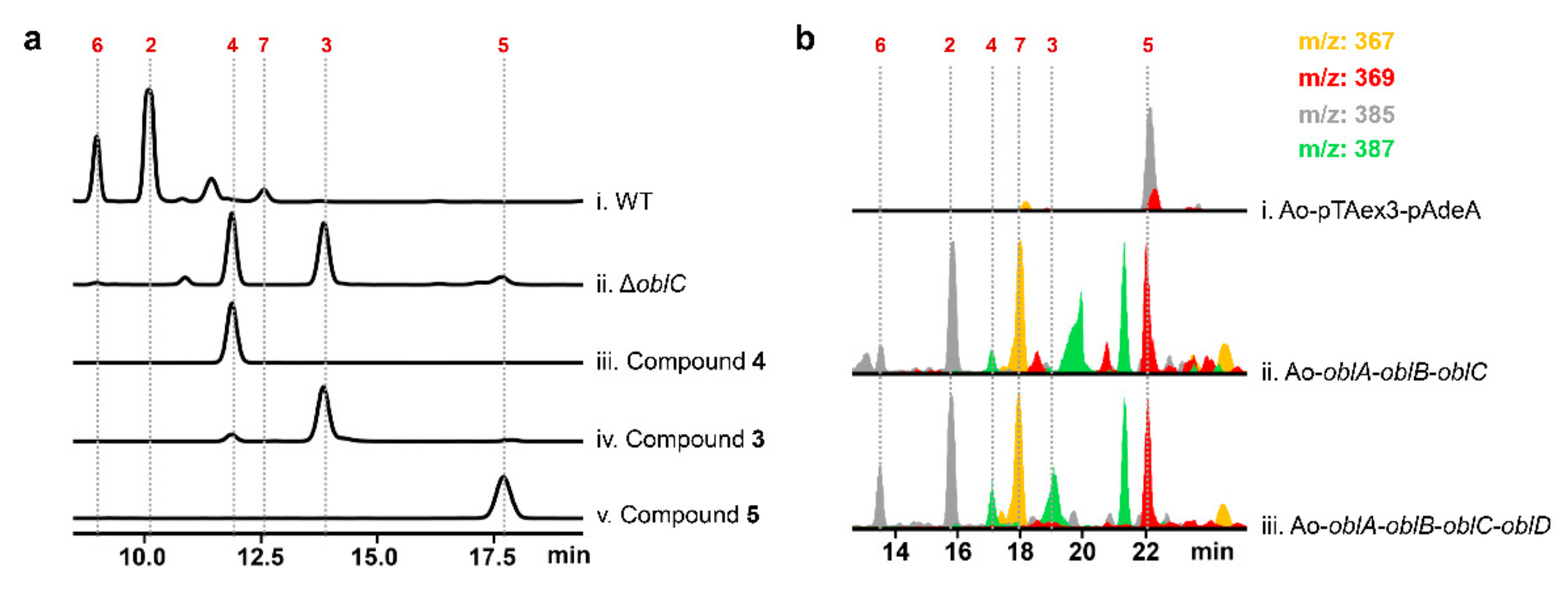
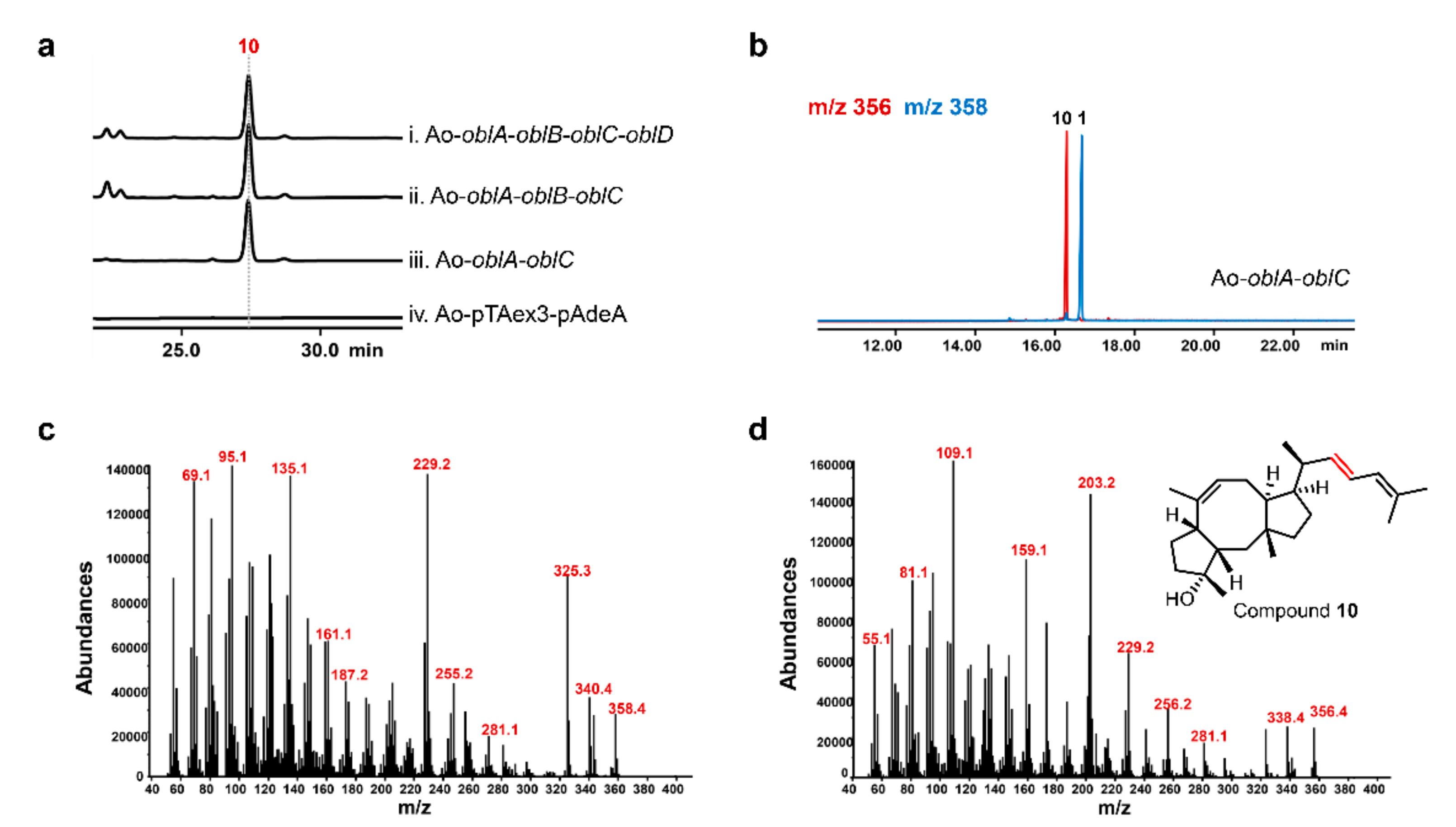
2.2. Unclustered FAD-Dependent Oxidoreductase OblCAu Is Involved in Ophiobolin Biosynthesis
2.3. OblCAu also Uses Ophiobolin F as the Substrate
2.4. Subcellular Localization Showed the Compartmentalize Biosynthesis of Ophiobolins
3. Discussion
4. Materials and Methods
4.1. General Experiential Materials
4.2. Strains and Culture Conditions
4.3. Software and Databases
4.4. Genomic DNA Extraction of A. ustus 094102
4.5. RNA Extraction of A. ustus 094102 and RNA Sequencing
4.6. qRT-PCR Analysis
4.7. Gene Inactivation in A. ustus 094102
4.8. Fermentation and Crude Extracts Analysis of Wild-Type Strain and Mutants
4.9. Heterologous Expression in A. oryzae
4.9.1. Construction of Plasmids
4.9.2. Protoplast Preparation and Transformation
4.9.3. Fermentation and Cultural Extracts Analysis of A. oryzae Transformants
4.10. Purification of Compounds 3–5 and 10
4.11. Feeding Experiments in ΔoblAAu Mutant
4.12. Florescence Observation
4.13. Inhibition Tests on A. oryzae
4.14. Cytotoxicity Assay
4.15. Homology Modeling and Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeng, T.; Liu, Z.; Zhuang, J.; Jiang, Y.; He, W.; Diao, H.; Lv, N.; Jian, Y.; Liang, D.; Qiu, Y.; et al. TeroKit: A Database-Driven Web Server for Terpenome Research. J. Chem. Inf. Model. 2020, 60, 2082–2090. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Li, D.S.; Ling, Y.; Liu, Y.C.; Zuo, Z.L.; Gan, L.S.; Luo, S.H.; Hua, J.; Chen, D.Y.; Xu, F.; et al. A cryptic plant terpene cyclase producing unconventional 18- and 14-membered macrocyclic C25 and C20 terpenoids with immunosuppressive activity. Angew. Chem. Int. Ed. Engl. 2021, 60, 25468–25476. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Huang, H.; Shao, C.; Xia, X.; Ma, L.; Huang, X.; Lu, Y.; Lin, Y.; Long, Y.; She, Z. Asperterpenols A and B, New Sesterterpenoids Isolated from a Mangrove Endophytic Fungus Aspergillus sp. 085242. Org. Lett. 2013, 15, 2522–2525. [Google Scholar] [CrossRef] [PubMed]
- Quan, Z.; Dickschat, J. S Biosynthetic gene cluster for asperterpenols A and B and the cyclization mechanism of asperterpenol A synthase. Org. Lett. 2020, 22, 7552–7555. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cai, Y.-S.; Cheng, F.; Yang, C.; Zhang, W.; Yu, W.; Yan, J.; Deng, Z.; Hong, K. Genome Mining Reveals a Multiproduct Sesterterpenoid Biosynthetic Gene Cluster in Aspergillus ustus. Org. Lett. 2021, 23, 1525–1529. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Deng, Z.; Hong, K. The Biological Activities of Sesterterpenoid-Type Ophiobolins. Mar. Drugs 2017, 15, 229. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Lu, Z.; Fan, J.; Wang, L.; Zhu, G.; Wang, Y.; Li, X.; Hong, K.; Piyachaturawat, P.; Chairoungdua, A.; et al. Ophiobolins from the Mangrove Fungus Aspergillus ustus. J. Nat. Prod. 2018, 81, 2–9. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Shen, L.; He, Y.; Liu, J.; Wang, J.; Hu, Z.; Zhang, Y. Bipolarolides A-G, Ophiobolin-Derived Sesterterpenes Representing Three New Carbon Skeletons from Bipolaris sp. TJ403-B1. Angew. Chem. Int. Ed. 2019, 58, 12091–12095. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Shen, L.; Hao, X.; Al Anbari, W.H.; Lin, S.; Li, H.; Gao, W.; Wang, J.; Hu, Z.; et al. Bipolaricins A–I, Ophiobolin-Type Tetracyclic Sesterterpenes from a Phytopathogenic Bipolaris sp. Fungus. J. Nat. Prod. 2019, 82, 2897–2906. [Google Scholar] [CrossRef]
- Liu, M.-T.; He, Y.; Shen, L.; Hu, Z.-X.; Zhang, Y.-H. Bipolarins A–H, eight new ophiobolin-type sesterterpenes with antimicrobial activity from fungus Bipolaris sp. TJ403-B1. Chin. J. Nat. Med. 2019, 17, 935–944. [Google Scholar] [CrossRef]
- Cai, R.; Jiang, H.; Mo, Y.; Guo, H.; Li, C.; Long, Y.; Zang, Z.; She, Z. Ophiobolin-Type Sesterterpenoids from the Mangrove Endophytic Fungus Aspergillus sp. ZJ-68. J. Nat. Prod. 2019, 82, 2268–2278. [Google Scholar] [CrossRef]
- Choi, B.-K.; Trinh, P.T.H.; Lee, H.-S.; Choi, B.-W.; Kang, J.S.; Ngoc, N.T.D.; Van, T.T.T.; Shin, H.J. New Ophiobolin Derivatives from the Marine Fungus Aspergillus flocculosus and Their Cytotoxicities against Cancer Cells. Mar. Drugs 2019, 17, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Liu, M.; He, Y.; Al Anbari, W.H.; Li, H.; Lin, S.; Chai, C.; Wang, J.; Hu, Z.; Zhang, Y. Novel Antimicrobial Compounds as Ophiobolin-Type Sesterterpenes and Pimarane-Type Diterpene from Bipolaris Species TJ403-B1. Front. Microbiol. 2020, 11, 856. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Tan, X.; Gu, L.; Liu, J.; Hao, X.; Tao, L.; Feng, H.; Cao, Y.; Shi, Z.; Duan, Y.; et al. New secondary metabolites with immunosuppressive activity from the phytopathogenic fungus Bipolaris maydis. Bioorg. Chem. 2020, 99, 103816. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Uvarani, C.; Wang, F.; Xue, Y.; Wu, N.; He, L.; Tian, D.; Chen, M.; Zhang, Y.; Hong, K.; et al. New ophiobolins from the deep-sea derived fungus Aspergillus sp. WHU0154 and their anti-inflammatory effects. Mar. Drugs. 2020, 18, 575. [Google Scholar] [CrossRef]
- Zatout, R.; Masi, M.; Sangermano, F.; Vurro, M.; Zonno, M.C.; Santoro, E.; Calabrò, V.; Superchi, S.; Evidente, A. Drophiobiolins A and B, Bioactive Ophiobolan Sestertepenoids Produced by Dreschslera gigantea. J. Nat. Prod. 2020, 83, 3387–3396. [Google Scholar] [CrossRef]
- Chi, L.-P.; Li, X.-M.; Wan, Y.-P.; Li, X.; Wang, B.-G. Ophiobolin Sesterterpenoids and Farnesylated Phthalide Derivatives from the Deep Sea Cold-Seep-Derived Fungus Aspergillus insuetus SD-512. J. Nat. Prod. 2020, 83, 3652–3660. [Google Scholar] [CrossRef]
- Fang, S.-T.; Liu, X.-H.; Yan, B.-F.; Miao, F.-P.; Yin, X.-L.; Li, W.-Z.; Ji, N.-Y. Terpenoids from the Marine-Derived Fungus Aspergillus sp. RR-YLW-12, Associated with the Red Alga Rhodomela confervoides. J. Nat. Prod. 2021, 84, 1763–1771. [Google Scholar] [CrossRef]
- Bladt, T.T.; Dürr, C.; Knudsen, P.B.; Kildgaard, S.; Frisvad, J.C.; Gotfredsen, C.H.; Seiffert, M.; Larsen, T.O. Bio-Activity and Dereplication-Based Discovery of Ophiobolins and Other Fungal Secondary Metabolites Targeting Leukemia Cells. Molecules 2013, 18, 14629–14650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Krasnoff, S.B.; Lu, S.-W.; Dunbar, C.D.; O’Neal, J.; Turgeon, B.G.; Yoder, O.C.; Gibson, D.M.; Hamann, M.T. Characterization of 6-epi-3-Anhydroophiobolin B from Cochliobolus heterostrophus. J. Nat. Prod. 1999, 62, 895–897. [Google Scholar] [CrossRef]
- Yang, T.; Lu, Z.; Meng, L.; Wei, S.; Hong, K.; Zhu, W.; Huang, C. The novel agent ophiobolin O induces apoptosis and cell cycle arrest of MCF-7 cells through activation of MAPK signaling pathways. Bioorg. Med. Chem. Lett. 2012, 22, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lv, C.; Zhu, T.; Yang, X.; Wei, S.; Sun, J.; Hong, K.; Zhu, W.; Huang, C. Ophiobolin-O reverses adriamycin resistance via cell cycle arrest and apoptosis sensitization in adriamycin-resistant human breast carcinoma (MCF-7/ADR) cells. Mar. Drugs. 2013, 11, 4570–4584. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhao, C.; Lu, J.; Wu, J.; Li, C.; Hu, Z.; Tian, W.; Yang, L.; Xiang, J.; Zhou, H.; et al. Sesterterpene MHO7 suppresses breast cancer cells as a novel estrogen receptor degrader. Pharmacol. Res. 2019, 146, 104294. [Google Scholar] [CrossRef]
- Tsuna, K.; Noguchi, N.; Nakada, M. Enantioselective Total Synthesis of (+)-Ophiobolin A. Chem. A Eur. J. 2013, 19, 5476–5486. [Google Scholar] [CrossRef]
- Brill, Z.G.; Grover, H.K.; Maimone, T.J. Enantioselective synthesis of an ophiobolin sesterterpene via a programmed radical cascade. Science 2016, 352, 1078–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thach, D.Q.; Brill, Z.G.; Grover, H.K.; Esguerra, K.V.; Thompson, J.K.; Maimone, T.J. Total synthesis of (+)-6-epi-ophiobolin A. Angew. Chem. Int. Ed. Engl. 2020, 59, 1532–1536. [Google Scholar] [CrossRef] [PubMed]
- Chiba, R.; Minami, A.; Gomi, K.; Oikawa, H. Identification of Ophiobolin F Synthase by a Genome Mining Approach: A Sesterterpene Synthase from Aspergillus clavatus. Org. Lett. 2013, 15, 594–597. [Google Scholar] [CrossRef]
- Narita, K.; Chiba, R.; Minami, A.; Kodama, M.; Fujii, I.; Gomi, K.; Oikawa, H. Multiple Oxidative Modifications in the Ophiobolin Biosynthesis: P450 Oxidations Found in Genome Mining. Org. Lett. 2016, 18, 1980–1983. [Google Scholar] [CrossRef]
- Chai, H.; Yin, R.; Liu, Y.; Meng, H.; Zhou, X.; Zhou, G.; Bi, X.; Yang, X.; Zhu, T.; Zhu, W.; et al. Sesterterpene ophiobolin biosynthesis involving multiple gene clusters in Aspergillus ustus. Sci. Rep. 2016, 6, 27181. [Google Scholar] [CrossRef] [Green Version]
- Mai, W.Y.; Hong, K. Heterologous expression of a fungal cytochrome P450 in Escherichia coli. Microbiol. China 2019, 46, 1092–1099. [Google Scholar]
- Yan, J.; Guo, J.; Yuan, W.; Mai, W.; Hong, K. Identification of Enzymes Involved in Sesterterpene Biosynthesis in Marine Fungi. Methods Enzymol. 2018, 604, 441–498. [Google Scholar] [CrossRef]
- Fujii, R.; Minami, A.; Tsukagoshi, T.; Sato, N.; Sahara, T.; Ohgiya, S.; Gomi, K.; Oikawa, H. Total biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase alpha: Heterologous expression of four biosynthetic genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2011, 75, 1813–1817. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Itoh, T.; Kinoshita, M.; Nakai, Y.; Kurotaki, M.; Kobayashi, M. Cytotoxic sesterterpenes, 6-epi-ophiobolin G and 6-epi-ophiobolin N, from marine derived fungus Emericella variecolor GF10. Tetrahedron 2004, 60, 6015–6019. [Google Scholar] [CrossRef]
- Ninkuu, V.; Zhang, L.; Yan, J.; Fu, Z.; Yang, T.; Zeng, H. Biochemistry of Terpenes and Recent Advances in Plant Protection. Int. J. Mol. Sci. 2021, 22, 5710. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hoefgen, S.; Valiante, V. Biosynthesis of fungal drimane-type sesquiterpene esters. Angew. Chem. Int. Ed. Engl. 2021, 60, 23763–23770. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Zhang, Y.; Guo, C.; Yang, Y.; Han, J.; Yu, B.; Yin, W.; Liu, H. Reconstitution of biosynthetic pathway for mushroom-derived cyathane diterpenes in yeast and generation of new “non-natural” analogues. Acta Pharm. Sin. B 2021, 11, 2945–2956. [Google Scholar] [CrossRef]
- Zhang, W.; Du, L.; Qu, Z.; Zhang, X.; Li, F.; Li, Z.; Qi, F.; Wang, X.; Jiang, Y.; Men, P.; et al. Compartmentalized biosynthesis of mycophenolic acid. Proc. Natl. Acad. Sci. USA 2019, 116, 13305–13310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.-J.; Yu, W.-L.; Yang, L.; Xie, B.-H.; Qin, K.-M.; Yin, Y.-P.; Yan, J.-J.; Gong, S.; Liu, T.-Y.; Zhou, H.-B.; et al. Design and synthesis of marine sesterterpene analogues as novel estrogen receptor α degraders for breast cancer treatment. Eur. J. Med. Chem. 2021, 229, 114081. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA Using TRIzol (TRI Reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb-prot5439. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Lv, S.; Chen, L.; Zhao, Y.; Deng, Z.; Hong, K. Production of sesterterpene ophiobolin by a bifunctional terpene synthase in Escherichia coli. Appl. Microbiol. Biotechnol. 2019, 103, 8785–8797. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| MCF-7 | MDA-MB-231 | MCF-7/ADR | |
|---|---|---|---|
| 3 | 4.88 ± 0.45 | 1.92 ± 0.67 | 0.77 ± 0.08 |
| 4 | 3.73 ± 0.16 | 1.52 ± 0.21 | 2.71 ± 0.31 |
| 5 | 3.64 ± 0.34 | 2.26 ± 0.15 | 1.82 ± 0.03 |
| 10 | >10 | 7.32 ± 0.82 | >20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, J.; Pang, J.; Liang, J.; Yu, W.; Liao, X.; Aobulikasimu, A.; Yi, X.; Yin, Y.; Deng, Z.; Hong, K. The Biosynthesis and Transport of Ophiobolins in Aspergillus ustus 094102. Int. J. Mol. Sci. 2022, 23, 1903. https://doi.org/10.3390/ijms23031903
Yan J, Pang J, Liang J, Yu W, Liao X, Aobulikasimu A, Yi X, Yin Y, Deng Z, Hong K. The Biosynthesis and Transport of Ophiobolins in Aspergillus ustus 094102. International Journal of Molecular Sciences. 2022; 23(3):1903. https://doi.org/10.3390/ijms23031903
Chicago/Turabian StyleYan, Jingjing, Jiamin Pang, Jianjia Liang, Wulin Yu, Xuequn Liao, Ayikaimaier Aobulikasimu, Xinrui Yi, Yapeng Yin, Zixin Deng, and Kui Hong. 2022. "The Biosynthesis and Transport of Ophiobolins in Aspergillus ustus 094102" International Journal of Molecular Sciences 23, no. 3: 1903. https://doi.org/10.3390/ijms23031903
APA StyleYan, J., Pang, J., Liang, J., Yu, W., Liao, X., Aobulikasimu, A., Yi, X., Yin, Y., Deng, Z., & Hong, K. (2022). The Biosynthesis and Transport of Ophiobolins in Aspergillus ustus 094102. International Journal of Molecular Sciences, 23(3), 1903. https://doi.org/10.3390/ijms23031903