GFP–Margatoxin, a Genetically Encoded Fluorescent Ligand to Probe Affinity of Kv1.3 Channel Blockers
Abstract
1. Introduction
2. Results and Discussion
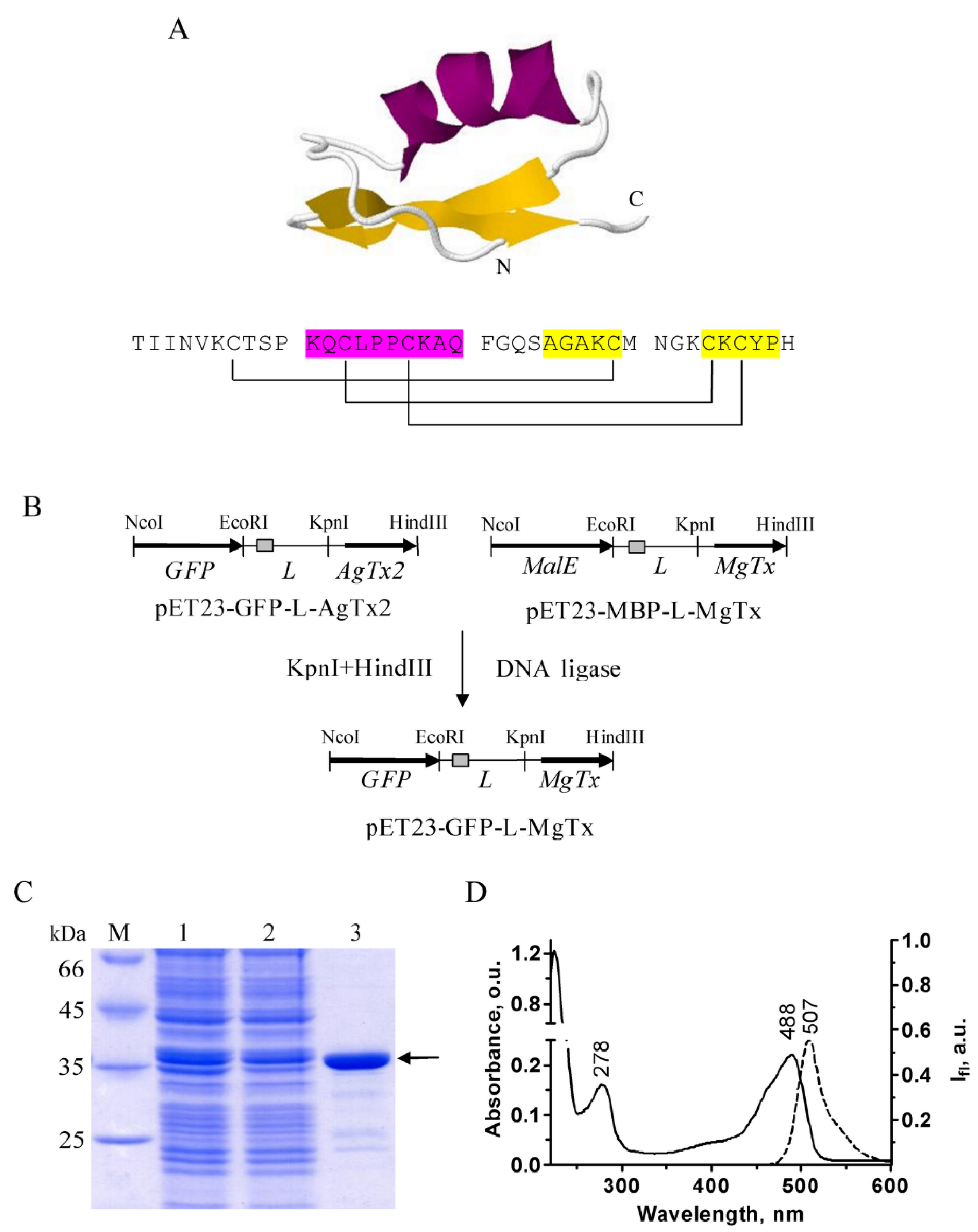
2.1. Design and Properties of GFP–MgTx
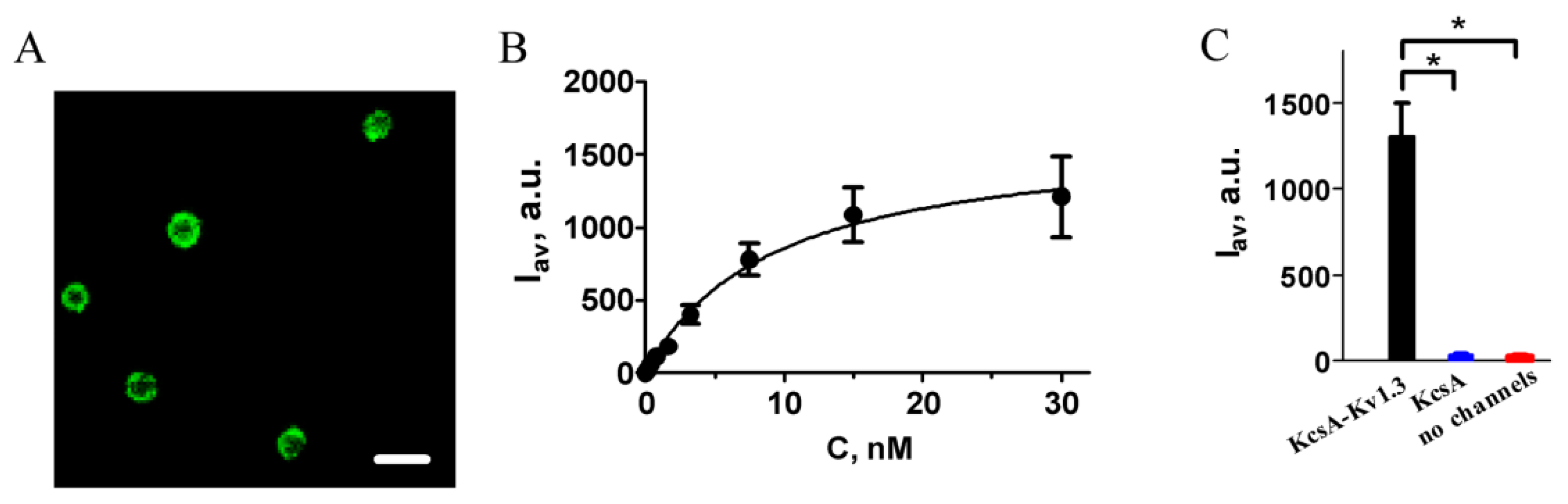
2.2. GFP–MgTx Interaction with the Kv1.3 Binding Site
2.3. GFP–MgTx as a Fluorescent Probe to Recognize Kv1.3 Blockers
3. Materials and Methods
3.1. Reagents and Cells
3.2. Gene Cloning, Expression and Purification of GFP–MgTx
3.3. Recombinant Toxins
3.4. Preparation of Spheroplasts and Binding Protocol
3.5. Microscopy and Ligand–Channel Interaction Analysis
3.6. Optical Spectroscopy
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AgTx2 | agitoxin 2 |
| GFP | green fluorescent protein |
| MgTx | margatoxin |
| HgTx1 | hongotoxin 1 |
| ChTx | charybdotoxin |
| ScyTx | scyllatoxin |
| TEA | tetraethylammonium |
| IC50 | a concentration of a competing ligand that displaces 50% of the fluorescent probe from the complex with the receptor |
| Kd | dissociation constant of GFP–MgTx |
| Kap | apparent dissociation constant of non-labeled ligand determined in the competitive binding experiment |
| Iav | average fluorescence intensity of GFP–MgTx bound at a surface of a spheroplast expressing KcsA–Kv1.3 |
| Iavs | Iav value at saturated binding of GFP–MgTx |
| Im | Iav value at zero concentration of a competing ligand in the competitive binding experiment |
References
- Beeton, C.; Wulff, H.; Barbaria, J.; Clot-Faybesse, O.; Pennington, M.; Bernard, D.; Cahalan, M.D.; Chandy, K.G.; Béraud, E. Selective blockade of T lymphocyte K(+) channels ameliorates experimental autoimmune encephalomyelitis, a model for multiple sclerosis. Proc. Natl. Acad. Sci. USA 2001, 98, 13942–13947. [Google Scholar] [CrossRef] [PubMed]
- Wulff, H.; Beeton, C.; Chandy, K.G. Potassium channels as therapeutic targets for autoimmune disorders. Curr. Opin. Drug Discov. Devel. 2003, 6, 640–647. [Google Scholar] [PubMed]
- Fomina, A.F.; Nguyen, H.M.; Wulff, H. Kv1.3 inhibition attenuates neuroinflammation through disruption of microglial calcium signaling. Channels 2021, 15, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Tajti, G.; Panyi, G. The Kv1.3 K + channel in the immune system and its “precision pharmacology” using peptide toxins. Biol. Futur. 2021, 72, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lu, G.; Chiang, E.Y.; Chernov-Rogan, T.; Grogan, J.L.; Chen, J. High-throughput electrophysiological assays for voltage gated ion channels using SyncroPatch 768PE. PLoS ONE 2017, 12, e0180154. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jurman, M.E.; Yellen, G. Dynamic Rearrangement of the Outer Mouth of a K+ Channel during Gating. Neuron 1996, 16, 859–867. [Google Scholar] [CrossRef]
- Linley, J.E. Perforated whole-cell patch-clamp recording. Methods Mol. Biol. 2013, 998, 149–157. [Google Scholar] [CrossRef]
- Ader, C.; Schneider, R.; Hornig, S.; Velisetty, P.; Wilson, E.M.; Lange, A.; Giller, K.; Ohmert, I.; Martin-Eauclaire, M.F.; Trauner, D.; et al. A structural link between inactivation and block of a K+ channel. Nat. Struct. Mol. Biol. 2008, 15, 605–612. [Google Scholar] [CrossRef]
- Dunlop, J.; Bowlby, M.; Peri, R.; Tawa, G.; LaRocque, J.; Soloveva, V.; Morin, J. Ion channel screening. Comb. Chem. High Throughput Screen. 2008, 11, 514–522. [Google Scholar] [CrossRef]
- Wolff, C.; Fuks, B.; Chatelain, P. Comparative study of membrane potential-sensitive fluorescent probes and their use in ion channel screening assays. J. Biomol. Screen. 2003, 8, 533–543. [Google Scholar] [CrossRef]
- Slack, M.; Kirchhoff, C.; Smoller, C.; Winkler, D.; Netzer, R. Identification of novel Kv1.3 blockers using a fluorescent cell-based ion channel assay. J. Biomol. Screen. 2006, 11, 57–64. [Google Scholar] [CrossRef]
- Liu, K.; Samuel, M.; Tillett, J.; Hennan, J.K.; Mekonnen, B.; Soloveva, V.; Harrison, R.K.; Paslay, J.W.; Larocque, J. High-throughput screening for Kv1.3 channel blockers using an improved FLIPR-based membrane-potential assay. J. Biomol. Screen. 2010, 15, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Beacham, D.W.; Blackmer, T.; O’Grady, M.; Hanson, G.T. Cell-based potassium ion channel screening using the FluxOR assay. J. Biomol. Screen. 2010, 15, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Gaston, J.P.; Baker, M.A.B. Fluorescence Approaches for Characterizing Ion Channels in Synthetic Bilayers. Membranes 2021, 11, 857. [Google Scholar] [CrossRef] [PubMed]
- Pragl, B.; Koschak, A.; Trieb, M.; Obermair, G.; Kaufmann, W.A.; Gerster, U.; Blanc, E.; Hahn, C.; Prinz, H.; Schütz, G.; et al. Synthesis, Characterization, and Application of Cy-Dye- and Alexa-Dye-Labeled Hongotoxin 1 Analogues. The First High Affinity Fluorescence Probes for Voltage-Gated K + Channels. Bioconjug. Chem. 2002, 13, 416–425. [Google Scholar] [CrossRef]
- Beeton, C.; Wulff, H.; Singh, S.; Botsko, S.; Crossley, G.; Gutman, G.A.; Cahalan, M.D.; Pennington, M.; Chandy, K.G. A novel fluorescent toxin to detect and investigate Kv1.3 channel up-regulation in chronically activated T lymphocytes. J. Biol. Chem. 2003, 278, 9928–9937. [Google Scholar] [CrossRef]
- Kuzmenkov, A.I.; Vassilevski, A.A.; Kudryashova, K.S.; Nekrasova, O.V.; Peigneur, S.; Tytgat, J.; Feofanov, A.V.; Kirpichnikov, M.P.; Grishin, E.V. Variability of potassium channel blockers in Mesobuthus eupeus scorpion venom with focus on Kv1.1: An integrated transcriptomic and proteomic study. J. Biol. Chem. 2015, 290, 12195–12209. [Google Scholar] [CrossRef]
- Kuzmenkov, A.I.; Nekrasova, O.V.; Kudryashova, K.S.; Peigneur, S.; Tytgat, J.; Stepanov, A.V.; Kirpichnikov, M.P.; Grishin, E.V.; Feofanov, A.V.; Vassilevski, A.A. Fluorescent protein-scorpion toxin chimera is a convenient molecular tool for studies of potassium channels. Sci. Rep. 2016, 6, 33314. [Google Scholar] [CrossRef]
- Nekrasova, O.V.; Primak, A.L.; Ignatova, A.A.; Novoseletsky, V.N.; Geras’kina, O.V.; Kudryashova, K.S.; Yakimov, S.A.; Kirpichnikov, M.P.; Arseniev, A.S.; Feofanov, A.V. N-Terminal Tagging with GFP Enhances Selectivity of Agitoxin 2 to Kv1.3-Channel Binding Site. Toxins 2020, 12, 802. [Google Scholar] [CrossRef]
- Bartok, A.; Toth, A.; Somodi, S.; Szanto, T.G.; Hajdu, P.; Panyi, G.; Varga, Z. Margatoxin is a non-selective inhibitor of human Kv1.3 K+ channels. Toxicon 2014, 87, 6–16. [Google Scholar] [CrossRef]
- Ramirez-Navarro, A.; Glazebrook, P.A.; Kane-Sutton, M.; Padro, C.; Kline, D.D.; Kunze, D.L. Kv1.3 channels regulate synaptic transmission in the nucleus of solitary tract. J. Neurophysiol. 2011, 105, 2772–2780. [Google Scholar] [CrossRef] [PubMed]
- Meneses, D.; Vega, A.V.; Torres-Cruz, F.M.; Barral, J. KV1 and KV3 Potassium Channels Identified at Presynaptic Terminals of the Corticostriatal Synapses in Rat. Neural Plast. 2016, 2016, 8782518. [Google Scholar] [CrossRef] [PubMed]
- Al Koborssy, D.; Palouzier-Paulignan, B.; Canova, V.; Thevenet, M.; Fadool, D.A.; Julliard, A.K. Modulation of olfactory-driven behavior by metabolic signals: Role of the piriform cortex. Brain Struct. Funct. 2019, 224, 315–336. [Google Scholar] [CrossRef] [PubMed]
- Doczi, M.A.; Morielli, A.D.; Damon, D.H. Kv1.3 channels in postganglionic sympathetic neurons: Expression, function, and modulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R733–R740. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, S.N.; Chen, B.S.; Lo, Y.C. Evidence for aconitine-induced inhibition of delayed rectifier K(+) current in Jurkat T-lymphocytes. Toxicology 2011, 289, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Pennington, M.; Jiang, Q.; Whartenby, K.A.; Calabresi, P.A. Characterization of the functional properties of the voltage-gated potassium channel Kv1.3 in human CD4+ T lymphocytes. J. Immunol. 2007, 179, 4563–4570. [Google Scholar] [CrossRef] [PubMed]
- Koo, G.C.; Blake, J.T.; Talento, A.; Nguyen, M.; Lin, S.; Sirotina, A.; Shah, K.; Mulvany, K.; Hora, D.; Cunningham, P.; et al. Blockade of the voltage-gated potassium channel Kv1.3 inhibits immune responses in vivo. J. Immunol. 1997, 158, 5120–5128. [Google Scholar] [PubMed]
- Wang, T.; Lee, M.H.; Johnson, T.; Allie, R.; Hu, L.; Calabresi, P.A.; Nath, A. Activated T-cells inhibit neurogenesis by releasing granzyme B: Rescue by Kv1.3 blockers. J. Neurosci. 2010, 30, 5020–5027. [Google Scholar] [CrossRef]
- Charolidi, N.; Schilling, T.; Eder, C. Microglial Kv1.3 Channels and P2Y12 Receptors Differentially Regulate Cytokine and Chemokine Release from Brain Slices of Young Adult and Aged Mice. PLoS ONE 2015, 10, e0128463. [Google Scholar] [CrossRef] [PubMed]
- Schilling, T.; Eder, C. Amyloid-β-induced reactive oxygen species production and priming are differentially regulated by ion channels in microglia. J. Cell. Physiol. 2011, 226, 3295–3302. [Google Scholar] [CrossRef]
- Legros, C.; Pollmann, V.; Knaus, H.G.; Farrell, A.M.; Darbon, H.; Bougis, P.E.; Martin-Eauclaire, M.F.; Pongs, O. Generating a high affinity scorpion toxin receptor in KcsA-Kv1.3 chimeric potassium channels. J. Biol. Chem. 2000, 275, 16918–16924. [Google Scholar] [CrossRef] [PubMed]
- Kudryashova, K.S.; Nekrasova, O.V.; Kuzmenkov, A.I.; Vassilevski, A.A.; Ignatova, A.A.; Korolkova, Y.V.; Grishin, E.V.; Kirpichnikov, M.P.; Feofanov, A. V Fluorescent system based on bacterial expression of hybrid KcsA channels designed for Kv1.3 ligand screening and study. Anal. Bioanal. Chem. 2013, 405, 2379–2389. [Google Scholar] [CrossRef]
- Kudryashova, K.S.; Nekrasova, O.V.; Kirpichnikov, M.P.; Feofanov, A.V. Chimeras of KcsA and Kv1 as a bioengineering tool to study voltage-gated potassium channels and their ligands. Biochem. Pharmacol. 2021, 190, 114646. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Stevens, S.P.; Williamson, J.M. Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry 1994, 33, 15061–15070. [Google Scholar] [CrossRef] [PubMed]
- Dauplais, M.; Gilquin, B.; Possani, L.D.; Gurrola-Briones, G.; Roumestand, C.; Menez, A. Determination of the three-dimensional solution structure of noxiustoxin: Analysis of structural differences with related short-chain scorpion toxins. Biochemistry 1995, 34, 16563–16573. [Google Scholar] [CrossRef] [PubMed]
- Nekrasova, O.; Kudryashova, K.; Fradkov, A.; Yakimov, S.; Savelieva, M.; Kirpichnikov, M.; Feofanov, A. Straightforward approach to produce recombinant scorpion toxins—Pore blockers of potassium channels. J. Biotechnol. 2017, 241, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Savelieva, M.V.; Kudryashova, K.; Nekrasova, O.V.; Feofanov, A.V. Fluorescent Ligands on the Basis of Hongotoxin 1: eGFP-Hongotoxin 1. Microsc. Microanal. 2019, 25, 1262–1263. [Google Scholar] [CrossRef]
- Helms, L.M.H.; Felix, J.P.; Bugianesi, R.M.; Garcia, M.L.; Stevens, S.; Leonard, R.J.; Knaus, H.G.; Koch, R.; Wanner, S.G.; Kaczorowski, G.J.; et al. Margatoxin binds to a homomultimer of K(V)1.3 channels in Jurkat cells. Comparison with K(V)1.3 expressed in CHO cells. Biochemistry 1997, 36, 3737–3744. [Google Scholar] [CrossRef]
- Shieh, C.C.; Kirsch, G.E. Mutational analysis of ion conduction and drug binding sites in the inner mouth of voltage-gated K+ channels. Biophys. J. 1994, 67, 2316–2325. [Google Scholar] [CrossRef]
- Andalib, P.; Consiglio, J.F.; Trapani, J.G.; Korn, S.J. The external TEA binding site and C-type inactivation in voltage-gated potassium channels. Biophys. J. 2004, 87, 3148–3161. [Google Scholar] [CrossRef][Green Version]
- Shakkottai, V.G.; Regaya, I.; Wulff, H.; Fajloun, Z.; Tomita, H.; Fathallah, M.; Cahalan, M.D.; Gargus, J.J.; Sabatier, J.M.; Chandy, K.G. Design and characterization of a highly selective peptide inhibitor of the small conductance calcium-activated K+ channel, SkCa2. J. Biol. Chem. 2001, 276, 43145–43151. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.L.; Garcia-Calvo, M.; Hidalgo, P.; Lee, A.; MacKinnon, R. Purification and Characterization of Three Inhibitors of Voltage-Dependent K+ Channels from Leiurus quinquestriatus var. hebraeus Venom. Biochemistry 1994, 33, 6834–6839. [Google Scholar] [CrossRef] [PubMed]
- Koschak, A.; Bugianesi, R.M.; Mitterdorfer, J.; Kaczorowski, G.J.; Garcia, M.L.; Knaus, H.G. Subunit composition of brain voltage-gated potassium channels determined by hongotoxin-1, a novel peptide derived from Centruroides limbatus venom. J. Biol. Chem. 1998, 273, 2639–2644. [Google Scholar] [CrossRef] [PubMed]
- Nekrasova, O.V.; Ignatova, A.A.; Nazarova, A.I.; Feofanov, A.V.; Korolkova, Y.V.; Boldyreva, E.F.; Tagvei, A.I.; Grishin, E.V.; Arseniev, A.S.; Kirpichnikov, M.P. Recombinant Kv channels at the membrane of escherichia coli bind specifically agitoxin2. J. Neuroimmune Pharmacol. 2009, 4, 83–91. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denisova, K.R.; Orlov, N.A.; Yakimov, S.A.; Kryukova, E.A.; Dolgikh, D.A.; Kirpichnikov, M.P.; Feofanov, A.V.; Nekrasova, O.V. GFP–Margatoxin, a Genetically Encoded Fluorescent Ligand to Probe Affinity of Kv1.3 Channel Blockers. Int. J. Mol. Sci. 2022, 23, 1724. https://doi.org/10.3390/ijms23031724
Denisova KR, Orlov NA, Yakimov SA, Kryukova EA, Dolgikh DA, Kirpichnikov MP, Feofanov AV, Nekrasova OV. GFP–Margatoxin, a Genetically Encoded Fluorescent Ligand to Probe Affinity of Kv1.3 Channel Blockers. International Journal of Molecular Sciences. 2022; 23(3):1724. https://doi.org/10.3390/ijms23031724
Chicago/Turabian StyleDenisova, Kristina R., Nikita A. Orlov, Sergey A. Yakimov, Elena A. Kryukova, Dmitry A. Dolgikh, Mikhail P. Kirpichnikov, Alexey V. Feofanov, and Oksana V. Nekrasova. 2022. "GFP–Margatoxin, a Genetically Encoded Fluorescent Ligand to Probe Affinity of Kv1.3 Channel Blockers" International Journal of Molecular Sciences 23, no. 3: 1724. https://doi.org/10.3390/ijms23031724
APA StyleDenisova, K. R., Orlov, N. A., Yakimov, S. A., Kryukova, E. A., Dolgikh, D. A., Kirpichnikov, M. P., Feofanov, A. V., & Nekrasova, O. V. (2022). GFP–Margatoxin, a Genetically Encoded Fluorescent Ligand to Probe Affinity of Kv1.3 Channel Blockers. International Journal of Molecular Sciences, 23(3), 1724. https://doi.org/10.3390/ijms23031724







