Evaluation of Controlled Ovarian Stimulation Protocols in Patients with Normal and Low Ovarian Reserve: Analyses of miRNAs and Selected Target Genes Involved in the Proliferation of Human Cumulus Cells and Oocyte Quality
Abstract
:1. Introduction
2. Results
2.1. Patients’ Data
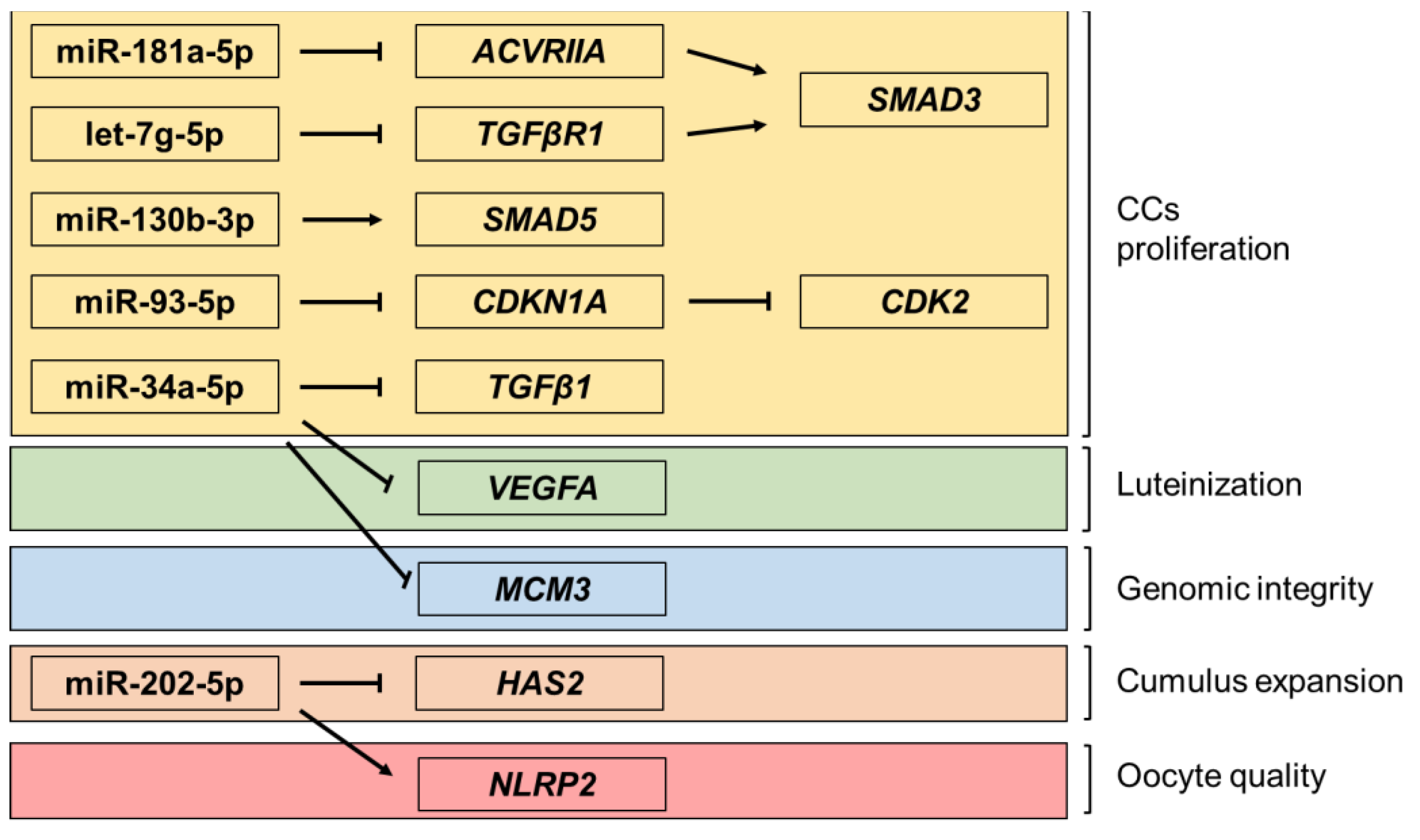
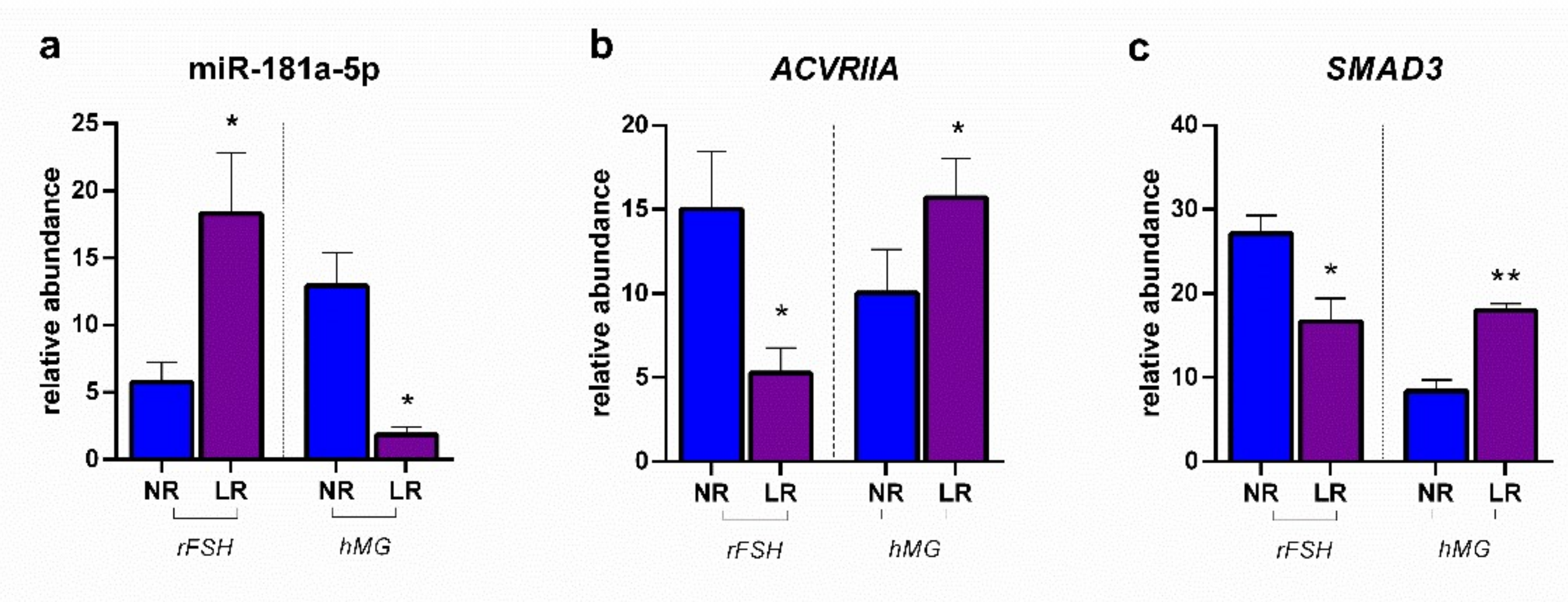
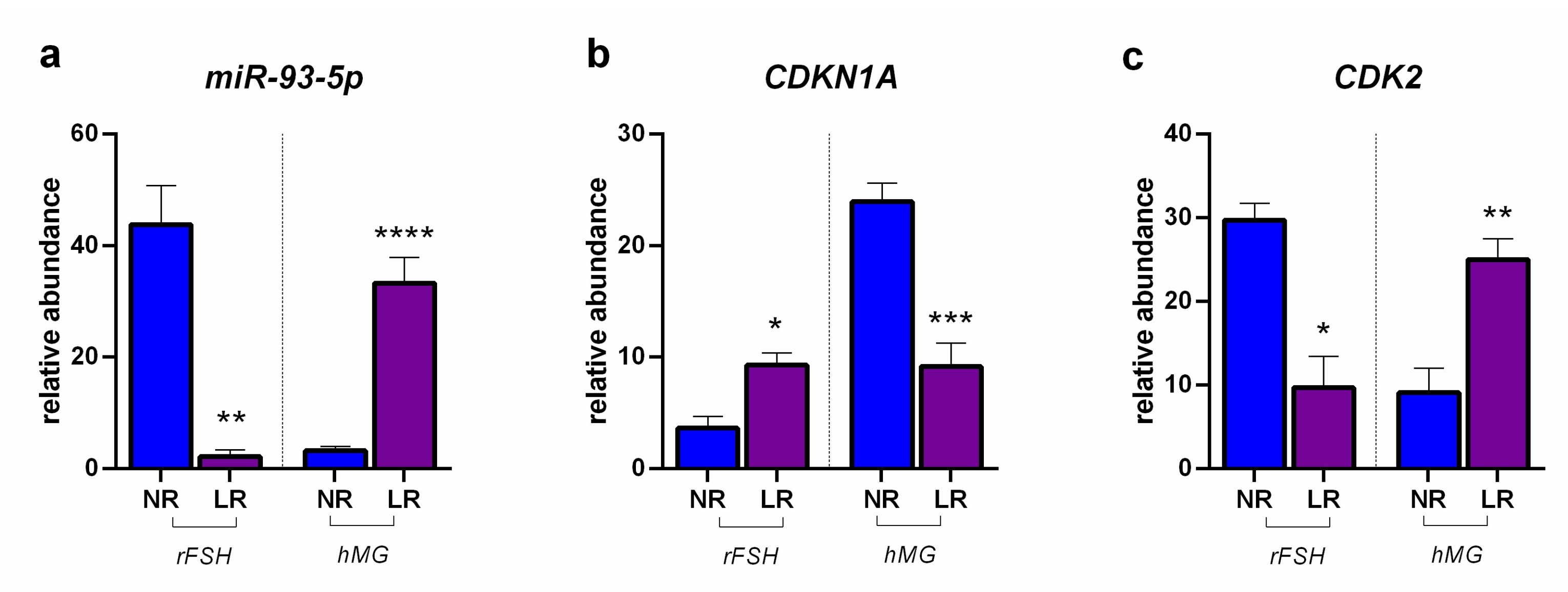
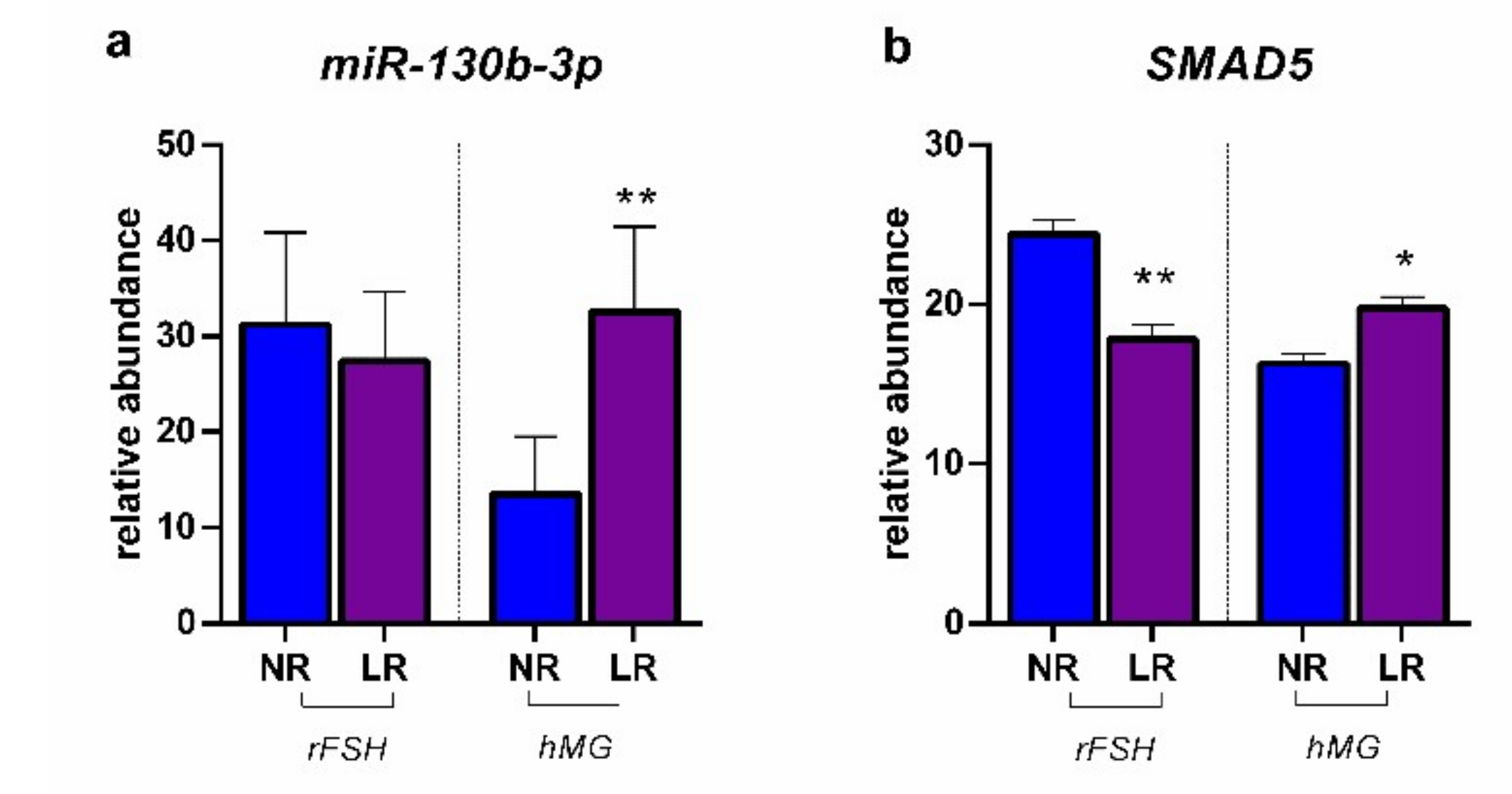
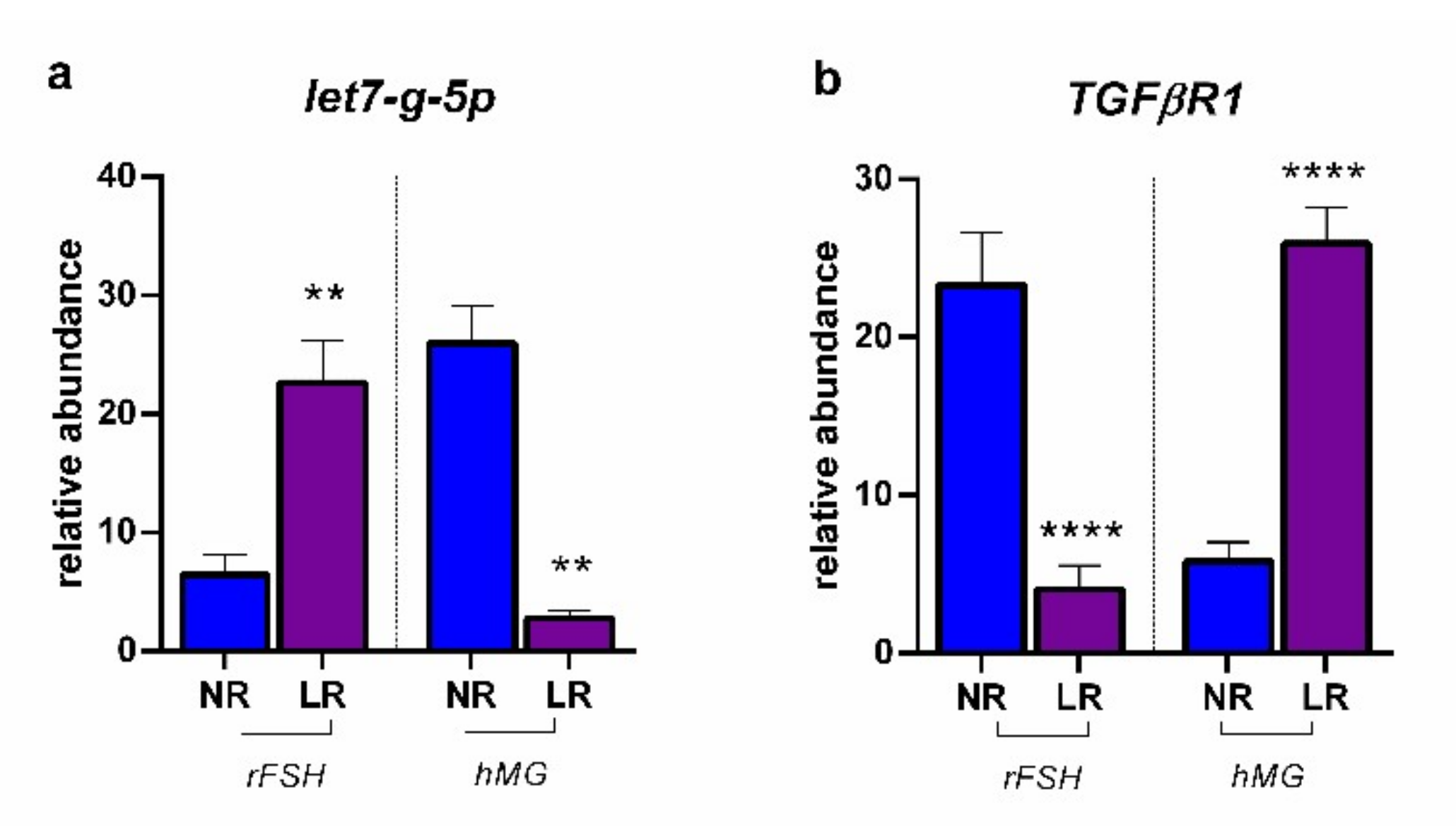
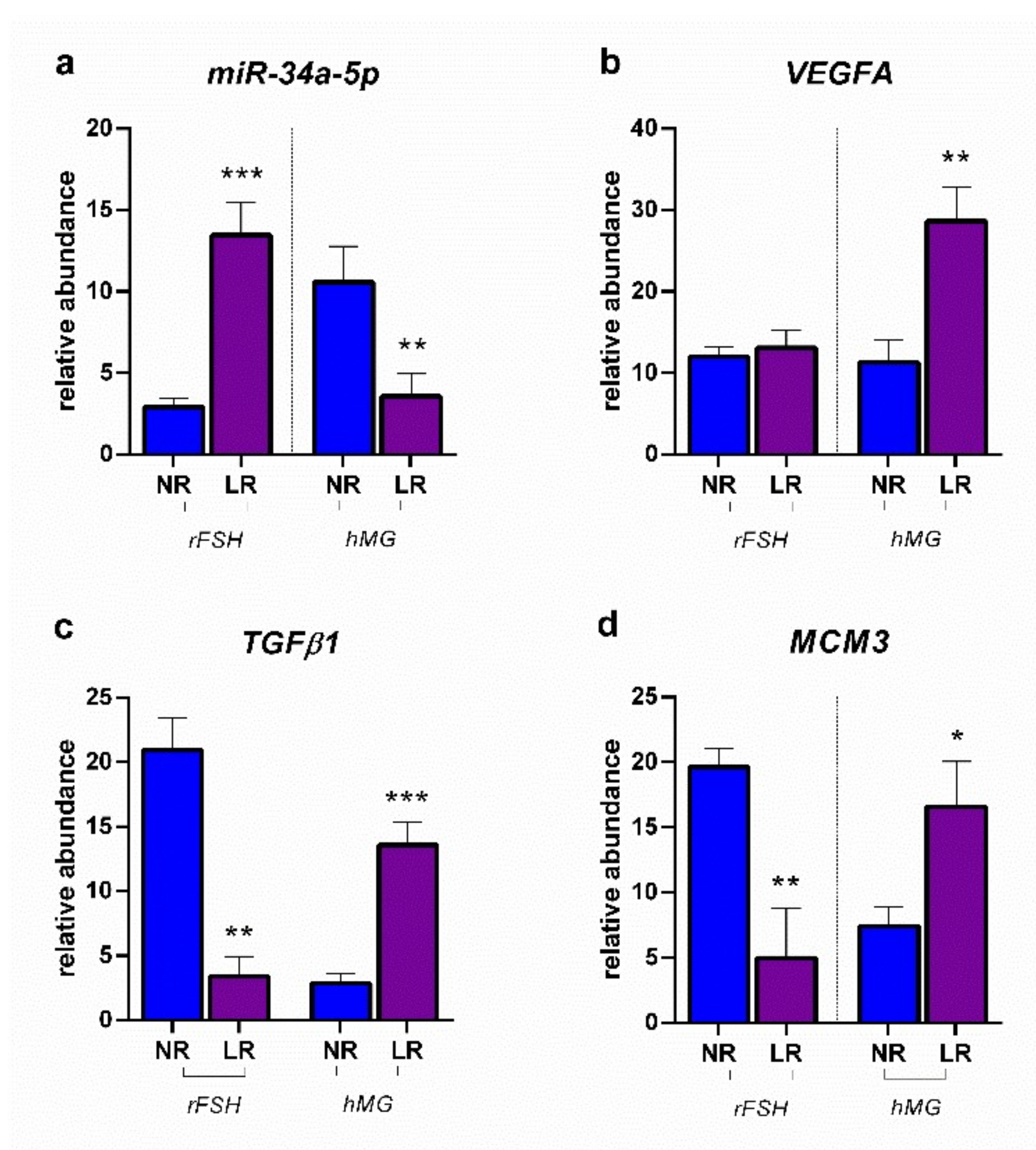
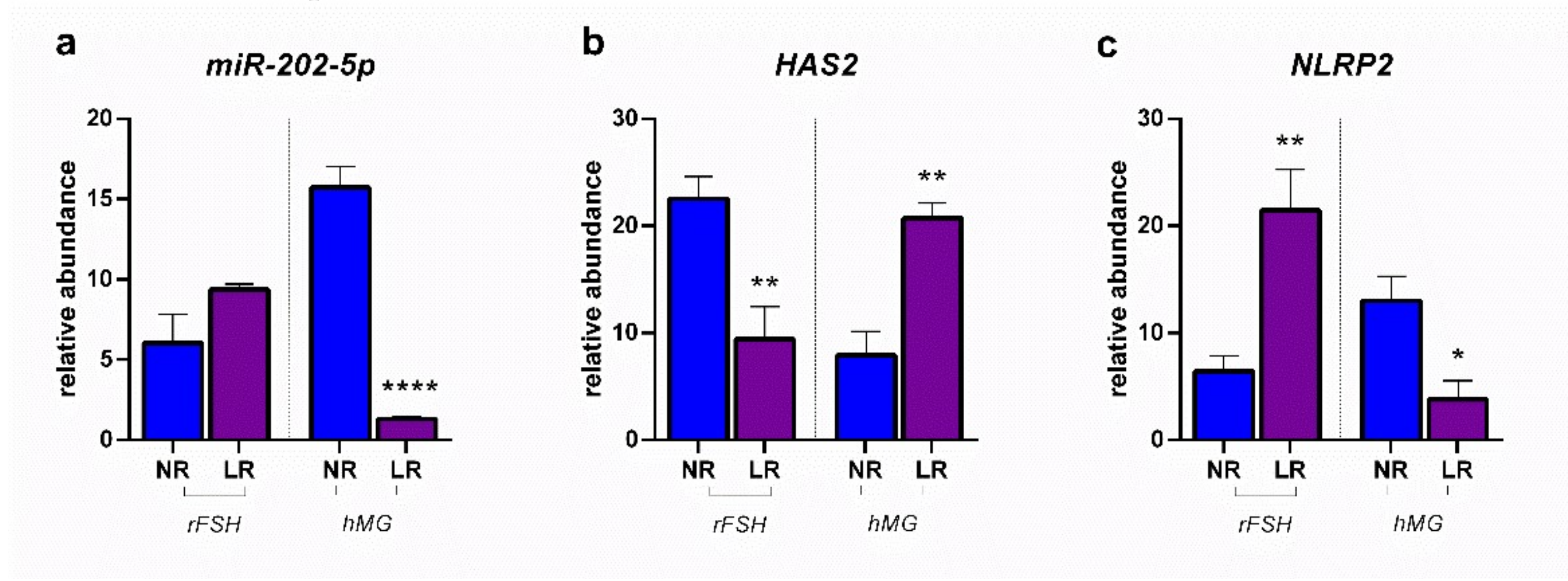
2.2. miRNA Expression and Gene Target Evaluation in CCs
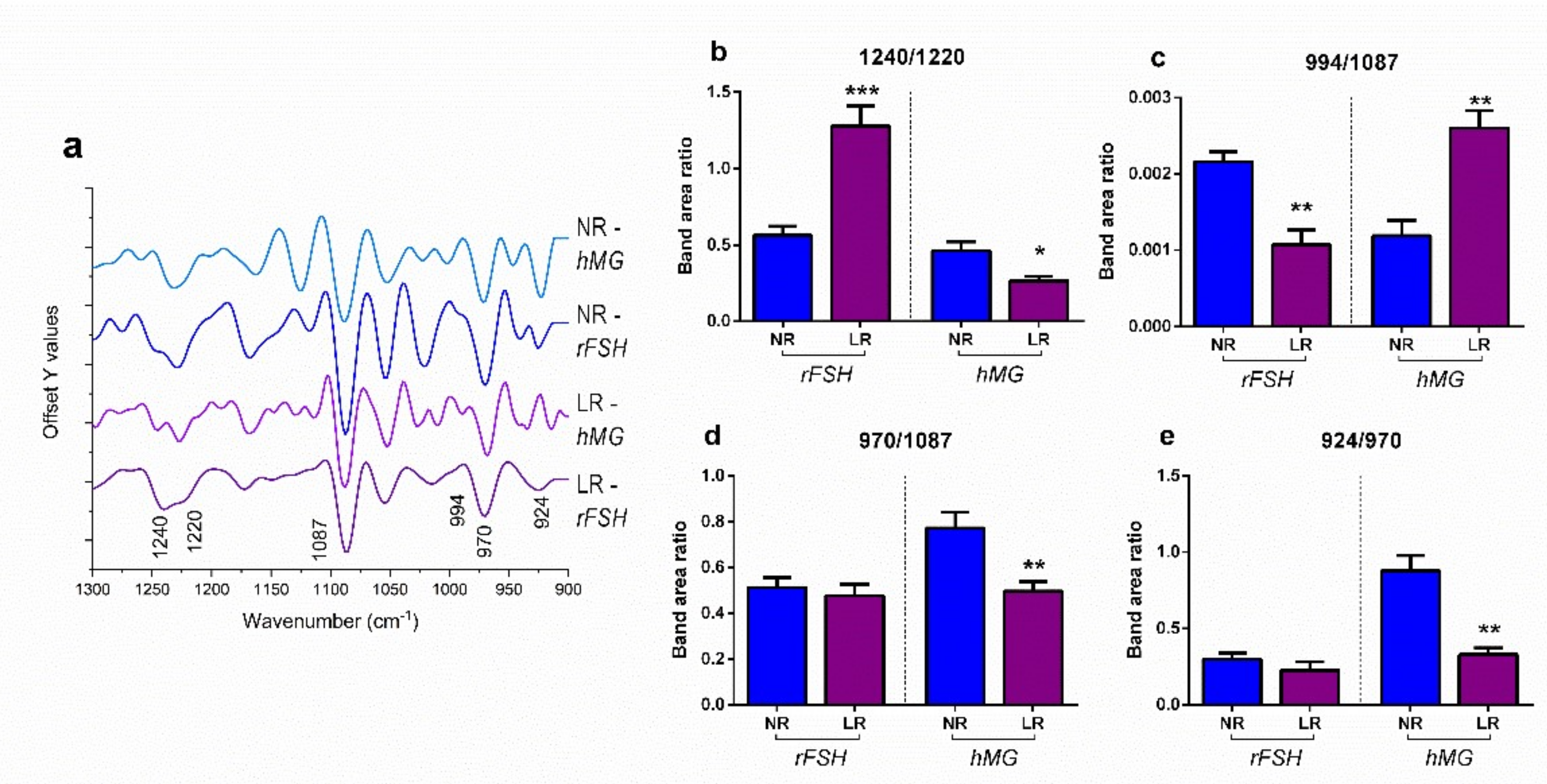
2.3. FTIRM Analysis
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Patient Enrolment and Experimental Design
- Twenty-seven patients (9 LR and 18 NR) treated with rFSH formulation → Follitropin α and β: these are recombinant molecules of the natural human follicle stimulating hormone (Puregon®, MSD, Roma, Italy; Gonal-F®, Merck, Darmstadt, Germany; Ovaleap®, Theramex, London, UK).
- Twenty-four patients (6 LR and 18 NR) treated with FSH + LH formulation → Menotropin (hMG): is a highly purified preparation of FSH and LH in a 1:1 ratio, extracted from the urine of postmenopausal women (Meropur®, Ferring, Sweden).
4.3. CCs Samples Collection and Preparation
4.4. Total RNA Extraction and cDNA Synthesis
4.5. Real-time Polymerase Chain Reaction
4.6. FTIRM Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kidder, G.M.; Vanderhyden, B.C. Bidirectional communication between oocytes and follicle cells: Ensuring oocyte developmental competence. Can. J. Physiol. Pharmacol. 2010, 88, 399–413. [Google Scholar] [CrossRef] [Green Version]
- Uyar, A.; Torrealday, S.; Seli, E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil. Steril. 2013, 99, 979–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Peng, X.; Mei, S. Autophagy in Ovarian Follicular Development and Atresia. Int. J. Biol. Sci. 2019, 15, 726–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assou, S.; Haouzi, D.; Dechaud, H.; Gala, A.; Ferrières, A.; Hamamah, S. Comparative Gene Expression Profiling in Human Cumulus Cells according to Ovarian Gonadotropin Treatments. Biomed Res. Int. 2013, 2013, 354582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, S.-Y.; Chang, D.C.; Lin, S.-L. The MicroRNA (miRNA): Overview of the RNA Genes that Modulate Gene Function. Mol. Biotechnol. 2008, 38, 257–268. [Google Scholar] [CrossRef]
- Imbar, T.; Eisenberg, I. Regulatory role of microRNAs in ovarian function. Fertil. Steril. 2014, 101, 1524–1530. [Google Scholar] [CrossRef]
- Tu, J.; Cheung, A.H.-H.; Chan, C.L.-K.; Chan, W.-Y. The Role of microRNAs in Ovarian Granulosa Cells in Health and Disease. Front. Endocrinol. 2019, 10, 174. [Google Scholar] [CrossRef] [Green Version]
- Pourteymour Fard Tabrizi, Z.; Miraj, S.; Tahmasebian, S.; Ghasemi, S. Plasma Levels of miR-27a, miR-130b, and miR-301a in Polycystic Ovary Syndrome. Int. J. Mol. Cell. Med. 2020, 9, 198–206. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, H.; Qian, Y.; Li, Y.; Chen, X.; Di, N.; Yang, D. miR-130b regulates gap junctional intercellular communication through connexin 43 in granulosa cells from patients with polycystic ovary syndrome. Mol. Hum. Reprod. 2020, 26, 576–584. [Google Scholar] [CrossRef]
- Sinha, P.B.; Tesfaye, D.; Rings, F.; Hossien, M.; Hoelker, M.; Held, E.; Neuhoff, C.; Tholen, E.; Schellander, K.; Salilew-Wondim, D. MicroRNA-130b is involved in bovine granulosa and cumulus cells function, oocyte maturation and blastocyst formation. J. Ovarian Res. 2017, 10, 37. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, J.; Pan, Z.; Du, X.; Li, X.; Ma, B.; Yao, W.; Li, Q.; Liu, H. The let-7g microRNA promotes follicular granulosa cell apoptosis by targeting transforming growth factor-β type 1 receptor. Mol. Cell. Endocrinol. 2015, 409, 103–112. [Google Scholar] [CrossRef]
- Zhang, J.-Q.; Gao, B.-W.; Guo, H.-X.; Ren, Q.-L.; Wang, X.-W.; Chen, J.-F.; Wang, J.; Zhang, Z.-J.; Ma, Q.; Xing, B.-S. miR-181a promotes porcine granulosa cell apoptosis by targeting TGFBR1 via the activin signaling pathway. Mol. Cell. Endocrinol. 2020, 499, 110603. [Google Scholar] [CrossRef] [PubMed]
- Krause, C.J.; Popp, O.; Thirunarayanan, N.; Dittmar, G.; Lipp, M.; Müller, G. MicroRNA-34a promotes genomic instability by a broad suppression of genome maintenance mechanisms downstream of the oncogene KSHV-vGPCR. Oncotarget 2016, 7, 10414–10432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, F.; Pan, Z.X.; Yao, Y.; Liu, H.L.; Liu, S.R.; Xie, Z.; Li, Q.F. miR-34a targets the inhibin beta B gene, promoting granulosa cell apoptosis in the porcine ovary. Genet. Mol. Res. 2014, 13, 2504–2512. [Google Scholar] [CrossRef] [PubMed]
- Broekmans, F.J.; Kwee, J.; Hendriks, D.J.; Mol, B.W.; Lambalk, C.B. A systematic review of tests predicting ovarian reserve and IVF outcome. Hum. Reprod. Update 2006, 12, 685–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broer, S.L.; Dólleman, M.; van Disseldorp, J.; Broeze, K.A.; Opmeer, B.C.; Bossuyt, P.M.M.; Eijkemans, M.J.C.; Mol, B.W.; Broekmans, F.J.M.; Broer, S.L.; et al. Prediction of an excessive response in in vitro fertilization from patient characteristics and ovarian reserve tests and comparison in subgroups: An individual patient data meta-analysis. Fertil. Steril. 2013, 100, 420–429. [Google Scholar] [CrossRef] [Green Version]
- Ferraretti, A.P.; La Marca, A.; Fauser, B.C.J.M.; Tarlatzis, B.; Nargund, G.; Gianaroli, L. ESHRE consensus on the definition of “poor response” to ovarian stimulation for in vitro fertilization: The Bologna criteria. Hum. Reprod. 2011, 26, 1616–1624. [Google Scholar] [CrossRef] [Green Version]
- La Marca, A.; Grisendi, V.; Griesinger, G. How Much Does AMH Really Vary in Normal Women? Int. J. Endocrinol. 2013, 2013, 959487. [Google Scholar] [CrossRef]
- Bosch, E.; Broer, S.; Griesinger, G.; Grynberg, M.; Humaidan, P.; Kolibianakis, E.; Kunicki, M.; La Marca, A.; Lainas, G.; Le Clef, N.; et al. ESHRE guideline: Ovarian stimulation for IVF/ICSI†. Hum. Reprod. Open 2020. [Google Scholar] [CrossRef]
- Massagué, J. TGF-β Signal Transduction. Annu. Rev. Biochem. 1998, 67, 753–791. [Google Scholar] [CrossRef]
- Naji, M.; Aleyasin, A.; Nekoonam, S.; Arefian, E.; Mahdian, R.; Amidi, F. Differential Expression of miR-93 and miR-21 in Granulosa Cells and Follicular Fluid of Polycystic Ovary Syndrome Associating with Different Phenotypes. Sci. Rep. 2017, 7, 14671. [Google Scholar] [CrossRef] [PubMed]
- Gioacchini, G.; Notarstefano, V.; Sereni, E.; Zacà, C.; Coticchio, G.; Giorgini, E.; Vaccari, L.; Carnevali, O.; Borini, A. Does the molecular and metabolic profile of human granulosa cells correlate with oocyte fate? New insights by Fourier transform infrared microspectroscopy analysis. Mol. Hum. Reprod. 2018, 24, 521–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.; Brown, C.W.; Matzuk, M.M. Genetic Analysis of the Mammalian Transforming Growth Factor-β Superfamily. Endocr. Rev. 2002, 23, 787–823. [Google Scholar] [CrossRef]
- Russell, D.; Salustri, A. Extracellular Matrix of the Cumulus-Oocyte Complex. Semin. Reprod. Med. 2006, 24, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Pascal, G.; Monget, P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol. Biol. 2009, 9, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Dixon, M.; Zucchelli, M.; Hambiliki, F.; Levkov, L.; Hovatta, O.; Kere, J. Expression Analysis of the NLRP Gene Family Suggests a Role in Human Preimplantation Development. PLoS ONE 2008, 3, e2755. [Google Scholar] [CrossRef] [PubMed]
- Kuchmiy, A.A.; D’Hont, J.; Hochepied, T.; Lamkanfi, M. NLRP2 controls age-associated maternal fertility. J. Exp. Med. 2016, 213, 2851–2860. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Chang, B.; Lu, C.; Su, J.; Wu, Y.; Lv, P.; Wang, Y.; Liu, J.; Zhang, B.; Quan, F.; et al. Nlrp2, a Maternal Effect Gene Required for Early Embryonic Development in the Mouse. PLoS ONE 2012, 7, e30344. [Google Scholar] [CrossRef] [Green Version]
- Matthäus, C.; Bird, B.; Miljković, M.; Chernenko, T.; Romeo, M.; Diem, M. Chapter 10 Infrared and Raman Microscopy in Cell Biology. Methods Cell Biol. 2008, 89, 275–308. [Google Scholar] [CrossRef] [Green Version]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [Green Version]
- Giorgini, E.; Sabbatini, S.; Rocchetti, R.; Notarstefano, V.; Rubini, C.; Conti, C.; Orilisi, G.; Mitri, E.; Bedolla, D.E.; Vaccari, L. In vitro FTIR microspectroscopy analysis of primary oral squamous carcinoma cells treated with cisplatin and 5-fluorouracil: A new spectroscopic approach for studying the drug–cell interaction. Analyst 2018, 143, 3317–3326. [Google Scholar] [CrossRef] [PubMed]
- Notarstefano, V.; Sabbatini, S.; Pro, C.; Belloni, A.; Orilisi, G.; Rubini, C.; Byrne, H.J.; Vaccari, L.; Giorgini, E. Exploiting fourier transform infrared and Raman microspectroscopies on cancer stem cells from oral squamous cells carcinoma: New evidence of acquired cisplatin chemoresistance. Analyst 2020, 145, 8038–8049. [Google Scholar] [CrossRef] [PubMed]
- Notarstefano, V.; Belloni, A.; Sabbatini, S.; Pro, C.; Orilisi, G.; Monterubbianesi, R.; Tosco, V.; Byrne, H.J.; Vaccari, L.; Giorgini, E. Cytotoxic Effects of 5-Azacytidine on Primary Tumour Cells and Cancer Stem Cells from Oral Squamous Cell Carcinoma: An In Vitro FTIRM Analysis. Cells 2021, 10, 2127. [Google Scholar] [CrossRef]
- Gioacchini, G.; Giorgini, E.; Vaccari, L.; Ferraris, P.; Sabbatini, S.; Bianchi, V.; Borini, A.; Carnevali, O. A new approach to evaluate aging effects on human oocytes: Fourier transform infrared imaging spectroscopy study. Fertil. Steril. 2014, 101, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Notarstefano, V.; Gioacchini, G.; Byrne, H.J.H.J.; Zacà, C.; Sereni, E.; Vaccari, L.; Borini, A.; Carnevali, O.; Giorgini, E. Vibrational characterization of granulosa cells from patients affected by unilateral ovarian endometriosis: New insights from infrared and Raman microspectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 212, 206–214. [Google Scholar] [CrossRef]
- Notarstefano, V.; Gioacchini, G.; Giorgini, E.; Montik, N.; Ciavattini, A.; Polidori, A.R.; Candela, F.A.; Vaccari, L.; Cignitti, M.; Carnevali, O. The Impact of Controlled Ovarian Stimulation Hormones on the Metabolic State and Endocannabinoid System of Human Cumulus Cells. Int. J. Mol. Sci. 2020, 21, 7124. [Google Scholar] [CrossRef]
- Whelan, D.R.; Bambery, K.R.; Heraud, P.; Tobin, M.J.; Diem, M.; McNaughton, D.; Wood, B.R. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acids Res. 2011, 39, 5439–5448. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, D.; Dey, S.K.; Saha, C. Mutation Induced Conformational Changes in Genomic DNA from Cancerous K562 Cells Influence Drug-DNA Binding Modes. PLoS ONE 2014, 9, e84880. [Google Scholar] [CrossRef] [Green Version]
- Mignolet, A.; Derenne, A.; Smolina, M.; Wood, B.R.; Goormaghtigh, E. FTIR spectral signature of anticancer drugs. Can drug mode of action be identified? Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 85–101. [Google Scholar] [CrossRef]
- Herbert, A.; Rich, A. The biology of left-handed Z-DNA. J. Biol. Chem. 1996, 271, 11595–11598. [Google Scholar] [CrossRef] [Green Version]
- Temiz, N.A.; Donohue, D.E.; Bacolla, A.; Luke, B.T.; Collins, J.R. The Role of Methylation in the Intrinsic Dynamics of B- and Z-DNA. PLoS ONE 2012, 7, e035558. [Google Scholar] [CrossRef] [PubMed]
- Alviggi, C.; Humaidan, P.; Ezcurra, D. Hormonal, functional and genetic biomarkers in controlled ovarian stimulation: Tools for matching patients and protocols. Reprod. Biol. Endocrinol. 2012, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Sun, H.; Jiang, Y.; Ding, L.; Wu, S.; Fang, T.; Yan, G.; Hu, Y. MicroRNA-181a Suppresses Mouse Granulosa Cell Proliferation by Targeting Activin Receptor IIA. PLoS ONE 2013, 8, e59667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, W. The human corpus luteum: Remodelling during luteolysis and maternal recognition of pregnancy. Rev. Reprod. 2000, 5, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Rothenburg, S.; Koch-Nolte, F.; Haag, F. DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles. Immunol. Rev. 2001, 184, 286–298. [Google Scholar] [CrossRef]
- Dovbeshko, G.I.; Chegel, V.I.; Gridina, N.Y.; Repnytska, O.P.; Shirshov, Y.M.; Tryndiak, V.P.; Todor, I.M.; Solyanik, G.I. Surface enhanced IR absorption of nucleic acids from tumor cells: FTIR reflectance study. Biopolymers 2002, 67, 470–486. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Sun, Y.-H.; Zhang, X.-H.; Chiang, V.L. Poly(T) Adaptor RT-PCR. In Next-Generation MicroRNA Expression Profiling Technology; Fan, J.-B., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 53–66. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [Green Version]
- Vaccari, L.; Birarda, G.; Businaro, L.; Pacor, S.; Grenci, G. Infrared Microspectroscopy of Live Cells in Microfluidic Devices (MD-IRMS): Toward a Powerful Label-Free Cell-Based Assay. Anal. Chem. 2012, 84, 4768–4775. [Google Scholar] [CrossRef]
- Weniger, M.; Honselmann, K.C.; Liss, A.S. The Extracellular Matrix and Pancreatic Cancer: A Complex Relationship. Cancers 2018, 10, 316. [Google Scholar] [CrossRef] [Green Version]
- Zucchiatti, P.; Mitri, E.; Kenig, S.; Billè, F.; Kourousias, G.; Bedolla, D.E.; Vaccari, L. Contribution of Ribonucleic Acid (RNA) to the Fourier Transform Infrared (FTIR) Spectrum of Eukaryotic Cells. Anal. Chem. 2016, 88, 12090–12098. [Google Scholar] [CrossRef] [PubMed]
- Notarstefano, V.; Sabbatini, S.; Conti, C.; Pisani, M.; Astolfi, P.; Pro, C.; Rubini, C.; Vaccari, L.; Giorgini, E. Investigation of human pancreatic cancer tissues by Fourier Transform Infrared Hyperspectral Imaging. J. Biophotonics 2020, 13, e201960071. [Google Scholar] [CrossRef]
- Notarstefano, V.; Sabbatini, S.; Sabbatini, M.; Arrais, A.; Belloni, A.; Pro, C.; Vaccari, L.; Osella, D.; Giorgini, E. Hyperspectral characterization of the MSTO-211H cell spheroid model: A FPA–FTIR imaging approach. Clin. Spectrosc. 2021, 3, 100011. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Banyay, M.; Sarkar, M.; Gräslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Gioacchini, G.; Giorgini, E.; Olivotto, I.; Maradonna, F.; Merrifield, D.L.; Carnevali, O. The Influence of Probiotics on Zebrafish Danio Rerio Innate Immunity and Hepatic Stress. Zebrafish 2014, 11, 98–106. [Google Scholar] [CrossRef]
| Treatment | # Patients | Ovarian Reserve | Retrieved Oocytes | % Immature Oocytes | % Mature Oocytes | % Fertilized Oocytes |
|---|---|---|---|---|---|---|
| rFSH | 9 | low | 3.44 ± 1.88 a | 21.00% a | 79.00% a | 69.45% a |
| 18 | normal | 8.06 ± 4.22 b | 22.06% a | 78.00% a | 78.64% b | |
| hMG | 6 | low | 3.00 ± 1.41 a | 8.83% b | 91.17% b | 91.67% c |
| 18 | normal | 6.33 ± 3.70 ab | 28.72% a | 71.28% a | 70.00% a |
| miRNA | Forward Primer (5′–3′) | Universal Reverse Primer (5′–3′) | miRBase Accession Number |
|---|---|---|---|
| miR-181a-5p | AACATTCAACGCTGTCGGTGAGT | GCGAGCACAGAATTAATACG | MI0000269 |
| miR-93-5p | CAAAGTGCTGTTCGTGCAGGTAG | GCGAGCACAGAATTAATACG | MI0000095 |
| miR-23a-5p | GGGGTTCCTGGGGATGGGATTT | GCGAGCACAGAATTAATACG | MI0000079 |
| miR-202-5p | TTCCTATGCATATACTTCTTTG | GCGAGCACAGAATTAATACG | MI0003130 |
| miR-130b-3p | CAGTGCAATGATGAAAGGGCAT | GCGAGCACAGAATTAATACG | MI0000748 |
| let-7g-5p | TGAGGTAGTAGTTTGTACAGTT | GCGAGCACAGAATTAATACG | MI0000433 |
| miR-34a-5p | TGGCAGTGTCTTAGCTGGTTGT | GCGAGCACAGAATTAATACG | MI0000268 |
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | NCBI Accession Number |
| ACTB | GCAGAAGGAGATCACATCCCTGGC | CATTGCCGTCACCTTCACCGTTC | NM_001101.5 |
| RPL13A | TCTGGAGGACTGTAAGAGGTATGC | AGACGCACAATCTTGAGAGCAG | NM_012423.3 |
| ACVRIIA | CCTCGGACTTTAGGTGTCTGG | CCAAGTATAGCACCTGAAGAACA | NM_001278579.2 |
| SMAD3 | GCCTGTGCTGGAACATCATC | TTGCCCTCATGTGTGCTCTT | NM_005902.4 |
| CDKN1A | AGGTGGACCTGGAGACTCTCAG | TCCTCTTGGAGAAGATCAGCCG | NM_000389.5 |
| CDK2 | ATGGATGCCTCTGCTCTCACTG | CCCGATGAGAATGGCAGAAAGC | NM_001798.5 |
| HAS2 | GTCATGTACACAGCCTTCAGAGC | ACAGATGAGGCTGGGTCAAGCA | NM_005328.3 |
| NLRP2 | CATTCTGCGTCAAGCACTGTCG | CCGTCCAGAAAGGAAGCATGTG | NM_001348003.2 |
| SMAD5 | CAGGAGTTTGCTCAGCTTCTGG | GGTGCTGGTTACATCCTGCCG | NM_005903.7 |
| TGFBR1 | GACAACGTCAGGTTCTGGCTCA | CCGCCACTTTCCTCTCCAAACT | NM_004612.4 |
| VEGFA | TTGCCTTGCTGCTCTACCTCCA | GATGGCAGTAGCTGCGCTGATA | NM_001025366.3 |
| TGFB1 | GCCGTGGAGGGGAAATTGAG | TGAACCCGTTGATGTCCACTT | NM_000660.7 |
| MCM3 | CGAGACCTAGAAAATGGCAGCC | GCAGTGCAAAGCACATACCGCA | NM_002388.6 |
| Wavenumber | Vibrational Mode and Biochemical Assignment | References |
|---|---|---|
| ~1240 | Asymmetric stretching vibrations of phosphate moieties of A-form DNA | [32,37,38,53] |
| ~1220 | Asymmetric stretching vibrations of phosphate moieties of B-form DNA | [32,37,38,53] |
| ~1087 | Symmetric stretching vibrations of phosphate moieties in nucleic acids | [54,55,56] |
| ~994 | C–C, C–O ring breathing of RNA ribose | [31,32,54,57] |
| ~970 | Backbone vibrations of nucleic acids, mainly double-strand DNA | [32,35,58] |
| ~924 | Left-handed helix DNA vibrations (Z-DNA) | [33,46,58,59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russo, G.; Notarstefano, V.; Montik, N.; Gioacchini, G.; Giorgini, E.; Polidori, A.R.; Candela, F.A.; Ciavattini, A.; Cignitti, M.; Carnevali, O. Evaluation of Controlled Ovarian Stimulation Protocols in Patients with Normal and Low Ovarian Reserve: Analyses of miRNAs and Selected Target Genes Involved in the Proliferation of Human Cumulus Cells and Oocyte Quality. Int. J. Mol. Sci. 2022, 23, 1713. https://doi.org/10.3390/ijms23031713
Russo G, Notarstefano V, Montik N, Gioacchini G, Giorgini E, Polidori AR, Candela FA, Ciavattini A, Cignitti M, Carnevali O. Evaluation of Controlled Ovarian Stimulation Protocols in Patients with Normal and Low Ovarian Reserve: Analyses of miRNAs and Selected Target Genes Involved in the Proliferation of Human Cumulus Cells and Oocyte Quality. International Journal of Molecular Sciences. 2022; 23(3):1713. https://doi.org/10.3390/ijms23031713
Chicago/Turabian StyleRusso, Giulia, Valentina Notarstefano, Nina Montik, Giorgia Gioacchini, Elisabetta Giorgini, Anna Rita Polidori, Fulvia Antonia Candela, Andrea Ciavattini, Maurizio Cignitti, and Oliana Carnevali. 2022. "Evaluation of Controlled Ovarian Stimulation Protocols in Patients with Normal and Low Ovarian Reserve: Analyses of miRNAs and Selected Target Genes Involved in the Proliferation of Human Cumulus Cells and Oocyte Quality" International Journal of Molecular Sciences 23, no. 3: 1713. https://doi.org/10.3390/ijms23031713
APA StyleRusso, G., Notarstefano, V., Montik, N., Gioacchini, G., Giorgini, E., Polidori, A. R., Candela, F. A., Ciavattini, A., Cignitti, M., & Carnevali, O. (2022). Evaluation of Controlled Ovarian Stimulation Protocols in Patients with Normal and Low Ovarian Reserve: Analyses of miRNAs and Selected Target Genes Involved in the Proliferation of Human Cumulus Cells and Oocyte Quality. International Journal of Molecular Sciences, 23(3), 1713. https://doi.org/10.3390/ijms23031713









