Nitrogen Uptake and Distribution in Different Chinese Cabbage Genotypes under Low Nitrogen Stress
Abstract
1. Introduction
2. Results
2.1. Response to LN of Different Chinese Cabbage Varieties
2.1.1. Field Experiment of Chinese Cabbage under Different N Supplying Levels
2.1.2. Hydroponic Experiments of Chinese Cabbage under CK and LN Conditions
2.2. N-RELATED Physiological Indexes under LN Condition in Different Chinese Cabbage Varieties
2.3. Transcriptome Analysis of the Effects of LN Stress on Chinese Cabbage
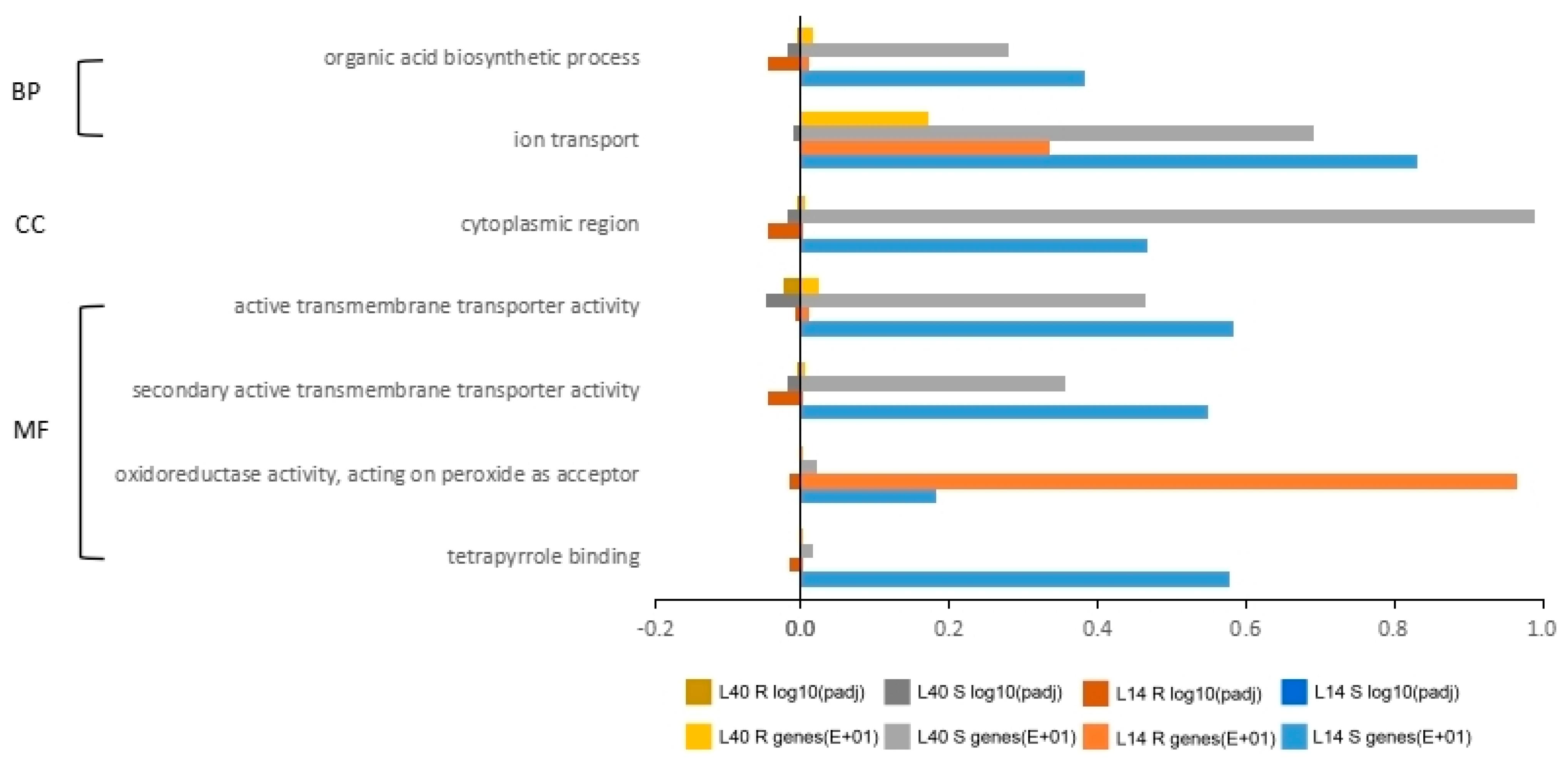
2.4. Gene Ontology (GO) and Pathway Analysis for Differentially Expressed Genes under LN Stress
2.5. DEGs Analysis Involved in the N Responsive Process
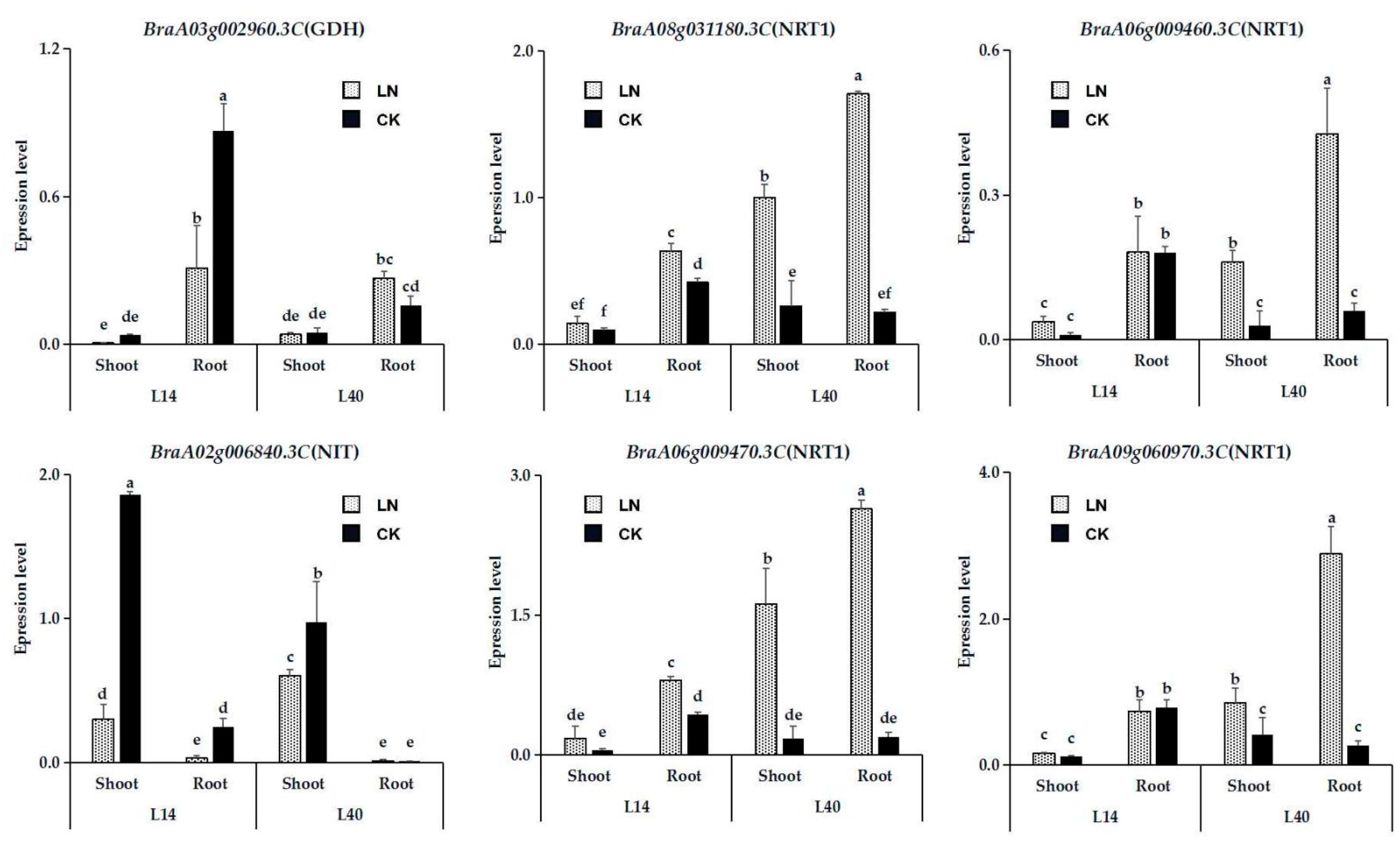
2.6. Expression Analysis of Essential Genes in N Metabolism Pathway in Response to LN Stress
3. Discussion
4. Materials and Methods
4.1. Test Materials
4.2. Experimental Design
4.2.1. Field Experiment
4.2.2. Solution Culture Experiment
4.3. Sampling and Measurement Methods
4.3.1. Investigation of Agronomic Traits
4.3.2. Root Morphology Analysis
4.3.3. Determination of Related Indexes of Nitrogen Metabolism
4.4. RNAseq Materials and Methods
4.4.1. RNA Isolation
4.4.2. cDNA Library Construction, Sequencing, and Data Processing
4.4.3. Analysis of Gene Expression Profile of Chinese Cabbage in Response to LN Stress
4.4.4. Quantitative RT-PCR Analysis
4.4.5. Data Processing and Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kamh, M.; Wiesler, F.; Ulas, A.; Horst, W.J. Root growth and N-uptake aactivity of oilseed rape (Brassica napus L.) cultivars differing in nitrogen efficiency. J. Plant Nutr. Soil Sci. 2005, 168, 130–137. [Google Scholar] [CrossRef]
- Pessemier, J.D.; Chardon, F.; Juraniec, M.; Delaplace, P.; Hermans, C. Natural variation of the root morphological response to nitrate supply in Arabidopsis thaliana. Mech. Dev. 2013, 130, 45–53. [Google Scholar] [CrossRef]
- Ikram, S.; Bedu, M.; Da Niel-Vedele, F.; Chaillou, S.; Chardon, F. Natural variation of Arabidopsis response to nitrogen availability. J. Exp. Bot. 2012, 63, 91. [Google Scholar] [CrossRef]
- Muraro, D.; Byrne, H.; King, J.; Bennett, M. The role of auxin and cytokinin signalling in specifying the root architecture of Arabidopsis thaliana. J. Theor. Biol. 2013, 317, 71–86. [Google Scholar] [CrossRef]
- Grsic-Rausch, S.; Kobelt, P.; Siemens, J.M.; Bischoff, M.; Ludwig-Müller, J. Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiol. 2000, 122, 369–378. [Google Scholar] [CrossRef]
- Kotur, Z.; Glass, A. A 150kda Plasma Membrane Complex of AtNRT2.5 and AtNAR2.1 is the major contributor to constitutive high-affinity nitrate influx in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 1490–1502. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef]
- Karen, S.S.; Heven, S. Decrease of pH gradients in tonoplast vesicles by NO3− and Cl−: Evidence for H+-coupled anion transport. Plant Physiol. 1987, 83, 490–496. [Google Scholar]
- Seki, M.; Umezawa, T.; Kim, J.M.; Matsui, A.; To, T.K.; Shinozaki, K. Transcriptome Analysis of Plant Drought and Salt Stress Response; Springer: Dordrecht, The Netherlands, 2007; pp. 261–283. [Google Scholar]
- Taji, T.; Seki, M.; Satou, M.; Sakurai, T.; Kobayashi, M.; Ishiyama, K.; Narusaka, Y.; Narusaka, M.; Zhu, J.K.; Shinozaki, K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004, 135, 1697–1709. [Google Scholar] [CrossRef]
- Xiong, S.; Wang, Y.; Wang, X.; Ding, S.; Yanpeng, W.U.; Wang, X. Analysis on activity of nitrogen transformation microorganism and enzyme in rhizosphere soil among winter wheat varieties. J. Triticeae Crops 2014, 34, 782–786. [Google Scholar]
- Jiao, J.J.; Wei, Q.R.; Pei, J.N.; Guo, Y.X.; Tang, L.Y.; Shi, Y. Difference of nitrogen response in different genotypes of potato. Acta Agric. Boreali-Sin. 2018, 5, 203–208. [Google Scholar]
- Zhang, P.P.; Zhang, G.T.; Huang, L.; Qiao, J.F.; Li, C.; Zhang, M.W.; Zhao, X.; Liu, J.B. Nitrogen efficiency and physiological characters of different maize genotypes under nitrogen fertilization reduction. J. Henan Agric. Sci. 2021, 5, 13–23. [Google Scholar]
- Zhang, X.; Tian, Z.W.; Hu, J.L.; Xiu, M.; Jiang, D.; Dai, T.B. Agronomic and physiological characteristics of high efficeint nitrogen utilization in Wheat. J. Triticeae Crops 2016, 10, 1315–1322. [Google Scholar]
- Chen, C.; Gong, H.; Zhang, J.; Yujun, X.U.; Gao, H. Evaluation of nitrogen nutrition characteristics of different rice cultivars at seedling stage. Chin. J. Eco-Agric. 2016, 10, 1347–1355. [Google Scholar]
- Qin, N.; Ma, C.Y.; Zhu, C.C.; Dai, S.T.; Song, Y.H.; Wang, C.Y.; Rui, Z.X.; Li, J.X. Screening of foxtail millet genotype with high nitrogen use efficiency and analysis of related characters. J. Henan Agric. Sci. 2019, 5, 22–29. [Google Scholar]
- Rahn, C.R. Nitrogen and field production of vegetable crops. Acta Hortic. 2000, 533, 361–370. [Google Scholar] [CrossRef]
- Jin, Y.T.; Miao, T.L. Investigation on the current situation of nitrate pollution to vegetables. Chem. Enterp. Manag. 2019, 12, 186–187. [Google Scholar]
- Zao, H.D.; Ge, M.Y.; Wang, H.D.; Wu, J.Y. Analysis on the utilization of nitrogen fertilizer in three main vegetables in China. North. Hortic. 2017, 5, 151–155. [Google Scholar]
- Dong, C.Y.; Niu, M.L.; Yao, Y.; Chang, X.Y.; Lu, L.F.; Li, N. Research on the construction and application of a big data platform for the whole vegetable industry chain: The case of Chinese cabbage. J. Agric. Big Data 2021, 1, 66–72. [Google Scholar]
- Li, J.L.; Chen, X.P.; Li, X.L.; Zhang, F.S. Effect of N fertilization on yield, nitrate content and N apparent loss Chinese cabbage. Acta Pedol. Sin. 2003, 2, 261–266. [Google Scholar]
- Staugaitis, G.; Viškelis, P. Amounts of nitrogen, phosphorus, potassium, calcium and magnesium in the heads and plant residues of Chinese cabbage crop fertilised with different rates of nitrogen. Sodininkyste Ir Darzininkyste 2005, 24, 98–106. [Google Scholar]
- Wu, C.Y.; Song, T.Y.; Zhang, X.M.; Wang, Z.; Tian, J.F.; Guo, L. Effects of nitrogen fertilizer on growth and yield of Chinese cabbage. J. Jilin Agric. Sci. 2014, 39, 80–83. [Google Scholar]
- Zhang, X.T. Identification of Chinese Cabbage Nitrogen Efficiency Genotypes and Screening and Analysis of Related Indexes. Master’s Thesis, The Hebei Agricultural University, Baoding, China, 2014. [Google Scholar]
- Liu, S.G.; Han, X.R.; Liu, X.H. Effects of different application methods of nitrogenous fertilizer on Chinese cabbage yield. J. Anhui Agric. Sci. 2015, 28, 105–106. [Google Scholar]
- Deng, X.; Fan, G. Effects of nitrogen fertilizer application amount on economic traits, yield and nitrogen utilization of Chinese cabbage. Guizhou Agric. Sci. 2015, 1, 117–119. [Google Scholar]
- He, Y.L.; Zi, F.; Liu, X.P. Effects of different nitrogen application levels on yield and benefit of Chinese cabbage. Hunan Agric. Sci. 2016, 2, 48–50. [Google Scholar]
- Wang, S.F.; Lin, L. Effects of different nitrogen amounts on Chinese cabbage yield and nitrogen agronomic efficiency. Chin. Hortic. Abstr. 2017, 6, 20–21. [Google Scholar]
- Chen, Y.N.; Lü, X.H.; Ding, Q.; Zheng, H.; Yang, N.; Song, Q.Q.; Wang, J.F.; Li, J.J.; Gao, J.W. Genotypic difference and characteristic analysis of nitrogen response in Chinese cabbage. Acta Agric. Boreali-Sin. 2020, 35, 131–140. [Google Scholar]
- Zörb, C.; Ludewig, U.; Hawkesford, M.J. Perspective on wheat yield and quality with reduced nitrogen supply. Trends Plant Sci. 2018, 23, 1029–1037. [Google Scholar] [CrossRef]
- Elena, A.V.; Rodrigo, A.G. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 2008, 11, 521–529. [Google Scholar]
- Ma, H.Y.; Wang, Q.L.; Qi, Y.L.; Fu, D.; Huo, E.W.; Shen, G.H.; Guo, G.Y. Screening and screening of rice nitrogen utilization genotypes. Guangdong Agric. Sci. 2011, 21, 31–46. [Google Scholar]
- Garnett, T.; Plett, D.; Conn, V.; Conn, S.; Rabie, H.; Rafalski, J.A.; Dhugga, K.; Tester, M.A.; Kaiser, B.N. Variation for n uptake system in maize: Genotypic response to N supply. Front. Plant Sci. 2015, 6, 936. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xu, F.C.; Dong, J.; Dong, J.H.; Qin, D.D.; Lu, M.Y.; Li, M.F. Genotypic difference of nitrogen use efficiency of wheat and correlation analysis of the related character. Sci. Agric. Sin. 2017, 50, 2647–2657. [Google Scholar]
- Hu, X.Y.; Guo, J.X.; Tian, G.L.; Gao, L.M.; Shen, Q.R.; Guo, S.W. Effects of different nitrogen supply patterns on root mological and physiological characteristics of rice. Chin. J. Rice Sci. 2017, 31, 72–80. [Google Scholar]
- Tao, S.; Hua, X.Y.; Wang, Y.N.; Guo, N.; Yan, X.F.; Lian, J.X. Research advance in effects of different nitrogen forms on growth and physiology of plants. Guizhou Agric. Ences 2017, 12, 64–68. [Google Scholar]
- Tsay, Y.F.; Chiu, C.C.; Tsai, C.B.; Ho, C.H.; Hsu, P.K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Gulati, A.; Jaiwal, P.K. Effect of NaCI on nitrate reductase, glutamate dehydrogenase and glutamate synthase in Vigna radiata calli. Biol. Plant. 1996, 38, 177–183. [Google Scholar] [CrossRef]
- Robinson, S.A.; Slade, A.P.; Fox, G.G.; Phillips, R.; Stewart, R.G.R. The role of glutamate dehydrogenase in plant nitrogen metabolism. Plant Physiol. 1991, 95, 509–516. [Google Scholar] [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, fegulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef]
- Labboun, S.; Tercé-Laforgue, T.; Roscher, A.; Bedu, M.; Restivo, F.M.; Velanis, C.N.; Skopelitis, D.S.; Moschou, P.N.; Roubelakis-Angelakis, K.A.; Suzuki, A.; et al. Resolving the role of plant glutamate dehydrogenase. I. in vivo real time nuclear magnetic resonance spectroscopy experiments. Plant Cell Physiol. 2009, 50, 1761–1773. [Google Scholar] [CrossRef][Green Version]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Development of nitrogen-efficient prolific hybrids of maize. Crop Sci. 2010, 27, 181–186. [Google Scholar] [CrossRef]
- Yan, Z.; Nie, J.Y.; Cheng, Y.; Guan, D.K.; Li, Z.X. Determination of chlorophyll content in fruits, vegetables and their products. China Fruits 2018, 2, 59–62. [Google Scholar]
- Li, R.Q.; Yu, C.; Li, Y.R.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986. [Google Scholar] [CrossRef]

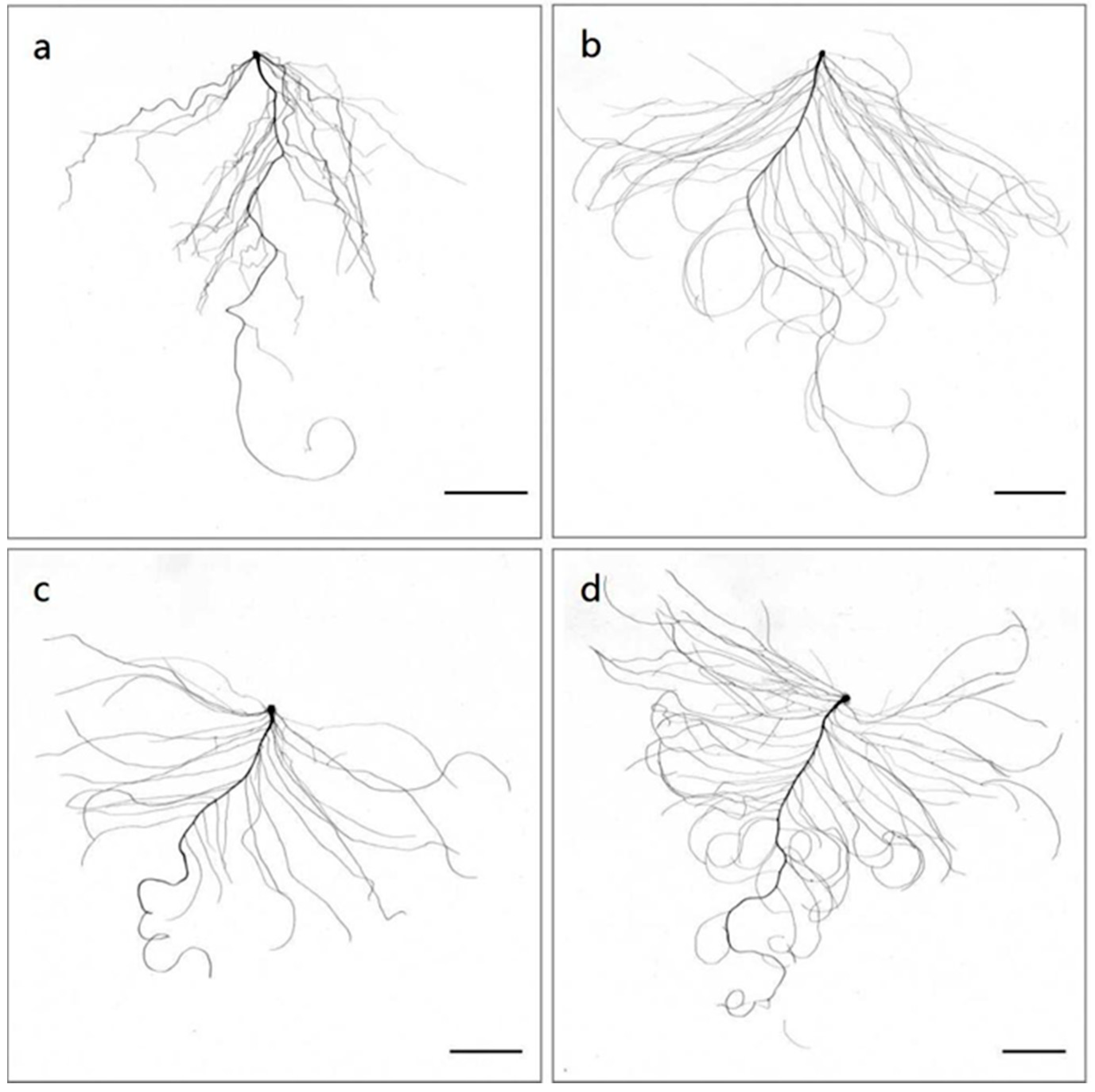
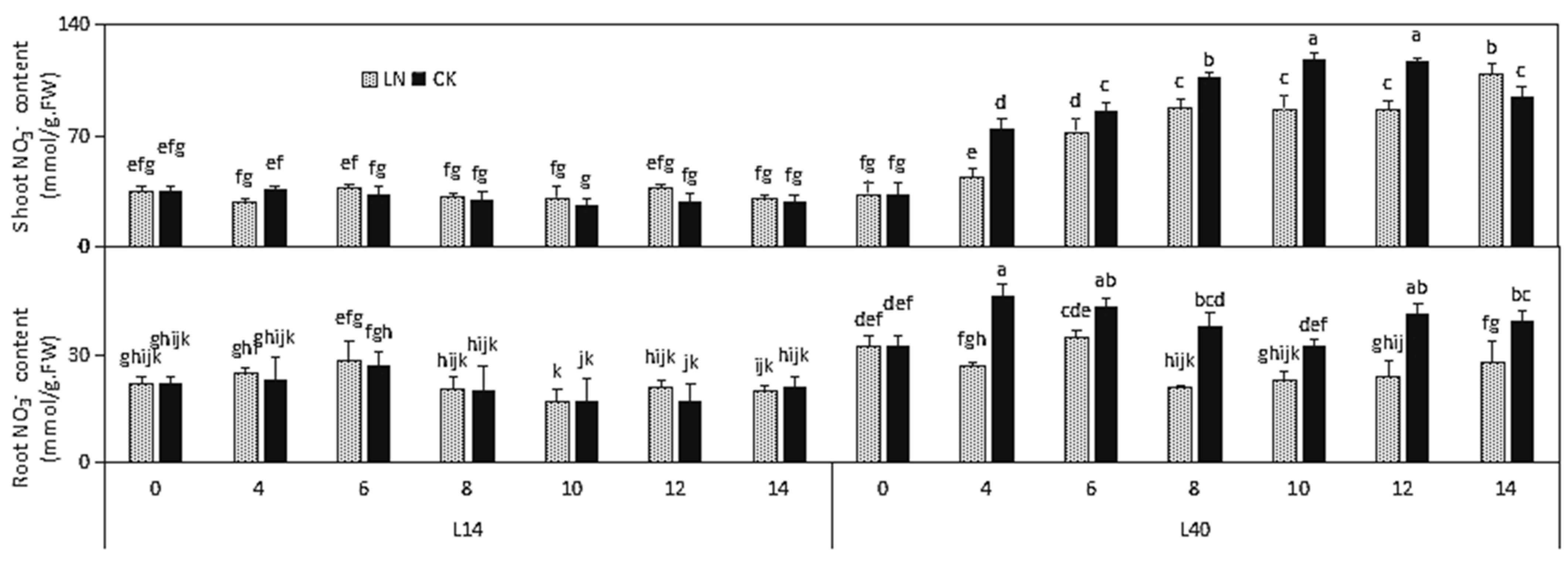
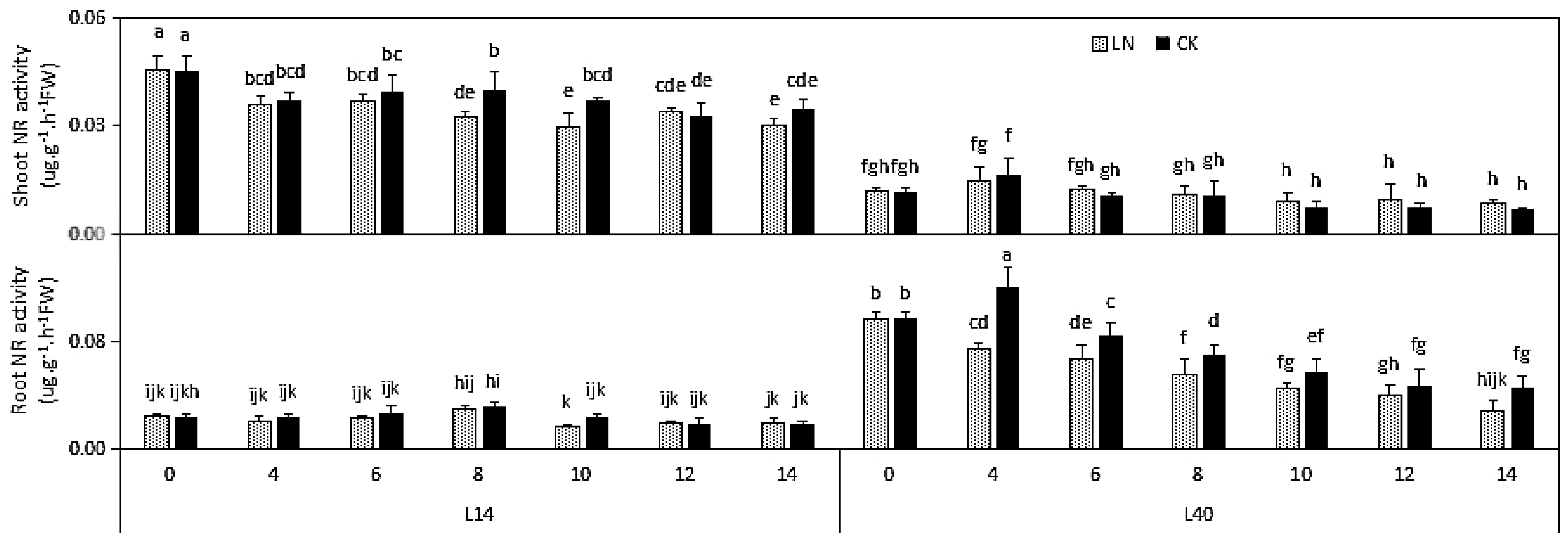
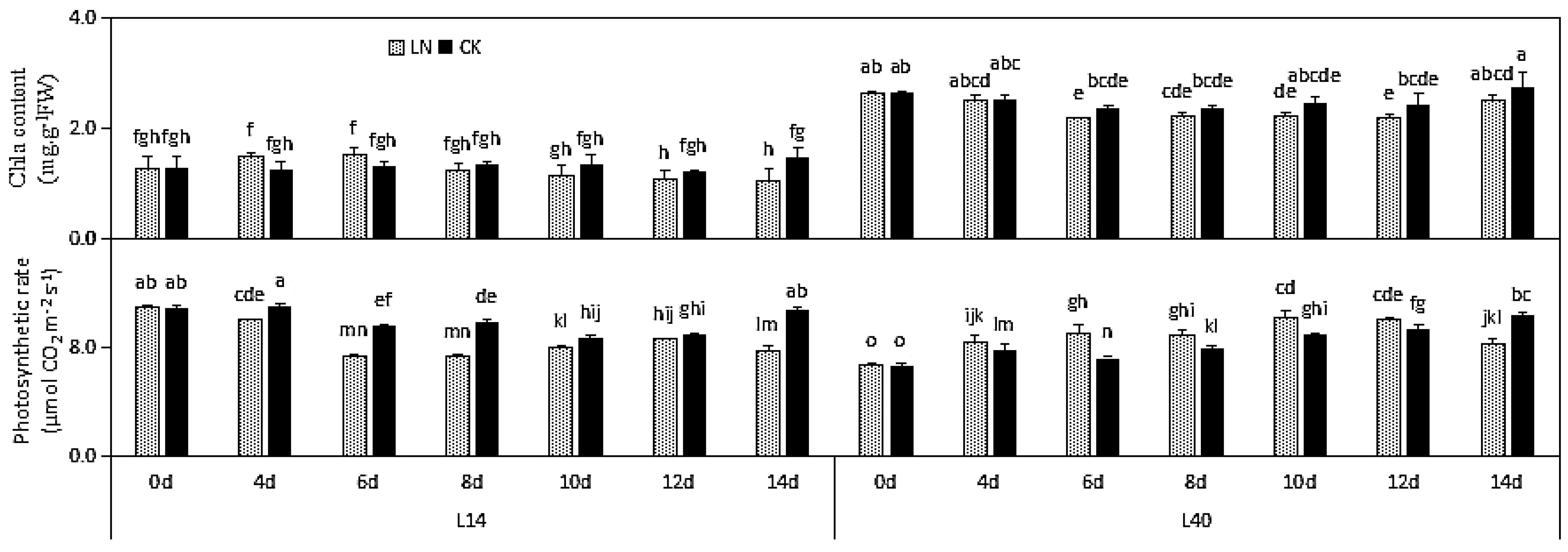

| Part | Trait | L14 | L40 | ||
|---|---|---|---|---|---|
| LN | CK | LN | CK | ||
| Shoot | SFW(g plant−1) | 0.75 ± 0.08 b | 0.93 ± 0.39 a | 0.72 ± 0.07 b | 0.74 ± 0.09 b |
| SDW(g plant−1) | 0.027 ± 0.001 a | 0.034 ± 0.007 a | 0.027 ± 0.001 a | 0.026 ± 0.005 a | |
| Root | RFW(g plant−1) | 0.097 ± 0.009 a | 0.064 ± 0.007 b | 0.103 ± 0.009 a | 0.087 ± 0.009 ab |
| RDW(g plant−1) | 0.0063 ± 0.0002 a | 0.0044 ± 0.0004 b | 0.0054 ± 0.0006 ab | 0.0047 ± 0.0005 b | |
| TRL(cm plant−1) | 19.23 ± 1.58 a | 18.67 ± 2.08 ab | 14.87 ± 0.91 b | 11.27 ± 2.61 c | |
| LRL(cm plant−1) | 363.41 ± 12.47 a | 169.1 ± 6.77 c | 279.78 ± 12.06 b | 188.41 ± 8.95 c | |
| RSA(cm2 plant−1) | 18.42 ± 1.86 a | 10.41 ± 1.36 b | 17.26 ± 0.97 a | 12.15 ± 1.62 b | |
| Sample | Total Raw Reads | Total Clean Reads | Q20 Percentage | Total Map | Unique Map | Total Genes | Novel Transcript |
|---|---|---|---|---|---|---|---|
| CK14S | 46,383,112 | 45,027,680 | 98.45 | 92.21% | 89.81% | 29,776 | 5324 |
| CK14R | 47,421,702 | 45,802,951 | 98.31 | 90.15% | 88.41% | 29,771 | 5319 |
| LN14S | 46,323,061 | 45,363,247 | 98.39 | 91.91% | 89.92% | 29,585 | 5133 |
| LN14R | 49,639,339 | 48,482,294 | 98.31 | 90.25% | 88.27% | 30,335 | 5883 |
| CK40S | 47,193,171 | 46,107,124 | 98.43 | 91.90% | 89.03% | 29,167 | 4786 |
| CK40R | 46,282,418 | 45,051,759 | 98.30 | 90.51% | 88.68% | 29,308 | 4927 |
| LN40S | 46,018,595 | 44,930,999 | 98.39 | 91.08% | 89.79% | 29,951 | 5570 |
| LN40R | 45,126,988 | 44,067,133 | 98.28 | 90.53% | 88.68% | 30,000 | 5619 |
| KEGGID | Description | LN14L vs. CK14L | LN14R vs. CK14R | LN40L vs. CK40L | LN40R vs. CK40R | ||||
|---|---|---|---|---|---|---|---|---|---|
| p-Value | GeneRatio | p-Value | GeneRatio | p-Value | GeneRatio | p-Value | GeneRatio | ||
| ath00053 | Ascorbate and aldarate metabolism | 0.006 | 4/62 | 0 | 0 | 0 | |||
| ath00195 | Photosynthesis | 0.000 | 10/62 | 0 | 0 | 0 | |||
| ath00196 | Photosynthesis—antenna proteins | 0.000 | 9/62 | 0.034 | 5/191 | 0.000 | 10/642 | 0 | |
| ath00220 | Arginine biosynthesis | 0 | 0.012 | 3/191 | 0 | 0 | |||
| ath00250 | Alanine, aspartate and glutamate metabolism | 0 | 0.000 | 9/191 | 0 | 0 | |||
| ath00280 | Valine, leucine and isoleucine degradation | 0 | 0.006 | 7/191 | 0 | 0 | |||
| ath00350 | Tyrosine metabolism | 0 | 0.030 | 5/191 | 0 | 0 | |||
| ath00380 | Tryptophan metabolism | 0 | 0 | 0.004 | 14/642 | 0 | |||
| ath00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 0 | 0.028 | 6/191 | 0 | 0.042 | 4/111 | ||
| ath00460 | Cyanoamino acid metabolism | 0 | 0.046 | 5/191 | 0 | 0.036 | 4/111 | ||
| ath00640 | Propanoate metabolism | 0 | 0.044 | 4/191 | 0 | 0 | |||
| ath00730 | Thiamine metabolism | 0.038 | 2/62 | 0 | 0 | 0 | |||
| ath00860 | Porphyrin and chlorophyll metabolism | 0.035 | 3/62 | 0 | 0 | 0 | |||
| ath00904 | Diterpenoid biosynthesis | 0 | 0 | 0 | 0.005 | 3/111 | |||
| ath00906 | Carotenoid biosynthesis | 0 | 0 | 0 | 0.007 | 4/111 | |||
| ath00908 | Zeatin biosynthesis | 0 | 0.013 | 4/191 | 0 | 0 | |||
| ath00910 | Nitrogen metabolism | 0 | 0.000 | 10/191 | 0 | 0 | |||
| ath00920 | Sulfur metabolism | 0 | 0.006 | 6/191 | 0.008 | 11/642 | 0 | ||
| ath00940 | Phenylpropanoid biosynthesis | 0 | 0.000 | 21/191 | 0 | 0.000 | 13/111 | ||
| ath00960 | Tropane, piperidine and pyridine alkaloid biosynthesis | 0 | 0.012 | 5/191 | 0 | 0 | |||
| ath00966 | Glucosinolate biosynthesis | 0 | 0.001 | 5/191 | 0.026 | 6/642 | 0 | ||
| ath04016 | MAPK signaling pathway—plant | 0 | 0 | 0.000 | 32/642 | 0.000 | 12/111 | ||
| ath04075 | Plant hormone signal transduction | 0 | 0 | 0 | 0.002 | 15/111 | |||
| ath04130 | SNARE interactions in vesicular transport | 0 | 0 | 0.008 | 11/642 | 0 | |||
| ath04626 | Plant-pathogen interaction | 0 | 0.018 | 13/191 | 0.034 | 31/642 | 0.000 | 14/111 | |
| ath04712 | Circadian rhythm—plant | 0.001 | 4/62 | 0 | 0 | 0 | |||
| Function | Gene Name | Gene ID | Chr. | Start | End | log2 Ratio (LN/CK) | |||
|---|---|---|---|---|---|---|---|---|---|
| Root | Shoot | ||||||||
| L14 | L40 | L14 | L40 | ||||||
| Nitrate transport | NRT1 | BraA06g009460.3C | A06 | 5,163,603 | 5,163,896 | 2.43 | 1.78 | 1.32 | |
| NRT1 | BraA06g009470.3C | A06 | 5,167,089 | 5,168,324 | 1.42 | 1.94 | 1.73 | ||
| NRT1 | BraA08g031180.3C | A08 | 20,956,829 | 20,960,518 | 1.13 | 1.55 | |||
| NRT1 | BraA09g060970.3C | A09 | 42,614,336 | 42,618,089 | 1.31 | ||||
| NRT2.1 | BraA06g005350.3C | A06 | 3,179,020 | 3,180,823 | 8.16 | 3.34 | |||
| NRT2.5 | BraA08g030690.3C | A08 | 20,761,898 | 20,763,666 | 8.37 | ||||
| NRT2.7 | BraA02g005540.3C | A02 | 2,623,331 | 2,624,856 | −2.01 | ||||
| Nitrate assimilation | GDH2 | BraA03g002960.3C | A03 | 1,321,750 | 1,323,677 | −2.11 | −1.15 | −1.03 | |
| GLU1 | BraA10g032170.3C | A10 | 19,964,582 | 19,972,116 | 1.02 | ||||
| GS2 | BraA08g009150.3C | A08 | 8,134,274 | 8,136,484 | 1.62 | ||||
| GSR1 | BraA04g010460.3C | A04 | 8,265,796 | 8,268,190 | 1.03 | 1.48 | |||
| Photosynthesis | ACA3 | BraA10g032140.3C | A10 | 19,954,817 | 19,956,218 | −2.45 | |||
| BCA3 | BraA07g014010.3C | A07 | 12,918,461 | 12,920,370 | 4.14 | −1.34 | 3.42 | ||
| BCA4 | BraA02g019760.3C | A02 | 11,362,629 | 11,365,101 | 1.21 | ||||
| CA1 | BraA01g044810.3C | A01 | 29,518,958 | 29,521,894 | −1.41 | ||||
| CA2 | BraA02g005650.3C | A02 | 2,674,642 | 2,677,016 | 4.37 | −3.24 | |||
| Auxin synthesis | NIT2 | BraA02g006840.3C | A02 | 3,245,216 | 3,246,972 | 1.28 | |||
| Chemical Element | Nutrient Solution | Treat | |
|---|---|---|---|
| CK(6.0 mM) | LN(0.2 mM) | ||
| ME | KNO3 | 3 mM | 0.1 mM |
| KCl | 0.0 mM | 3 mM | |
| KH2PO4 | 1.5 mM | 1.5 mM | |
| K2HPO4 | 1.5 mM | 1.5 mM | |
| (NH4)SO4 | 0.75 mM | 25 µM | |
| MgCl2 | 1 mM | 1 mM | |
| K2SO4 | 2 mM | 2 mM | |
| Ca(NO3)2 | 0.75 mM | 0.75mM | |
| CaCl2 | 3.25 mM | 3.25 mM | |
| Fe | Na2Fe-EDTA | 40 µM | 40 µM |
| TE | H3BO3 | 60 µM | 60 µM |
| MnSO4 | 14 µM | 14 µM | |
| ZnSO4 | 1 µM | 1 µM | |
| CuSO4 | 0.6 µM | 0.6 µM | |
| NiCl2 | 0.4 µM | 0.4 µM | |
| H2MoO4 | 0.3 µM | 0.3 µM | |
| CoCl2 | 20 nM | 20 nM | |
| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| BraA08g033360.3C (Bra-Action) | ATACCAGGCTTGAGCATACCG | GCCAAAGAGGCCATCAGACAA |
| BraA06g009460.3C | ATTGAGCACATTGGCGTTAGGA | ACCACTTGGAGAAAACGAGGAAA |
| BraA06g009470.3C | CTTCCTGAAACAAAATCTCAAACCC | AAGCCACCGAGGAGACAGAGC |
| BraA08g031180.3C | TTATTGCGGCGGAAGGCG | CATCGGTTAGTGTTGAAAGTGTCCA |
| BraA09g032690.3C | ATTTCCTCCTGGCTGCCTTGA | TGGGTCGCTCCTCGTAGTCGT |
| BraA02g006840.3C | CGGTGCTGCTATTTGCTG | CTTCGTGTTGCTCTGGGT |
| BraA03g002960.3C | TGAAACACAAAGAGGCAA | ATGGAGCAACAGTTCGTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, J.; Zhou, D.; Song, J.; Gao, J. Nitrogen Uptake and Distribution in Different Chinese Cabbage Genotypes under Low Nitrogen Stress. Int. J. Mol. Sci. 2022, 23, 1573. https://doi.org/10.3390/ijms23031573
Zhang Y, Li J, Zhou D, Song J, Gao J. Nitrogen Uptake and Distribution in Different Chinese Cabbage Genotypes under Low Nitrogen Stress. International Journal of Molecular Sciences. 2022; 23(3):1573. https://doi.org/10.3390/ijms23031573
Chicago/Turabian StyleZhang, Yihui, Jingjuan Li, Dandan Zhou, Jie Song, and Jianwei Gao. 2022. "Nitrogen Uptake and Distribution in Different Chinese Cabbage Genotypes under Low Nitrogen Stress" International Journal of Molecular Sciences 23, no. 3: 1573. https://doi.org/10.3390/ijms23031573
APA StyleZhang, Y., Li, J., Zhou, D., Song, J., & Gao, J. (2022). Nitrogen Uptake and Distribution in Different Chinese Cabbage Genotypes under Low Nitrogen Stress. International Journal of Molecular Sciences, 23(3), 1573. https://doi.org/10.3390/ijms23031573






