Expression Characterization of AtPDI11 and Functional Analysis of AtPDI11 D Domain in Oxidative Protein Folding
Abstract
1. Introduction
2. Results
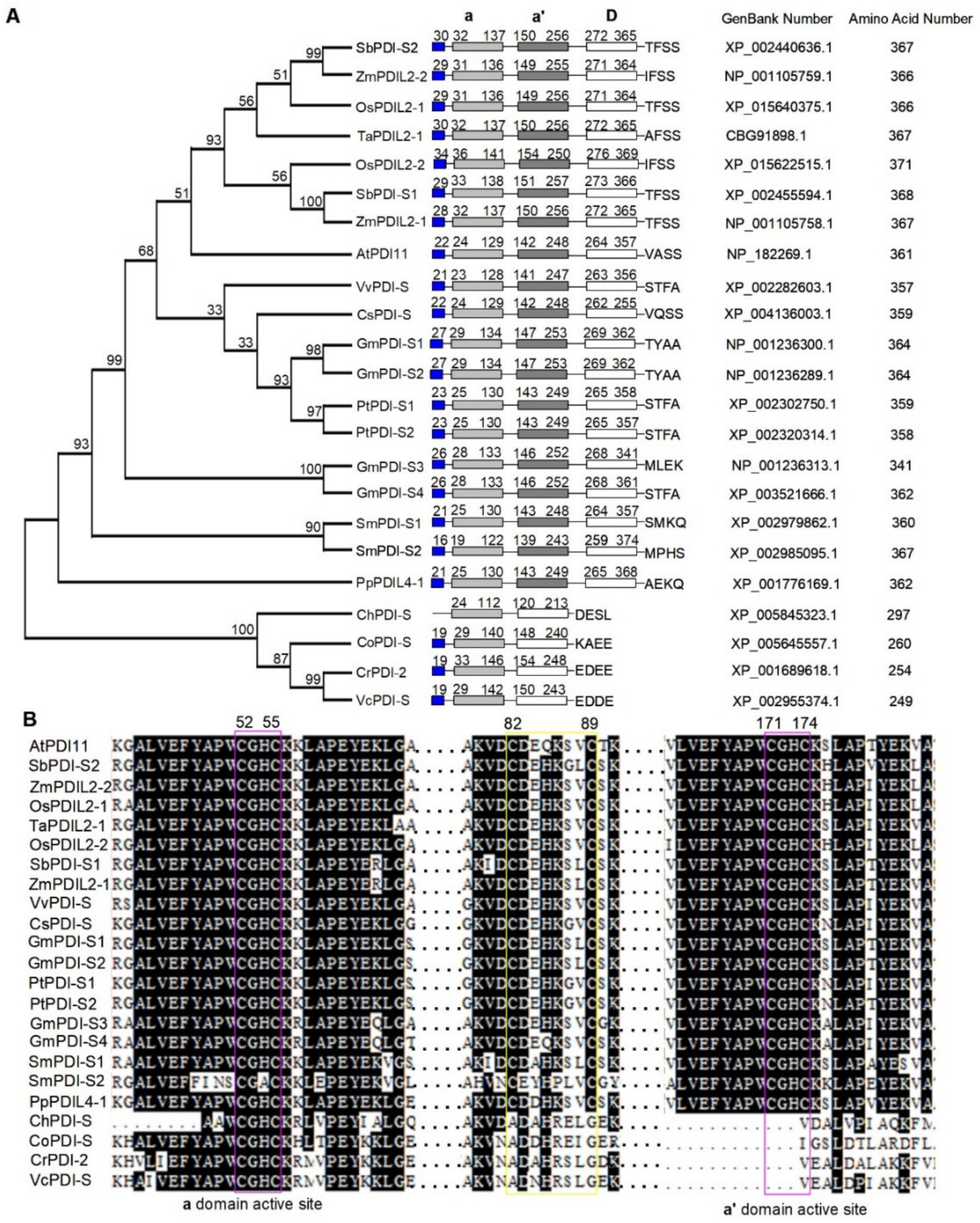
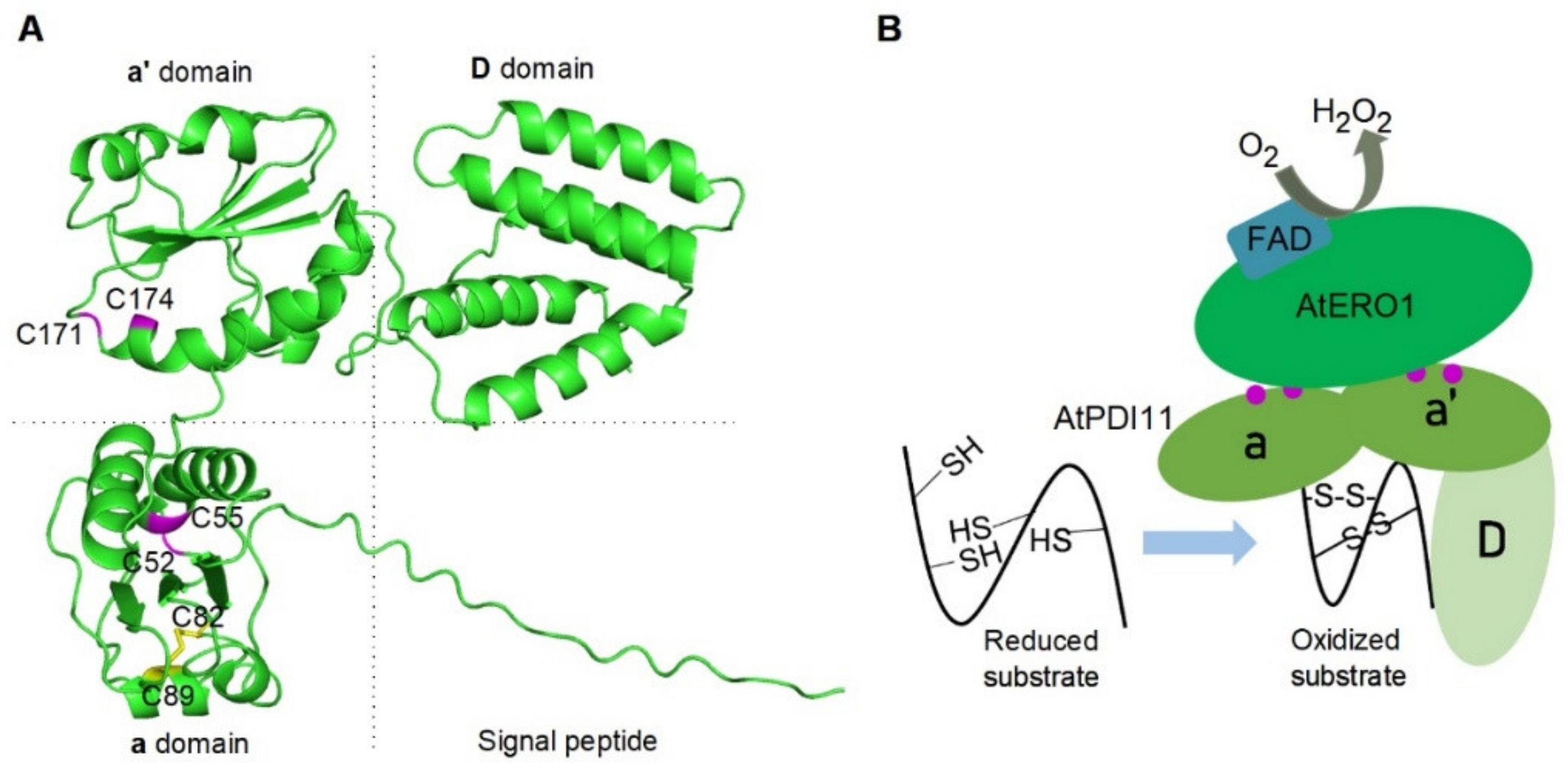
2.1. Sequence Analysis of AtPDI11 and AtPDI11 Homologs from Various Species
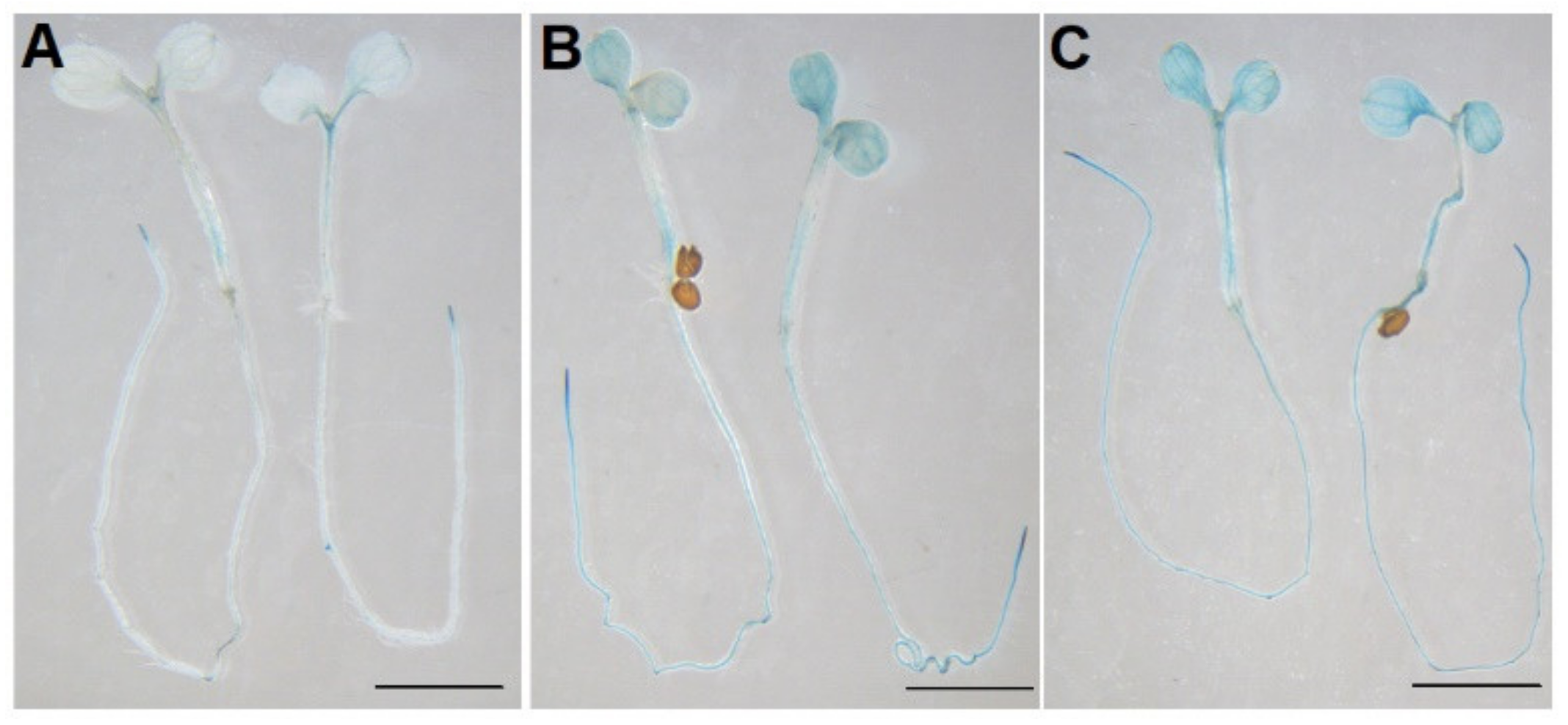
2.2. Histochemical Analysis Reveals Tissue-Specific Expression of AtPDI11 Transcripts
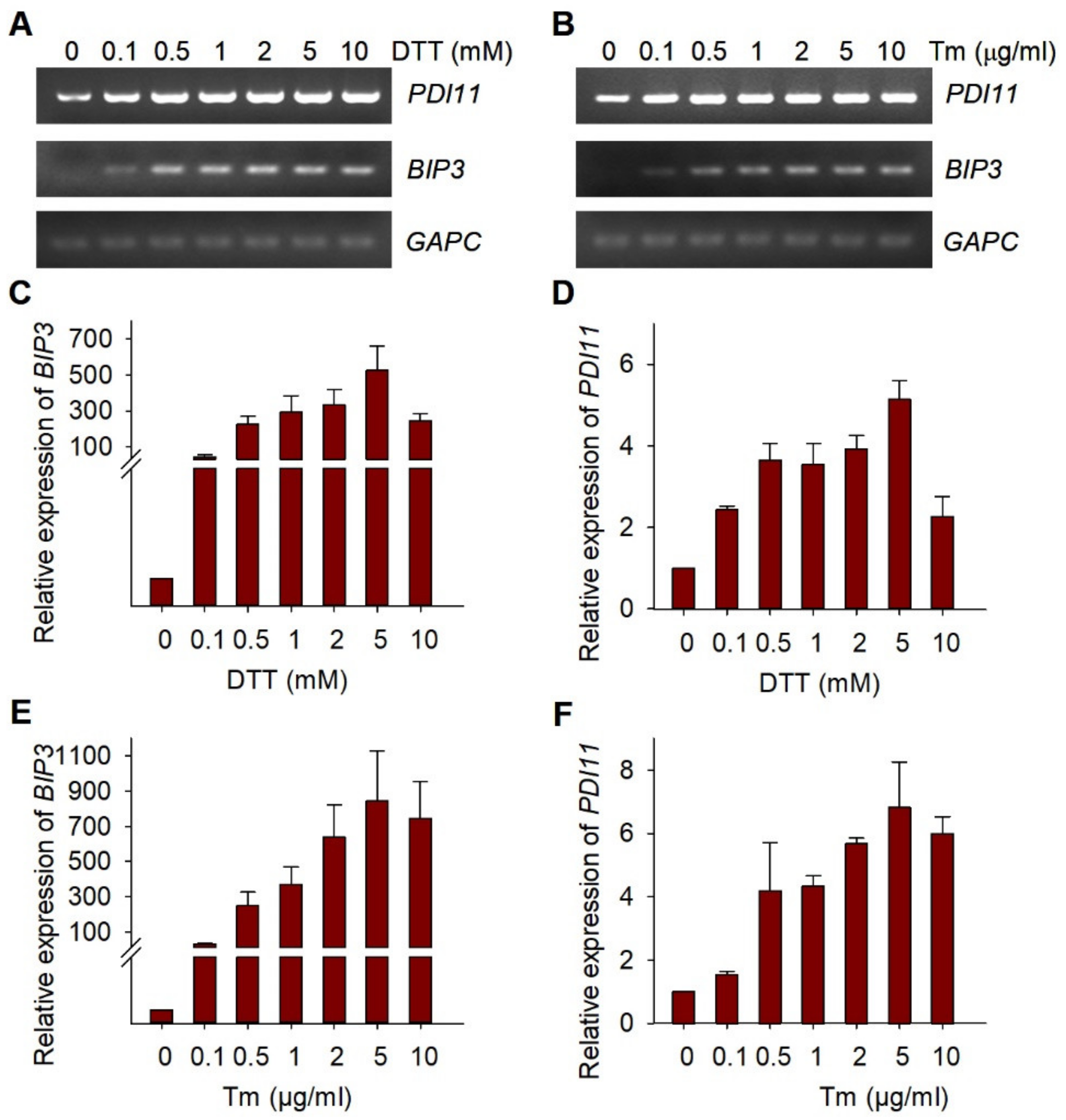
2.3. Expression of AtPDI11 Is Induced by ER Stress
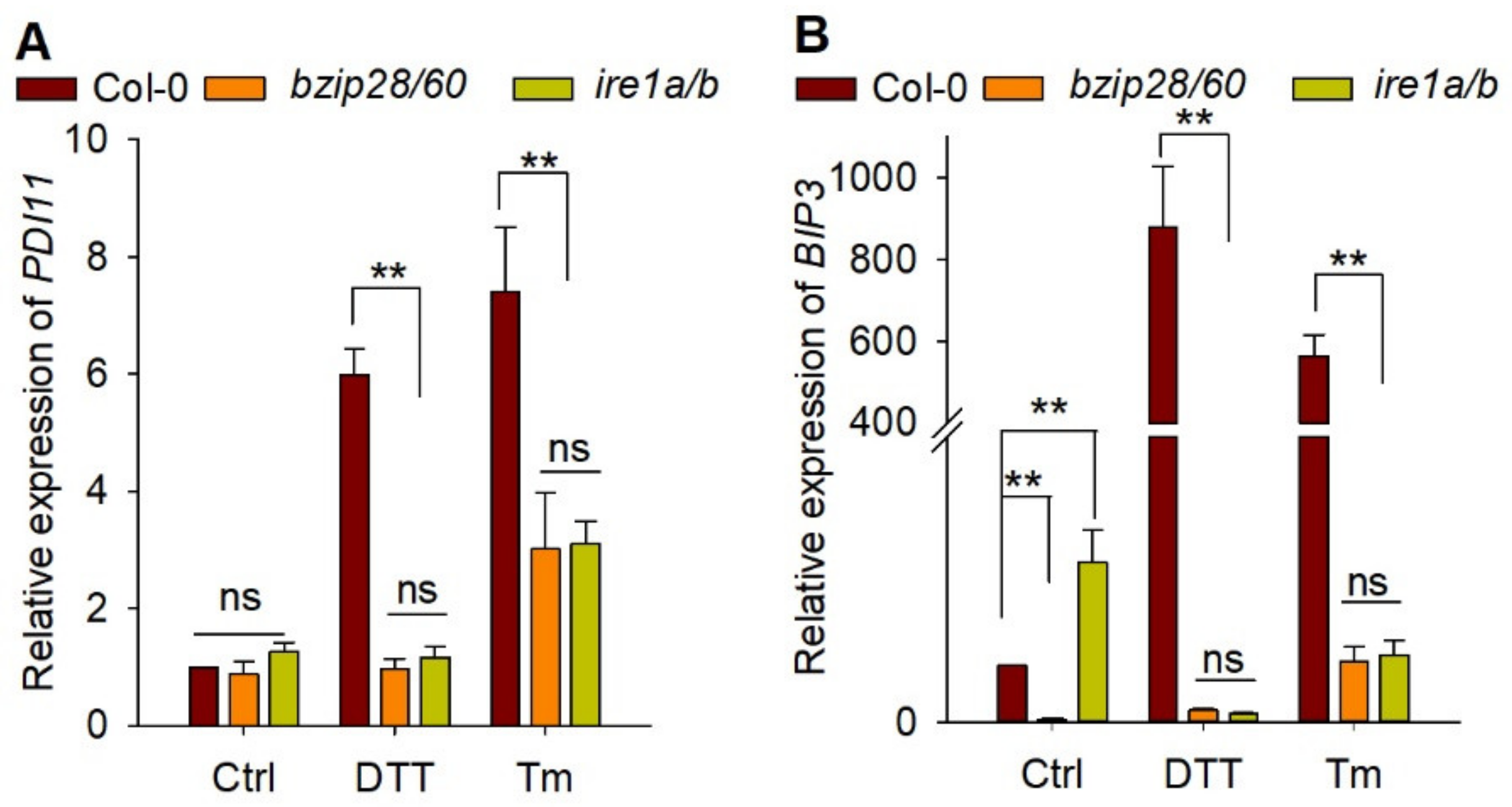
2.4. The Induction of AtPDI11 by ER Stress Is Governed by Key UPR Signaling Mediators
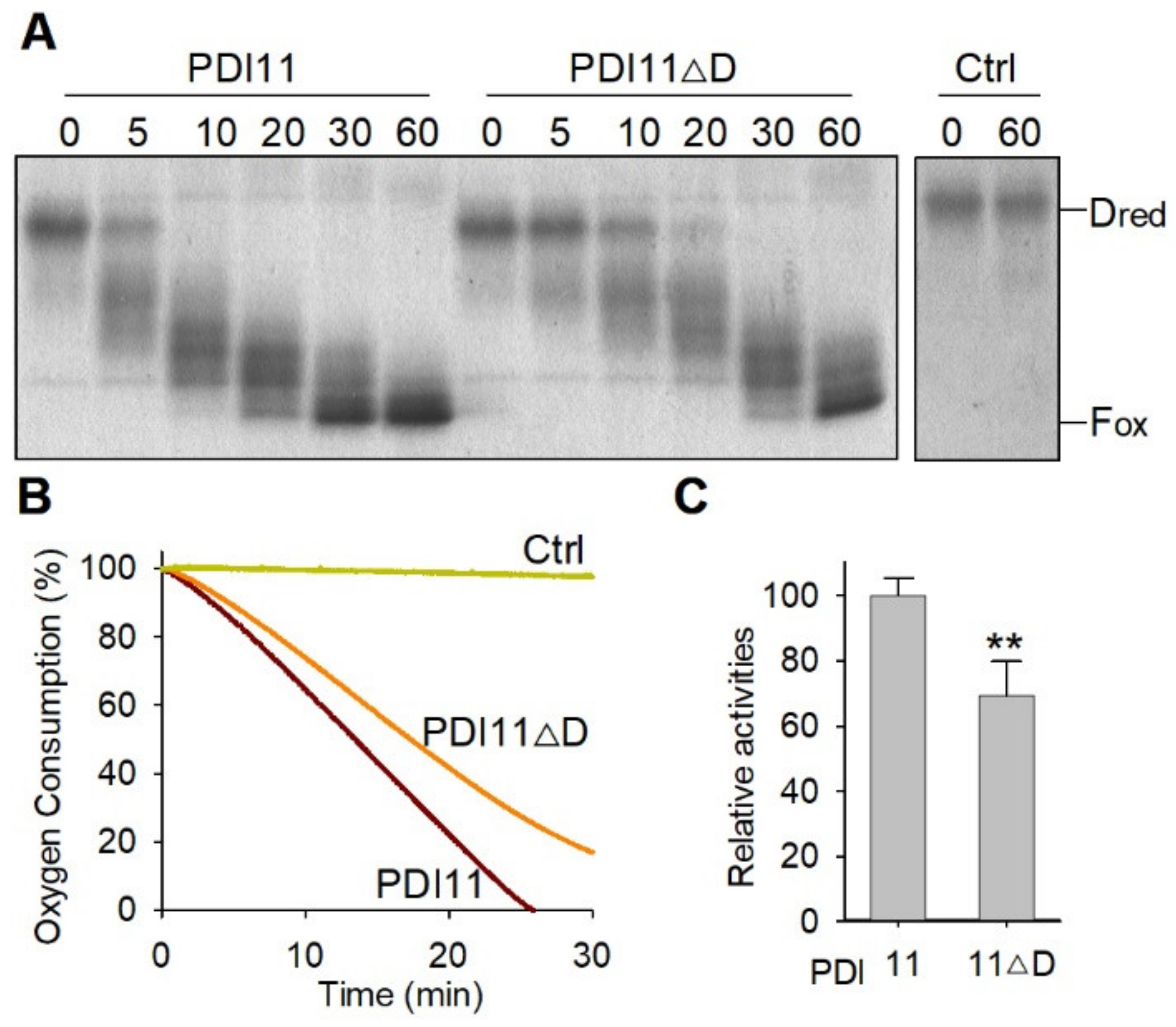
2.5. AtPDI11 D Domain Is Required for Its Oxidative Protein Folding Activity
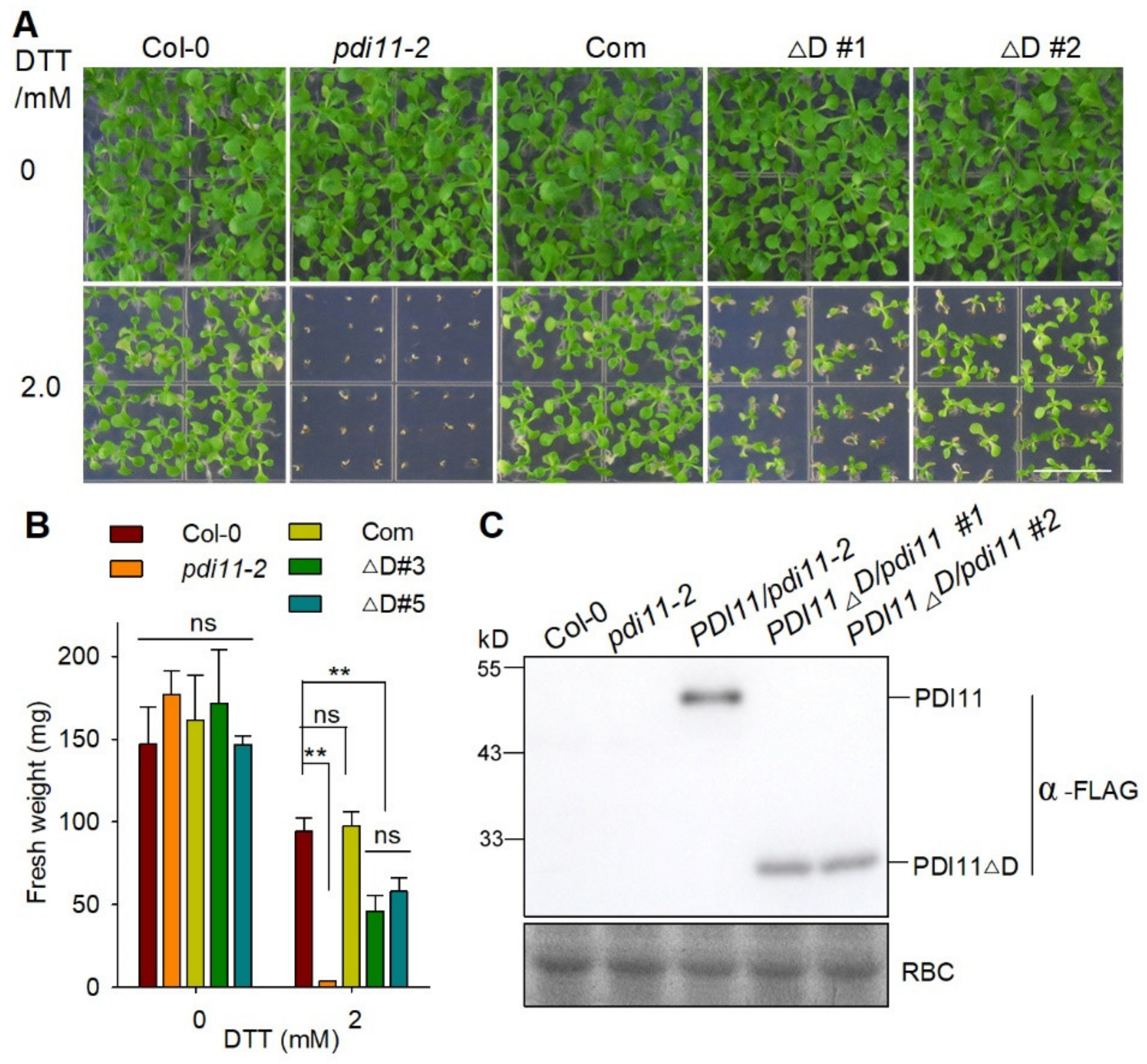
2.6. The D Domain Is Required for the Role of AtPDI11 When Plants Are Grown under Reducing Conditions
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction and Generation of Transgenic Plants
4.3. Recombinant Proteins Expression and Purification
4.4. Phylogenetic Analysis and Sequence Alignment
4.5. RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (RT-PCR)
4.6. Histochemical Staining
4.7. Oxygen Consumption Assay
4.8. Gel-Based Denatured and Reduced RNase A Reoxidation Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hwang, C.; Sinskey, A.J.; Lodish, H.F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 1992, 257, 1496–1502. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J.; Riemer, J.; Rouhier, N. Oxidative protein folding: State-of-the-art and current avenues of research in plants. New Phytol. 2019, 221, 1230–1246. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Iemura, S.; Kamiya, Y.; Ron, D.; Kato, K.; Natsume, T.; Nagata, K. Ero1-alpha and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J. Cell Biol. 2013, 202, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Vitale, A.; Boston, R.S. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 2008, 9, 1581–1588. [Google Scholar] [CrossRef]
- Deng, Y.; Srivastava, R.; Howell, S.H. Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int. J. Mol. Sci. 2013, 14, 8188–8212. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Liu, J.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428. [Google Scholar] [CrossRef]
- Sun, J.; Li, J.; Wang, M.; Song, Z.; Liu, J. Protein Quality Control in Plant Organelles: Current Progress and Future Perspectives. Mol. Plant 2021, 14, 95–114. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Wang, C.C. Protein disulfide-isomerase, a folding catalyst and a redox-regulated chaperone. Free. Radic. Biol. Med. 2015, 83, 305–313. [Google Scholar] [CrossRef]
- Tian, G.; Xiang, S.; Noiva, R.; Lennarz, W.J.; Schindelin, H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 2006, 124, 61–73. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Ren, J.; Fang, J.; Ke, H.; Gong, W.; Feng, W.; Wang, C.C. Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase. Antioxid. Redox Signal. 2013, 19, 36–45. [Google Scholar] [CrossRef]
- Selles, B.; Jacquot, J.P.; Rouhier, N. Comparative genomic study of protein disulfide isomerases from photosynthetic organisms. Genomics 2011, 97, 37–50. [Google Scholar] [CrossRef][Green Version]
- Yuen, C.Y.; Wong, K.; Christopher, D.A. Phylogenetic characterization and promoter expression analysis of a novel hybrid protein disulfide isomerase/cargo receptor subfamily unique to plants and chromalveolates. Mol. Genet. Genom. MGG 2016, 291, 455–469. [Google Scholar] [CrossRef]
- Lu, D.; Christopher, D.A. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genom. MGG 2008, 280, 199–210. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, L.; Zhou, Z.; Wang, M.; Wang, X.; Zhang, S.; Sun, Q. The thiol-disulfide exchange activity of AtPDI1 is involved in the response to abiotic stresses. BMC Plant Biol. 2021, 21, 557. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Li, R.; Yuan, L.; Dai, Y.; Wang, X. Identification and Functional Analysis of a Protein Disulfide Isomerase (AtPDI1) in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 913. [Google Scholar] [CrossRef]
- Kumar, M.N.; Hsieh, Y.F.; Verslues, P.E. At14a-Like1 participates in membrane-associated mechanisms promoting growth during drought in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 10545–10550. [Google Scholar] [CrossRef]
- Wittenberg, G.; Levitan, A.; Klein, T.; Dangoor, I.; Keren, N.; Danon, A. Knockdown of the Arabidopsis thaliana chloroplast protein disulfide isomerase 6 results in reduced levels of photoinhibition and increased D1 synthesis in high light. Plant J. Cell Mol. Biol. 2014, 78, 1003–1013. [Google Scholar] [CrossRef]
- Andème Ondzighi, C.; Christopher, D.A.; Cho, E.J.; Chang, S.-C.; Staehelin, L.A. Arabidopsis Protein Disulfide Isomerase-5 Inhibits Cysteine Proteases during Trafficking to Vacuoles before Programmed Cell Death of the Endothelium in Developing Seeds. Plant Cell 2008, 20, 2205–2220. [Google Scholar] [CrossRef]
- Cho, E.J.; Yuen, C.Y.; Kang, B.H.; Ondzighi, C.A.; Staehelin, L.A.; Christopher, D.A. Protein disulfide isomerase-2 of Arabidopsis mediates protein folding and localizes to both the secretory pathway and nucleus, where it interacts with maternal effect embryo arrest factor. Mol. Cells 2011, 32, 459–475. [Google Scholar] [CrossRef]
- Feldeverd, E.; Porter, B.W.; Yuen, C.Y.L.; Iwai, K.; Carrillo, R.; Smith, T.; Barela, C.; Wong, K.; Wang, P.; Kang, B.H.; et al. The Arabidopsis Protein Disulfide Isomerase Subfamily M Isoform, PDI9, Localizes to the Endoplasmic Reticulum and Influences Pollen Viability and Proper Formation of the Pollen Exine During Heat Stress. Front. Plant Sci. 2020, 11, 610052. [Google Scholar] [CrossRef]
- Yuen, C.Y.; Shek, R.; Kang, B.H.; Matsumoto, K.; Cho, E.J.; Christopher, D.A. Arabidopsis protein disulfide isomerase-8 is a type I endoplasmic reticulum transmembrane protein with thiol-disulfide oxidase activity. BMC Plant Biol. 2016, 16, 181. [Google Scholar] [CrossRef]
- Yuen, C.Y.L.; Wang, P.; Kang, B.-H.; Matsumoto, K.; Christopher, D.A. A Non-Classical Member of the Protein Disulfide Isomerase Family, PDI7 of Arabidopsis thaliana, Localizes to the cis-Golgi and Endoplasmic Reticulum Membranes. Plant Cell Physiol. 2017, 58, 1103–1117. [Google Scholar] [CrossRef]
- Fan, F.; Zhang, Y.; Huang, G.; Zhang, Q.; Wang, C.C.; Wang, L.; Lu, D. AtERO1 and AtERO2 Exhibit Differences in Catalyzing Oxidative Protein Folding in the Endoplasmic Reticulum. Plant Physiol. 2019, 180, 2022–2033. [Google Scholar] [CrossRef]
- Fan, F.; Zhang, Q.; Zhang, Y.; Huang, G.; Liang, X.; Wang, C.-C.; Wang, L.; Lu, D. Two Protein Disulfide Isomerase Subgroups Work Synergistically in Catalyzing Oxidative Protein Folding. Plant Physiol. 2022, 188, 241–254. [Google Scholar] [CrossRef]
- Matsusaki, M.; Okuda, A.; Masuda, T.; Koishihara, K.; Mita, R.; Iwasaki, K.; Hara, K.; Naruo, Y.; Hirose, A.; Tsuchi, Y.; et al. Cooperative Protein Folding by Two Protein Thiol Disulfide Oxidoreductases and 1 in Soybean. Plant Physiol. 2016, 170, 774–789. [Google Scholar] [CrossRef]
- Barak, N.N.; Neumann, P.; Sevvana, M.; Schutkowski, M.; Naumann, K.; Malesević, M.; Reichardt, H.; Fischer, G.; Stubbs, M.T.; Ferrari, D.M. Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29. J. Mol. Biol. 2009, 385, 1630–1642. [Google Scholar] [CrossRef]
- Wang, H.; Boavida, L.C.; Ron, M.; McCormick, S. Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell 2008, 20, 3300–3311. [Google Scholar] [CrossRef]
- Wadahama, H.; Kamauchi, S.; Ishimoto, M.; Kawada, T.; Urade, R. Protein disulfide isomerase family proteins involved in soybean protein biogenesis. FEBS J. 2007, 274, 687–703. [Google Scholar] [CrossRef]
- Fan, F.; Zhang, Y.; Wang, S.; Han, Y.; Wang, L.; Lu, D. Characterization of the oxidative protein folding activity of a unique plant oxidoreductase, Arabidopsis protein disulfide isomerase-11. Biochem. Biophys. Res. Commun. 2018, 495, 1041–1047. [Google Scholar] [CrossRef]
- Zhou, Y.; Fan, F.; Han, Y.; Lu, D. Arabidopsis PDI11 interacts with lectin molecular chaperons calreticulin 1 and 2 through its D domain. Biochem. Biophys. Res. Commun. 2022, 588, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Srivastava, R.; Howell, S.H. Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 19633–19638. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Kishigami, S.; Sone, M.; Inokuchi, H.; Mogi, T.; Ito, K. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA 1997, 94, 11857–11862. [Google Scholar] [CrossRef]
- Monnat, J.; Neuhaus, E.M.; Pop, M.S.; Ferrari, D.M.; Kramer, B.; Soldati, T. Identification of a novel saturable endoplasmic reticulum localization mechanism mediated by the C-terminus of a Dictyostelium protein disulfide isomerase. Mol. Biol. Cell 2000, 11, 3469–3484. [Google Scholar] [CrossRef]
- Lai, Y.S.; Renna, L.; Yarema, J.; Ruberti, C.; He, S.Y.; Brandizzi, F. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2018, 115, 5203–5212. [Google Scholar] [CrossRef]
- Korner, C.J.; Du, X.; Vollmer, M.E.; Pajerowska-Mukhtar, K.M. Endoplasmic Reticulum Stress Signaling in Plant Immunity—At the Crossroad of Life and Death. Int. J. Mol. Sci. 2015, 16, 26582–26598. [Google Scholar] [CrossRef]
- Ruberti, C.; Lai, Y.; Brandizzi, F. Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J. Cell Mol. Biol. 2018, 93, 155–165. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.J.; Sidhu, A.; Zhu, L.; Liang, Y.; Freedman, R.B.; Wang, C.C. Reconstitution of human Ero1-Lalpha/protein-disulfide isomerase oxidative folding pathway in vitro. Position-dependent differences in role between the a and a′ domains of protein-disulfide isomerase. J. Biol. Chem. 2009, 284, 199–206. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Sun, J.; Xu, Y.; Ye, S.; Jiang, H.; Chen, Q.; Liu, F.; Zhou, W.; Chen, R.; Li, X.; Tietz, O.; et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, F.; Zhang, H.; Wei, Q.; Wei, Y. Expression Characterization of AtPDI11 and Functional Analysis of AtPDI11 D Domain in Oxidative Protein Folding. Int. J. Mol. Sci. 2022, 23, 1409. https://doi.org/10.3390/ijms23031409
Fan F, Zhang H, Wei Q, Wei Y. Expression Characterization of AtPDI11 and Functional Analysis of AtPDI11 D Domain in Oxidative Protein Folding. International Journal of Molecular Sciences. 2022; 23(3):1409. https://doi.org/10.3390/ijms23031409
Chicago/Turabian StyleFan, Fenggui, Hao Zhang, Qian Wei, and Yahui Wei. 2022. "Expression Characterization of AtPDI11 and Functional Analysis of AtPDI11 D Domain in Oxidative Protein Folding" International Journal of Molecular Sciences 23, no. 3: 1409. https://doi.org/10.3390/ijms23031409
APA StyleFan, F., Zhang, H., Wei, Q., & Wei, Y. (2022). Expression Characterization of AtPDI11 and Functional Analysis of AtPDI11 D Domain in Oxidative Protein Folding. International Journal of Molecular Sciences, 23(3), 1409. https://doi.org/10.3390/ijms23031409





