Interplay between Selenium, Selenoproteins and HIV-1 Replication in Human CD4 T-Lymphocytes
Abstract
:1. Introduction
2. Results
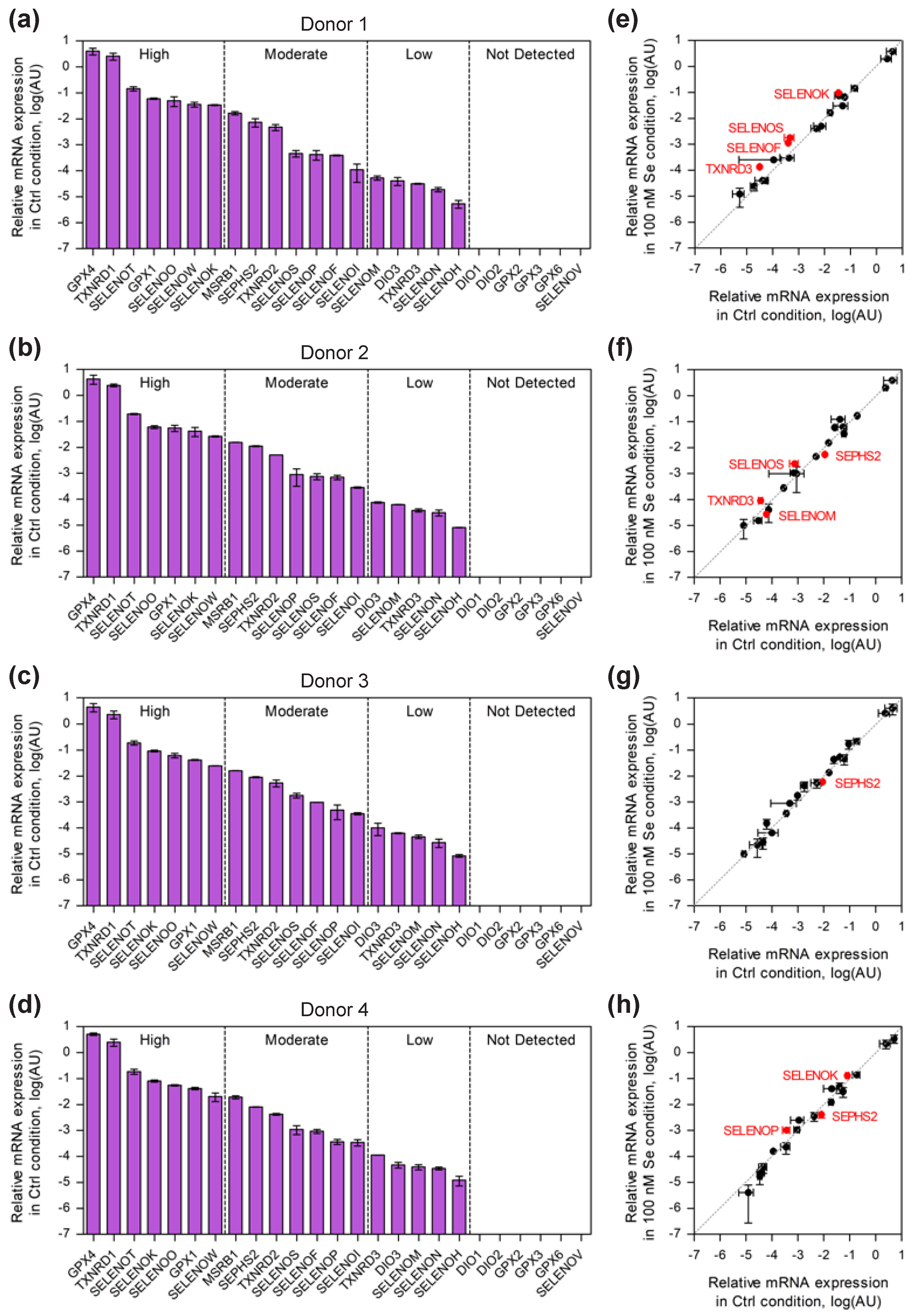
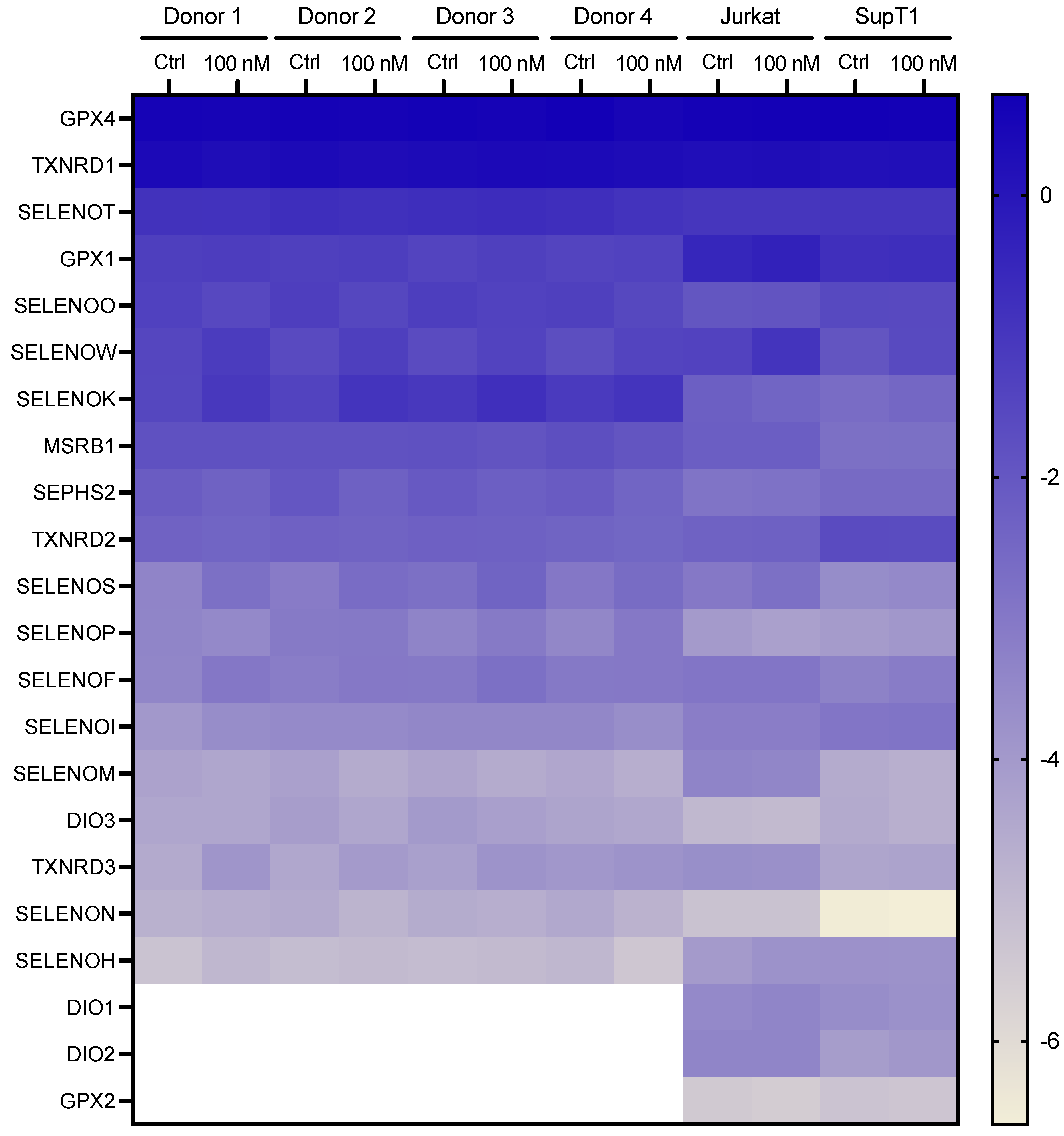
2.1. Comparison of Selenoprotein mRNA Expression Pattern in Established and Primary CD4 T-Cells
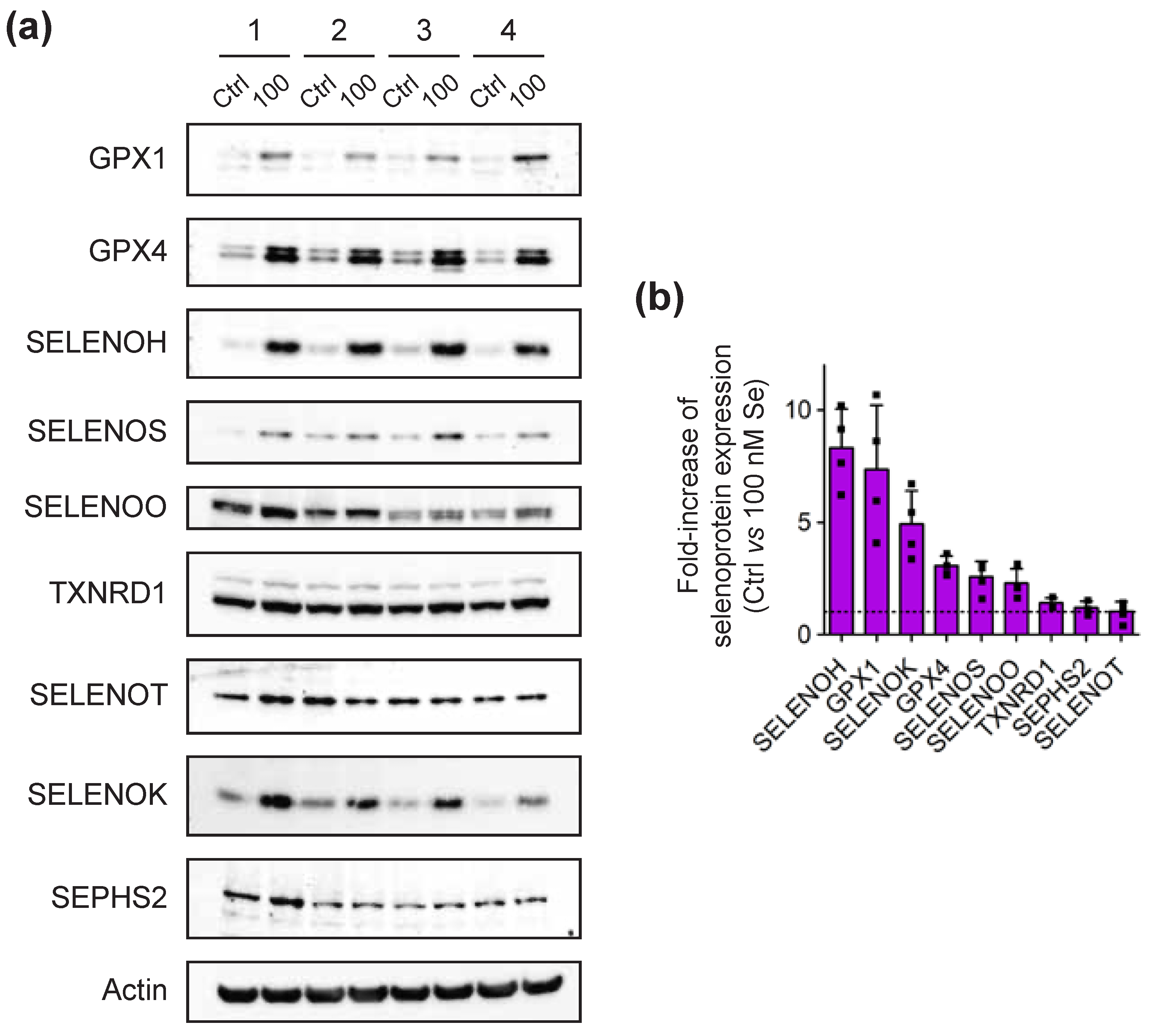
2.2. Selenium-Dependent Hierarchy of Selenoprotein Expression Levels in Established and Primary CD4 T-Cells
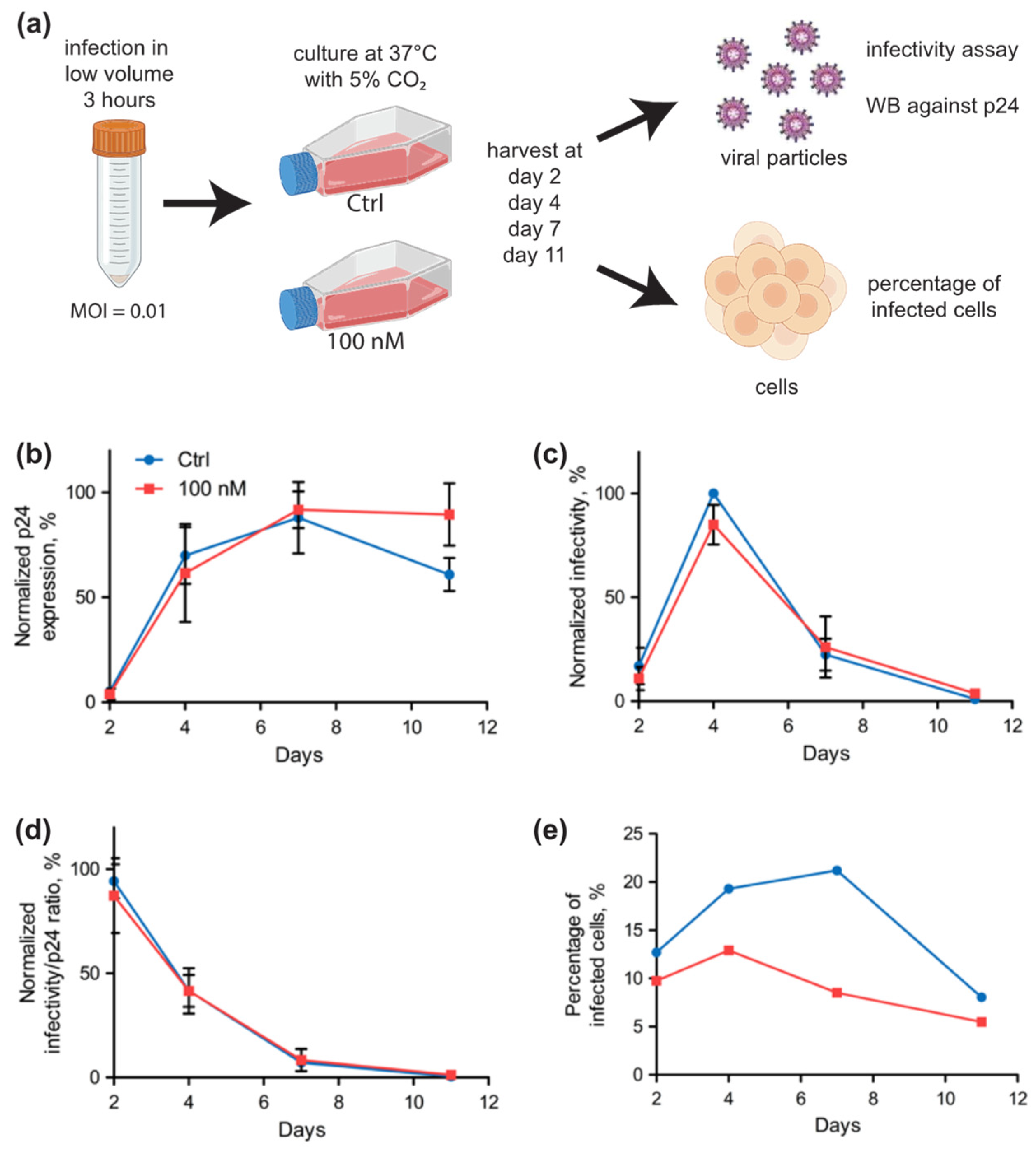
2.3. Selenium Levels Did Not Affect HIV-1 Replication in Jurkat Cells but Modified the Proportion of Infected Cells
2.4. Selenium Levels Did Not Affect HIV-1 Replication in Primary Cells but Modified the Proportion of Infected Cells
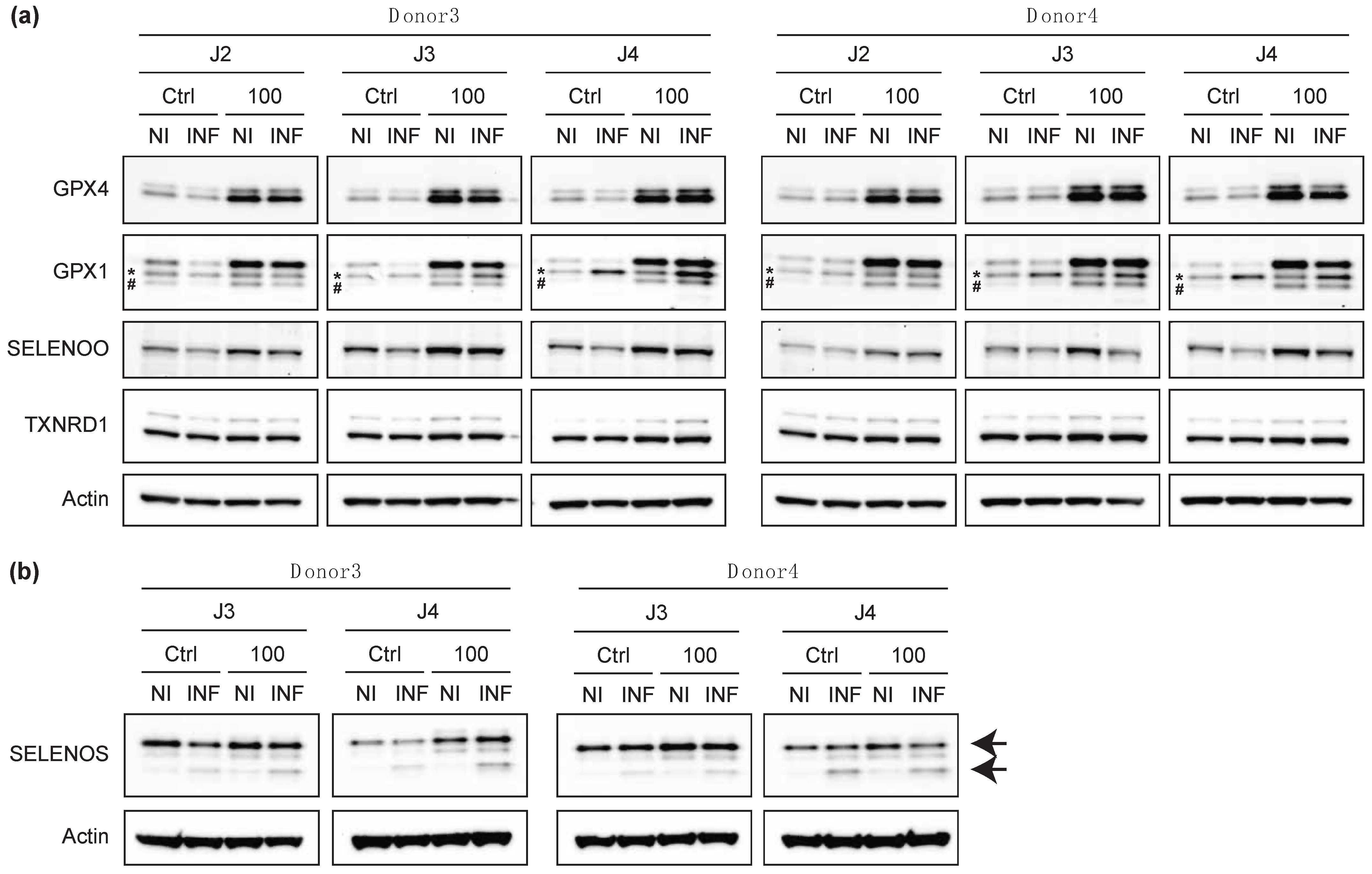
2.5. The Infection of Primary T Cells with HIV-1 Altered the Levels of Certain Selenoproteins
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Isolation and Culture of Primary CD4 T-Cells
4.4. HIV-1 Production in HEK293T Cells and Titration
4.5. Infection of CD4 T-Cells with HIV-1
4.6. Quantification of Infectious HIV-1 Produced by CD4 T Cells
4.7. Quantification of p24 Contained in Viral Particles
4.8. Total RNA Extraction and Analysis by RT-qPCR
4.9. Protein Extraction and Analysis by Western Blot
4.10. GPX and TXNRD Enzymatic Assays
4.11. Flow Cytometry
4.12. Ethics Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferguson, M.R.; Rojo, D.R.; von Lindern, J.J.; O’Brien, W.A. HIV-1 replication cycle. Clin. Lab. Med. 2002, 22, 611–635. [Google Scholar] [CrossRef]
- Nyamweya, S.; Hegedus, A.; Jaye, A.; Rowland-Jones, S.; Flanagan, K.L.; Macallan, D.C. Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev. Med. Virol. 2013, 23, 221–240. [Google Scholar] [CrossRef] [PubMed]
- Samji, H.; Cescon, A.; Hogg, R.S.; Modur, S.P.; Althoff, K.N.; Buchacz, K.; Burchell, A.N.; Cohen, M.; Gebo, K.A.; Gill, M.J.; et al. Closing the gap: Increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS ONE 2013, 8, e81355. [Google Scholar] [CrossRef] [PubMed]
- Palella, F.J., Jr.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Guillin, O.M.; Vindry, C.; Ohlmann, T.; Chavatte, L. Selenium, Selenoproteins and Viral Infection. Nutrients 2019, 11, 2101. [Google Scholar] [CrossRef] [Green Version]
- Pitney, C.L.; Royal, M.; Klebert, M. Selenium supplementation in HIV-infected patients: Is there any potential clinical benefit? J. Assoc. Nurses AIDS Care 2009, 20, 326–333. [Google Scholar] [CrossRef]
- Bogden, J.D.; Oleske, J.M. The essential trace minerals, immunity, and progression of HIV-1 infection. Nutr. Res. 2007, 27, 69–77. [Google Scholar] [CrossRef]
- Kamwesiga, J.; Mutabazi, V.; Kayumba, J.; Tayari, J.-C.K.; Uwimbabazi, J.C.; Batanage, G.; Uwera, G.; Baziruwiha, M.; Ntizimira, C.; Murebwayire, A.; et al. Effect of selenium supplementation on CD4+ T-cell recovery, viral suppression and morbidity of HIV-infected patients in Rwanda: A randomized controlled trial. AIDS 2015, 29, 1045–1052. [Google Scholar] [CrossRef] [Green Version]
- Baum, M.K.; Campa, A.; Lai, S.; Sales Martinez, S.; Tsalaile, L.; Burns, P.; Farahani, M.; Li, Y.; van Widenfelt, E.; Page, J.B.; et al. Effect of micronutrient supplementation on disease progression in asymptomatic, antiretroviral-naive, HIV-infected adults in Botswana: A randomized clinical trial. JAMA 2013, 310, 2154–2163. [Google Scholar] [CrossRef] [Green Version]
- Hurwitz, B.E.; Klaus, J.R.; Llabre, M.M.; Gonzalez, A.; Lawrence, P.J.; Maher, K.J.; Greeson, J.M.; Baum, M.K.; Shor-Posner, G.; Skyler, J.S.; et al. Suppression of human immunodeficiency virus type 1 viral load with selenium supplementation: A randomized controlled trial. Arch. Intern. Med. 2007, 167, 148–154. [Google Scholar] [CrossRef]
- de Menezes Barbosa, E.G.; Junior, F.B.; Machado, A.A.; Navarro, A.M. A longer time of exposure to antiretroviral therapy improves selenium levels. Clin. Nutr. 2015, 34, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Campa, A.; Sales Martinez, S.; Baum, M.K. Selenium in HIV/AIDS. In Selenium: Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Schweizer, U., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer Science + Business Media, LLC: New York, NY, USA, 2016; pp. 333–342. [Google Scholar]
- Schomburg, L. Selenium Deficiency Due to Diet, Pregnancy, Severe Illness, or COVID-19-A Preventable Trigger for Autoimmune Disease. Int. J. Mol. Sci. 2021, 22, 8532. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.A.; Levander, O.A.; Handy, J. Selenium deficiency and viral infection. J. Nutr. 2003, 133 (Suppl. 1), 1463S–1467S. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.A.; Nelson, H.K.; Shi, Q.; Van Dael, P.; Schiffrin, E.J.; Blum, S.; Barclay, D.; Levander, O.A. Selenium deficiency increases the pathology of an influenza virus infection. FASEB J. 2001, 15, 1481–1483. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.A. Rapid genomic evolution of a non-virulent coxsackievirus B3 in selenium-deficient mice. Biomed. Environ. Sci. 1997, 10, 307–315. [Google Scholar]
- Beck, M.A.; Shi, Q.; Morris, V.C.; Levander, O.A. Rapid genomic evolution of a non-virulent coxsackievirus B3 in selenium-deficient mice results in selection of identical virulent isolates. Nat. Med. 1995, 1, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Sonet, J.; Bulteau, A.-L.; Chavatte, L. Selenium and Selenoproteins in Human Health and Diseases. In Metallomics: Analytical Techniques and Speciation Methods; Michalke, B., Ed.; 2016 Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2016; pp. 364–381. [Google Scholar]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [Green Version]
- Papp, L.V.; Lu, J.; Holmgren, A.; Khanna, K.K. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid. Redox Signal. 2007, 9, 775–806. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigo, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [Green Version]
- Driscoll, D.M.; Copeland, P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003, 23, 17–40. [Google Scholar] [CrossRef]
- Brigelius-Flohe, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta 2012, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.S. Focus on mammalian thioredoxin reductases—Important selenoproteins with versatile functions. Biochim. Biophys. Acta 2009, 1790, 495–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiong, W.; Chen, L.L.; Huang, J.Q.; Lei, X.G. Selenoprotein V protects against endoplasmic reticulum stress and oxidative injury induced by pro-oxidants. Free Radic. Biol. Med. 2020, 160, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Pothion, H.; Jehan, C.; Tostivint, H.; Cartier, D.; Bucharles, C.; Falluel-Morel, A.; Boukhzar, L.; Anouar, Y.; Lihrmann, I. Selenoprotein T: An Essential Oxidoreductase Serving as a Guardian of Endoplasmic Reticulum Homeostasis. Antioxid. Redox Signal. 2020, 33, 1257–1275. [Google Scholar] [CrossRef] [PubMed]
- Rocca, C.; Pasqua, T.; Boukhzar, L.; Anouar, Y.; Angelone, T. Progress in the emerging role of selenoproteins in cardiovascular disease: Focus on endoplasmic reticulum-resident selenoproteins. Cell. Mol. Life Sci. 2019, 76, 3969–3985. [Google Scholar] [CrossRef]
- Pitts, M.W.; Hoffmann, P.R. Endoplasmic reticulum-resident selenoproteins as regulators of calcium signaling and homeostasis. Cell Calcium 2018, 70, 76–86. [Google Scholar] [CrossRef]
- Addinsall, A.B.; Wright, C.R.; Andrikopoulos, S.; van der Poel, C.; Stupka, N. Emerging roles of endoplasmic reticulum-resident selenoproteins in the regulation of cellular stress responses and the implications for metabolic disease. Biochem. J. 2018, 475, 1037–1057. [Google Scholar] [CrossRef]
- Avery, J.C.; Hoffmann, P.R. Selenium, Selenoproteins, and Immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef] [Green Version]
- Vindry, C.; Ohlmann, T.; Chavatte, L. Translation regulation of mammalian selenoproteins. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2480–2492. [Google Scholar] [CrossRef]
- Bulteau, A.-L.; Chavatte, L. Update on selenoprotein biosynthesis. Antioxid. Redox Signal. 2015, 23, 775–794. [Google Scholar] [CrossRef]
- Sonet, J.; Bulteau, A.L.; Touat-Hamici, Z.; Mosca, M.; Bierla, K.; Mounicou, S.; Lobinski, R.; Chavatte, L. Selenoproteome Expression Studied by Non-Radioactive Isotopic Selenium-Labeling in Human Cell Lines. Int. J. Mol. Sci. 2021, 22, 7308. [Google Scholar] [CrossRef] [PubMed]
- Touat-Hamici, Z.; Bulteau, A.L.; Bianga, J.; Jean-Jacques, H.; Szpunar, J.; Lobinski, R.; Chavatte, L. Selenium-regulated hierarchy of human selenoproteome in cancerous and immortalized cells lines. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2493–2505. [Google Scholar] [CrossRef] [PubMed]
- Hammad, G.; Legrain, Y.; Touat-Hamici, Z.; Duhieu, S.; Cornu, D.; Bulteau, A.L.; Chavatte, L. Interplay between Selenium Levels and Replicative Senescence in WI-38 Human Fibroblasts: A Proteomic Approach. Antioxidants 2018, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Touat-Hamici, Z.; Legrain, Y.; Sonet, J.; Bulteau, A.-L.; Chavatte, L. Alteration of selenoprotein expression during stress and in aging. In Selenium: Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Schweizer, U., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer Science + Business Media, LLC: New York, NY, USA, 2016; pp. 539–551. [Google Scholar]
- Touat-Hamici, Z.; Legrain, Y.; Bulteau, A.-L.; Chavatte, L. Selective up-regulation of human selenoproteins in response to oxidative stress. J. Biol. Chem. 2014, 289, 14750–14761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legrain, Y.; Touat-Hamici, Z.; Chavatte, L. Interplay between selenium levels, selenoprotein expression, and replicative senescence in WI-38 human fibroblasts. J. Biol. Chem. 2014, 289, 6299–6310. [Google Scholar] [CrossRef] [Green Version]
- Bock, A.; Forchhammer, K.; Heider, J.; Leinfelder, W.; Sawers, G.; Veprek, B.; Zinoni, F. Selenocysteine: The 21st amino acid. Mol. Microbiol. 1991, 5, 515–520. [Google Scholar] [CrossRef]
- Bock, A.; Forchhammer, K.; Heider, J.; Baron, C. Selenoprotein synthesis: An expansion of the genetic code. Trends Biochem. Sci. 1991, 16, 463–467. [Google Scholar] [CrossRef]
- Lejeune, F. Nonsense-mediated mRNA decay at the crossroads of many cellular pathways. BMB Rep. 2017, 50, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Vindry, C.; Guillin, O.; Mangeot, P.E.; Ohlmann, T.; Chavatte, L. A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA([Ser]Sec) Gene. Cells 2019, 8, 574. [Google Scholar] [CrossRef] [Green Version]
- Dalley, B.K.; Baird, L.; Howard, M.T. Studying Selenoprotein mRNA Translation Using RNA-Seq and Ribosome Profiling. Methods Mol. Biol. 2018, 1661, 103–123. [Google Scholar]
- Fradejas-Villar, N.; Seeher, S.; Anderson, C.B.; Doengi, M.; Carlson, B.A.; Hatfield, D.L.; Schweizer, U.; Howard, M.T. The RNA-binding protein Secisbp2 differentially modulates UGA codon reassignment and RNA decay. Nucleic Acids Res. 2017, 45, 4094–4107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, M.T.; Carlson, B.A.; Anderson, C.B.; Hatfield, D.L. Translational redefinition of UGA codons is regulated by selenium availability. J. Biol. Chem. 2013, 288, 19401–19413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gladyshev, V.N.; Stadtman, T.C.; Hatfield, D.L.; Jeang, K.T. Levels of major selenoproteins in T cells decrease during HIV infection and low molecular mass selenium compounds increase. Proc. Natl. Acad. Sci. USA 1999, 96, 835–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariotti, M.; Ridge, P.G.; Zhang, Y.; Lobanov, A.V.; Pringle, T.H.; Guigo, R.; Hatfield, D.L.; Gladyshev, V.N. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS ONE 2012, 7, e33066. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, A.V.; Fomenko, D.E.; Zhang, Y.; Sengupta, A.; Hatfield, D.L.; Gladyshev, V.N. Evolutionary dynamics of eukaryotic selenoproteomes: Large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome. Biol. 2007, 8, R198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlenius, T.C.; Shah, F.; Yu, W.C.; Hawkes, H.J.; Tinggi, U.; Clarke, F.M.; Tonissen, K.F. The selenium content of cell culture serum influences redox-regulated gene expression. Biotechniques 2011, 50, 295–301. [Google Scholar] [CrossRef]
- Brigelius-Flohe, R.; Kipp, A.P. Physiological functions of GPx2 and its role in inflammation-triggered carcinogenesis. Ann. N. Y. Acad. Sci. 2012, 1259, 19–25. [Google Scholar] [CrossRef]
- Stafford, W.C.; Peng, X.; Olofsson, M.H.; Zhang, X.; Luci, D.K.; Lu, L.; Cheng, Q.; Tresaugues, L.; Dexheimer, T.S.; Coussens, N.P.; et al. Irreversible inhibition of cytosolic thioredoxin reductase 1 as a mechanistic basis for anticancer therapy. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Sonet, J.; Mosca, M.; Bierla, K.; Modzelewska, K.; Flis-Borsuk, A.; Suchocki, P.; Ksiazek, I.; Anuszewska, E.; Bulteau, A.L.; Szpunar, J.; et al. Selenized Plant Oil Is an Efficient Source of Selenium for Selenoprotein Biosynthesis in Human Cell Lines. Nutrients 2019, 11, 1524. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, P.; Desfarges, S.; Bartha, I.; Joos, B.; Zangger, N.; Munoz, M.; Gunthard, H.F.; Beerenwinkel, N.; Telenti, A.; Ciuffi, A. 24 hours in the life of HIV-1 in a T cell line. PLoS Pathog. 2013, 9, e1003161. [Google Scholar] [CrossRef]
- Prelli Bozzo, C.; Kmiec, D.; Kirchhoff, F. When good turns bad: How viruses exploit innate immunity factors. Curr. Opin. Virol. 2021, 52, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Kluge, S.F.; Sauter, D.; Kirchhoff, F. SnapShot: Antiviral restriction factors. Cell 2015, 163, 774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandstrom, P.A.; Murray, J.; Folks, T.M.; Diamond, A.M. Antioxidant defenses influence HIV-1 replication and associated cytopathic effects. Free Radic. Biol. Med. 1998, 24, 1485–1491. [Google Scholar] [CrossRef]
- Naamati, A.; Williamson, J.C.; Greenwood, E.J.; Marelli, S.; Lehner, P.J.; Matheson, N.J. Functional proteomic atlas of HIV infection in primary human CD4+ T cells. eLife 2019, 8, e41431. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, E.J.; Matheson, N.J.; Wals, K.; van den Boomen, D.J.; Antrobus, R.; Williamson, J.C.; Lehner, P.J. Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. eLife 2016, 5, e18296. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, V.N.; Arner, E.S.; Berry, M.J.; Brigelius-Flohe, R.; Bruford, E.A.; Burk, R.F.; Carlson, B.A.; Castellano, S.; Chavatte, L.; Conrad, M.; et al. Selenoprotein Gene Nomenclature. J. Biol. Chem. 2016, 291, 24036–24040. [Google Scholar] [CrossRef] [Green Version]
- Sonet, J.; Bierla, K.; Bulteau, A.L.; Lobinski, R.; Chavatte, L. Comparison of analytical methods using enzymatic activity, immunoaffinity and selenium-specific mass spectrometric detection for the quantitation of glutathione peroxidase 1. Anal. Chim. Acta 2018, 1011, 11–19. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guillin, O.M.; Vindry, C.; Ohlmann, T.; Chavatte, L. Interplay between Selenium, Selenoproteins and HIV-1 Replication in Human CD4 T-Lymphocytes. Int. J. Mol. Sci. 2022, 23, 1394. https://doi.org/10.3390/ijms23031394
Guillin OM, Vindry C, Ohlmann T, Chavatte L. Interplay between Selenium, Selenoproteins and HIV-1 Replication in Human CD4 T-Lymphocytes. International Journal of Molecular Sciences. 2022; 23(3):1394. https://doi.org/10.3390/ijms23031394
Chicago/Turabian StyleGuillin, Olivia M., Caroline Vindry, Théophile Ohlmann, and Laurent Chavatte. 2022. "Interplay between Selenium, Selenoproteins and HIV-1 Replication in Human CD4 T-Lymphocytes" International Journal of Molecular Sciences 23, no. 3: 1394. https://doi.org/10.3390/ijms23031394
APA StyleGuillin, O. M., Vindry, C., Ohlmann, T., & Chavatte, L. (2022). Interplay between Selenium, Selenoproteins and HIV-1 Replication in Human CD4 T-Lymphocytes. International Journal of Molecular Sciences, 23(3), 1394. https://doi.org/10.3390/ijms23031394