Abstract
Autoimmune diseases are disorders that destruct or disrupt the body’s own tissues by its own immune system. Several studies have revealed that polymorphisms of multiple genes are involved in autoimmune diseases. Meanwhile, gene therapy has become a promising approach in autoimmune diseases, and clustered regularly interspaced palindromic repeats and CRISPR-associated protein 9 (CRISPR-Cas9) has become one of the most prominent methods. It has been shown that CRISPR-Cas9 can be applied to knock out proprotein convertase subtilisin/kexin type 9 (PCSK9) or block PCSK9, resulting in lowering low-density lipoprotein cholesterol. In other studies, it can be used to treat rare diseases such as ornithine transcarbamylase (OTC) deficiency and hereditary tyrosinemia. However, few studies on the treatment of autoimmune disease using CRISPR-Cas9 have been reported so far. In this review, we highlight the current and potential use of CRISPR-Cas9 in the management of autoimmune diseases. We summarize the potential target genes for immunomodulation using CRISPR-Cas9 in autoimmune diseases including rheumatoid arthritis (RA), inflammatory bowel diseases (IBD), systemic lupus erythematosus (SLE), multiple sclerosis (MS), type 1 diabetes mellitus (DM), psoriasis, and type 1 coeliac disease. This article will give a new perspective on understanding the use of CRISPR-Cas9 in autoimmune diseases not only through animal models but also in human models. Emerging approaches to investigate the potential target genes for CRISPR-Cas9 treatment may be promising for the tailored immunomodulation of some autoimmune diseases in the near future.
1. Introduction
Autoimmune diseases are disorders that destructs or disrupts the body’s own tissues by its own immune system [1]. Normally, the body’s immune system has immunologic tolerance so that it does not hurt its own body. However, in an autoimmune disease, the immune system attacks its own tissues and organs. Autoimmune diseases occur in up to 3–5% of the general population [1], and they decrease life expectancy. It has been shown that autoimmune diseases are associated with mood disorders [2] and impaired quality of life. Additionally, it has been known that various genetic factors account for autoimmune diseases, such as rheumatoid arthritis, systemic lupus erythematosus, type 1 diabetes mellitus, and multiple sclerosis [3].
Meanwhile, gene therapy has become a promising approach as our understanding of the immunological and molecular basis of autoimmune diseases advances [4]. The ultimate objective of gene therapy is regulation of the level of inflammatory cytokines and infiltration of lymphocytes to the affected sites (Figure 1) [4]. There has been a growing need for gene-editing therapeutic approaches, with clustered regularly interspaced palindromic repeats and CRISPR-associated protein 9 (CRIPSR-Cas9) being one of the most prominent methods [5]. CRISPR-Cas9, which has been widely renowned as genome-editing technology, is originally an adaptive immune system of bacteria and archaea and can be applicable in eukaryotic cells with single guide RNA (sgRNA) that contains complementary base sequence of a gene of interest [6]. By creating a synthetic sgRNA of a sequence complementary to a particular gene, Cas9, an RNA-guided DNA nuclease can knock out the gene (Figure 2) [7]. Gene knockout occurs during repair through non-homologous end joining (NHEJ) responding to the double strand breakage. In addition, knock-in can be performed via homologous direct recombination (HDR) if adequate template DNA is present [7,8]. The possibility of therapeutic application of CRISPR-Cas9 has been emerging based on various studies ranging from gene-editing in embryo with germline genetic variant to in vivo delivery in animal models of specific genetic diseases using viral or lipid vectors [5].
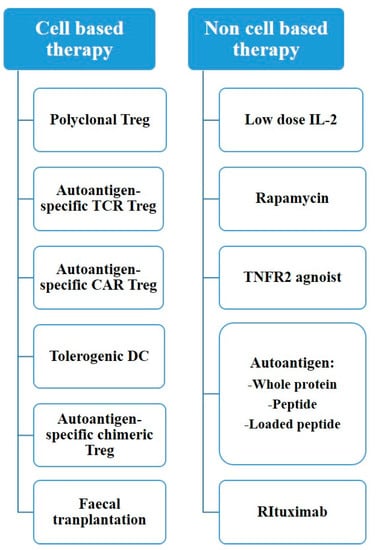
Figure 1.
Treatment options for autoimmune disease.
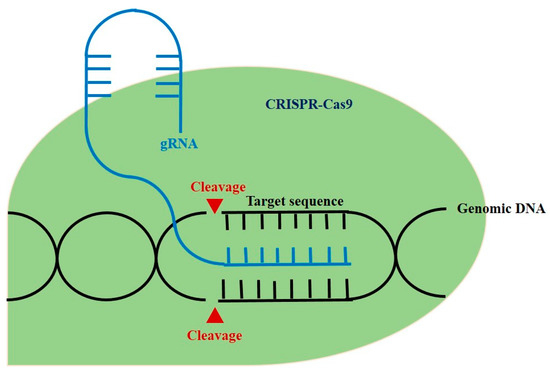
Figure 2.
The schematic diagram of CRISPR-Cas 9 in genomic engineering.
Application of CRISPR-Cas9 in genetic diseases has been reported in several investigations [9,10,11,12,13,14,15]. CRISPR-Cas9 can be applied to knock out proprotein convertase subtilisinkexin type 9 (PCSK9) or block PCSK9, resulting in lowering low-density lipoprotein cholesterol [9,10]. Possibilities are not limited to frequently occurring diseases, and it holds great promise to treat rare diseases such as Ornithinetranscarbamylase (OTC) deficiency and hereditary tyrosinemia [11,12]. However, limited studies on the treatment of autoimmune disease using CRISPR-Cas9 have been reported so far. In this review, we highlight the current and potential use of CRISPR-Cas9 in the management of autoimmune diseases.
2. CRISPR-Cas9 and Autoimmune Diseases
2.1. In Vivo Therapeutic Trials Using CRISPR-Cas9
Autoimmunity usually occurs through systemic immune reaction caused by the failure to regulate the immune response adequately. For in vivo genetic correction via CRISPR-Cas9, sgRNA and Cas9 must be penetrated into the nucleus of the cell to edit the targeted DNA site. There are two major CRISPR-Cas9 delivery systems: viral vectors and lipid nanoparticles [5]. When using the viral transfer vector, adeno-associated virus (AAV), which has low possibility of genomic integration into foreign genes and has been demonstrated to be safe, is most widely used [16]. Given that AAV is small in size, large-sized SpCas9 derived from Streptococcus pyogenes is infrequently used despite its high efficacy, thus smaller Cas9 proteins derived from other bacterial species are used [17]. Cas9 derived from Staphylococcus aureus (SaCas9) or Campylobacter jejuni (CjCas9) can be packed together with sgRNA in AAV and applied to in vivo gene correction with a decent efficiency [18,19].
PCSK9, which regulates low-density lipoprotein (LDL) and is known as a major risk factor for atherosclerosis and coronary heart disease, was knocked out in hepatocytes using NHEJ with SaCas9, resulting in a significant decrease in LDL [20]. Another study reported that in vivo delivery of meganuclease with AAV targeting PCSK9 showed reduced levels of serum cholesterol [9,10]. Rare metabolic diseases have also been studied. A mouse model of OTC deficiency was treated with AAV8 and SaCas9, and a limited but significant therapeutic effect was observed in the management of hereditary tyrosinemia [11,12]. In some studies of Duchenne muscular dystrophy (DMD), the pathogenic mutation within exon23 was knocked out, and improvement in skeletal muscle and myocardial function was observed [13,14,15].
2.2. Inflammatory Molecules, Immunogenetics and CRISPR-Cas9
Autoimmunity is based on the simple concept of an imbalance between pro-inflammatory and anti-inflammatory stimuli, leading to an abnormal immune response. Cytokines like interleukin-1 (IL-1) and tumor necrosis factor (TNF) play an important role in inflammation, thus reducing the expression of these cytokines eventually leads to abrogation of the autoimmune process [21]. For example, interleukin-36 (IL-36) cytokines (IL-36A, IL-36B, and IL-36G) and members of the IL-1 family can cause pro-inflammatory effects in the skin and other organs [22]. Studies have shown that cytokines belonging to the IL-36 family may play an important role in the development of autoimmune diseases such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), inflammatory bowel disease (IBD), Sjögren’s syndrome, and psoriasis vulgaris [23,24,25]. Myeloid differentiation primary response gene 88 (MyD88) adapter protein is known to be activated by IL-36 stimulation. Inactivation of MyD88 adapter protein by CRISPR-Cas9 reduced the activity of differentially expressed genes inducing the expression of IL-1B and IL-36G [26].
Tumor necrosis factor alpha-induced protein 3 (TNFAIP3) is induced by TNF-alpha and is known to inhibit NF-kappa B activation and TNF-mediated apoptosis [27]. The genetic variation of TNFAIP3 has been reported to be associated with susceptibility to develop SLE and RA [28,29]. In SLE, a genome-wide association study demonstrated that UBE2L3 could be a novel therapeutic target. UBE2L3 was identified as the key E2 enzyme for linear ubiquitin chain assembly complex (LUBAC) and essential for LUBAC-mediated activation of NF-kB [30]. In addition, association with familial Behçet-like auto inflammatory syndrome and infantile-onset intractable IBD was also reported [31,32]. A study on transcription activation-like effector nuclease (TALEN)-mediated knockout of TNFAIP3 demonstrated that correcting pathogenic TNFAIP3 variants can possibly reverse its related autoimmune phenotypes [33]. In addition, knockout of a candidate causal variant, rs6927172, by CRISPR-Cas9 gene editing influenced the expression of the IL-20RA and TNFAIP3 genes, potentially involved in the autoimmune response [34].
In addition to cytokines, many genetic variants that contribute to the development of autoimmune diseases are associated with T cells. For example, FoxP3 + regulatory T cells play an important role in immunological tolerance [35]. Immuno-dysregulation polyendocrinopathy, enteropathy X-linked (IPEX) syndrome caused by mutation of FoxP3 in humans can lead to a variety of autoimmune disorders in early childhood [36]. In IPEX, FoxP3 is decreased and the function of Tregs is remarkably degraded. In fact, epigenetic editing of the promoter and conserved non-coding sequence 2(CNS2) in the FoxP3 gene showed a 20–30% reduction in the effector T cell division [37] and thus CRISPR-Cas9 may be a potential therapeutic approach to manage IPEX cases.
2.3. CRISPR-Cas9 and iPSC
In autoimmunity, various treatment methods have been studied using CRISPR-Cas9. Induced pluripotent stem cells (iPSCs) were produced by adding interleukin-1 receptor antagonist (IL1Ra) or soluble TNF receptor gene by using CRISPR-Cas9 gene editing and differentiated into articular cartilage. This showed that inflammatory response was attenuated by initiating dynamic negative feedback upon stimulation with inflammatory cytokines [38]. Other studies have reported that articular cartilage is resistant to IL-1alpha-mediated tissue degradation by the deletion of interleukin-1 receptor 1 (IL1r1) in murine iPSCs [39].
Gene editing in mesenchymal stem cells (MSCs) was utilized with CRISPR-Cas9 to target the endogenous activation of pancreatic transcription factors and MSC chemokine receptors. MSCs were differentiated into surrogate insulin-producing cells and may be transplanted through ex vivo expansion and transplantation while maintaining their immunomodulatory properties [40]. Other studies have successfully activated endogenous human insulin transcription using sgRNAs targeting multiple insulin promoter and a nuclease-deficient Cas9-virion protein 160 gene (dCas9-VP160) in human embryonic kidney 293 T cell (HEK293T), Hela, and human fibroblasts [41].
3. CRISPR-Cas9 and Rheumatoid Arthritis
Three studies tested genetic therapy in human cell models in RA. From these studies, the MYC, FOXO1 gene, SNP rs6927172, TNFAIP3, OLIG3 gene, and miR-155 have been suggested as appropriate candidates for CRISPR-Cas9 treatment.
Yang et al. suggested that MYC and FOXO1 genes are associated with rheumatoid arthritis [42]. It has been thought that in RA patients, CD4+ T-cells express higher autophagy, and MYC was thought to be the regulator of the pathway [42]. FOXO1 has also been thought to be correlated with RA activity [42]. This study, by collecting ATAC-seq, Hi-C, Capture Hi-C, and nuclear RNA-seq data in stimulated CD4+ T cells over 24 hours, provides evidence that MYC and FOXO1 genes may be causal factors of RA [42]. In the study by Yu et al., an intergenic SNP rs6927172 on chromosome 6q23 region was found to be associated with RA disease course according to a genome-wide association study [43]. According to the study, many genes such as IL20RA, IL22RA2, IFNGR1, OLIG3, and TNFAIP3 flank the SNP area, and when they disrupt the SNP area using CRISPR-Cas9, only TNFAIP3 and OLIG3 showed decreased expressivity [43]. It shows that the SNP rs6927172, TNFAIP3 and OLIG3 are importantly associated with RA disease course [43]. Jing et al. demonstrated that microRNA 155 (miR-155) could be an important pro-inflammatory factor in RA patients [44]. They used an miR-155 knockout RAW 264.7 macrophage cell line and found that in the cell line, SHP1 was up-regulated and pro-inflammatory cytokine making process was impaired [44]. Thus, they suggest that genome editing of miR-155 can be a potential therapeutic strategy for RA [44].
4. CRISPR-Cas9 and Inflammatory Bowel Disease
Four studies in human cell models and another four studies in mouse models tested genetic therapy in IBD. From these studies, JAK2, TL1A, SGK2, PTPN2, c-MYC, HDAC7, and IFN- γ genes and miR-125a have been suggested as appropriate candidates for CRISPR-Cas9 treatment.
4.1. Studies on CRISPR-Cas9 and IBD Using Human Immune Cells
Analysis of rs1887428, located in the promoter region of the Janus kinase 2 [JAK2] gene, by Cardinale et al. revealed target genes affected by this polymorphism [45]. Authors found out that the risk allele of rs1887428 is bound by the transcription factor (TF) RBPJ [45]. According to the paper, rs1887428 did not have very large impacts on JAK2 expression but its impact was on STAT5B amplified downwards [45]. The limitation of this study was that it could not find the TF binding motif and expression quantitative trait locus [45]. Mokhtar et al. demonstrated that the expression of SGK2 part of the PI3K/Akt pathway affects IBD disease course [46]. Immunohistochemistry was performed on SW480 cells and SGK2 gene knockdown was performed by CRISPR gRNA [46]. As a result, SGK2 protein was localized in the cytoplasm of colonic epithelial cells both in long and short duration ulcerative colitis [46]. This indicates that SGK2 gene may have a crucial role in IBD disease course [46]. In the study by Li et al., PTPN2 gene variants occurred in Crohn’s disease patients [46]. In these patients, PTPN2 had loss of function mutation and it was associated with rs7234029 SNP [46]. They used subepithelial myofibroblast cells (SEMF) [46]. The result showed that PTPN2 expression was increased in the affected ileum compared to normal ileum and, in contrast, CRISPR-Cas9 mediated PTPN2 deletion resulted in higher levels of STAT3 and Erk1/2 phosphorylation and proliferation [47]. It indicates that PTPN2 gene variants and SNP rs7234029 have a crucial role in Crohn’s disease [47]. Matthews et al. investigated rs6651252 SNP on chromosome 8 [48]. The study demonstrated that rs6651252 resides within a Wnt responsive DNA enhancer element and disease-associated allele increases binding of the TCF7L2 transcription factor to the region [48]. Using CRISPR-Cas9, they found that rs6651252 enhancer regulates expression of the c-MYC, and they revealed that MYC expression levels are elevated in the Crohn’s disease patients [48].
4.2. Studies on CRISPR-Cas9 and IBD Using Non-Human Cells
Pai et al. focused on a tumor necrosis factor super family member TL1A [49]. According to the study, spontaneous ileitis was present in TL1A transgenic mice [49]. When neutralizing anti-TL1A was used, brush border fanning and bacterial endocytosis caused by MLCK reduced [49]. Lastly, the expression of TL1A, IFNγ, and both MLCK1 and 2 was upregulated in the mucosa of IBD patients. [48] Thus, the study demonstrates that the flare-up of TL1A, IFNγ, and MLCK is associated with IBD diseases course [49]. Friedrich et al. used primary colonic epithelial cells (CEC), human T84, and murine CMT93 to characterize functions of HDAC7 [50]. Knockout mice were generated using CRISPR-Cas9 system [50]. The result showed that HDAC function was reduced in IBD patients, and according to the knockout mice, HDAC was found to play a crucial role in maintenance of the intestinal barrier [50]. Eftychi et al. crossed NEMOIEC-KO with IFNγ −/− mice generated by CRISPR-Cas9 to demonstrate that IFN-γ can have a crucial role in colon inflammation [51]. They found that many pro-inflammatory cytokines such as Tnf, IL1b, and IL6 showed decreased expressivity in the knockout mice and suggested that IFN-γ is essential for colon inflammation [51]. Ge et al. showed that microRNA-125a can suppress intestinal mucosal inflammation [52]. According to their results, miR-125a expression was reduced in mucosa and peripheral blood of IBD patients [52]. MiR-125a suppresses Th1/Th17 cell differentiation and TNF-α production. In addition, miR-125a knockout mice showed more severe forms of colitis [52].
5. CRISPR-Cas9 and Systemic Lupus Erythematosus
Two studies tested genetic therapy in human cell models in SLE. From these studies, the A20 DUB and CXorf21 gene have been suggested as appropriate candidates for CRISPR-Cas9 treatment. Odqvist et al. aimed to assess whether TNFAIP3 (A20) deubiquitinase (DUB) increases the risk of SLE [53]. They used CRISPR-Cas9 to make human U937 monocytes with A20 DUB inactivating C103A knock-in mutation [53]. Results showed that A20 C103A cells or cells with rs2230926 polymorphism drew increased neutrophil extracellular trap and increased frequency of auto antibodies to citrullinated epitopes [53]. They concluded that interrupting the A20 DUB domain increases susceptibility to SLE [53]. Harris et al. focused on the involvement of Chromosome X open reading frame 21 (CXorf21) genes in SLE disease course [54]. They conducted in vitro CRISPR-Cas9 knockdown experiments and found that CXorf21 knockdown resulted in a decreased expression of TNF-alpha and IL-6 [54]. They concluded that sexually dimorphic expression of CXorf21 could be a risk factor for SLE [54].
6. CRISPR-Cas9 and Multiple Sclerosis
Four studies tested genetic therapy in human cell models in MS. From these studies, the IR7R gene, the RNA helicase DEAD box polypeptide 39B, the IL2RA gene, and the TNFRSF1A gene have been suggested as appropriate candidates for CRISPR-Cas9 treatment. Gregory et al. demonstrated that the IL7Rα-related immune response pathway is crucial in the pathogenesis of MS [55]. The rs6897932 SNP in exon 6 of IL7R affects gene expression of the soluble versus membrane-bound form of the protein and increases the risk of MS [55]. The soluble form of IL7Rα is a driver of increased MS risk. The ‘C’ allele of rs6897932 is associated with MS, and it enhances skipping of exon 6 of the IL7R gene by augmenting an exon splicing silencer [55]. The finding suggests that rs6897932 may affect the amount of the soluble versus membrane-bound form of the protein [55]. Isoforms are significant to regulate IL7 signaling pathway [55]. As a result, there is a direct relationship between alternative rs6897932 alleles and MS risk. Galarza-Munoz et al. demonstrated that human epistatic interaction is associated with MS risk. RNA helicase DDX39B, which is a potent activator of IL7R exon 6 and repressor of the soluble form of IL7R, strongly correlates with MS risk [56]. It was shown that epistatic interaction between rs2523506 in DDX39B and rs6897932 in IL7R regulates IL7R exon 6 splicing and increases risk of MS [56]. Not only local mutations in the IL7R gene, but also genetic and functional epistasis with IR7R gene is related to the risk of MS. In another study, Maier et al. showed that the IL-2RA gene has many variants that increase the risk of MS, and several variants were associated with sIL-2RA levels independently [57]. Maier et al. investigated IL-2RA genetic heterogeneity in MS and type 1 diabetes mellitus (T1DM) together, which are related to share alleles. IL2RA variants contributed to the risk of MS and T1DM respectively [57]. rs2104286 SNP located in intron 1 of IL2RA was mainly associated with susceptibility of MS [56]. Gregory et al. showed that TNF blocking agents are effective treatment options for non-MS autoimmune diseases. The investigation suggests that rs1800693 SNP in the TNFRSF1A gene, which encodes tumor necrosis factor receptor 1 (TNFR1), is associated with MS as the causal variant [58]. TNF blocking agents have side effects that promote the onset of MS and other autoimmune diseases such as RA. However, the study by Gregory et al. reported a disease signal for rs1800693 predictive of side effects [58].
7. CRISPR-Cas9 and Type 1 Diabetes Mellitus
One study tested genetic therapy in human cell models in Type 1 Diabetes mellitus (DM), and the other study tested this in mice models. From these studies, the AID/RAD51, and SNP rs10914542 of LCK gene have been suggested as appropriate candidates for CRISPR-Cas9 treatment. Zhu et al. attempted to reveal the role of SNPs of the Lymphocyte-specific protein tyrosine kinase (LCK) gene [59]. They gained blood samples from type 1 DM patients and used CRISPR-Cas9 to find the role of LCK SNP [59]. Among SNPs, only SNP rs10914542 showed meaningful association [59]. Thus, they demonstrate that G allele of rs10914542 SNP in LCK increases the risk of Type 1 DM [59]. Jeremy et al. used activation-induced cytidine deaminase (AID) gene knockout mice and found that AID/RAD51 could be a target for treatment of Type 1 DM patients [60].
8. CRISPR-Cas9 and Psoriasis
One study tested genetic therapy in human cell models in psoriasis and the other study tested this in mouse models. From these studies, desmoglein 1 and ERAP1 genes have been suggested as appropriate candidates for CRISPR-Cas9 treatment. Arakawa et al. demonstrated that ERAP1 has a crucial role in the development of psoriasis [61]. They generated ERAP1 knockout melanoma cell line and revealed that epistasis between HLA-C*06:02 and ERAP1 variants affects psoriasis [61]. The course includes ADAMTS-like protein 5 (ADAMTSL5) and immunogenicity of melanocytes [61]. Roth-Carter et al. used desmoglein 1 knockout mice and found that the knockout mice exhibited barrier impairment [62]. Analysis of E18.5 skin of knockout mice revealed that inhibition of Dsg1 leads to increase in pathways to psoriatic process [62]. These results support a role of Dsg1 for differentiation of epidermis, formation of barriers, and regulation of inflammatory responses [62].
9. CRISPR-Cas9 and Type 1 Coeliac Disease
One study tested genetic therapy in cell models regarding coeliac disease [63]. From the study, the α- or γ-gliadin genes have been suggested as appropriate candidates for CRISPR-Cas9 treatment. According to Jouanin et al., wheat grains contain gluten proteins in which immunogenic epitopes can trigger coeliac disease [63]. They analyzed α- and γ-gliadin gene sequences and generated CRISPR-Cas9 constructs targeting α- or γ-gliadins [63]. The result showed that it is possible to use CRISPR-Cas9 to edit α- or γ-gliadins and make safe forms of grains [63].
10. Conclusions
In this study, we tried to widen our understanding of the role of CRISPR-Cas9 on the treatment of autoimmune diseases through a comprehensive review of literatures. We also reviewed target genes and SNPs of various autoimmune diseases manipulated by the CRISPR-Cas9 method. The review of target genes and SNPs is organized in Table 1 and Table 2. As autoimmunity is based on an imbalance between pro-inflammatory and anti-inflammatory stimuli, cytokines such as IL1, IL36, and TNF alpha and T cell related factors are important in autoimmune diseases. By creating a synthetic sgRNA of a sequence complementary to a particular gene, Cas9, an RNA-guided DNA nuclease can knock out the genes related to the cytokines and T cell factors. Therefore, CRISPR-Cas9 may be a potential therapeutic approach to manage autoimmune diseases.

Table 1.
Human cell models using CRISPR-Cas9 in autoimmune diseases.

Table 2.
Non-human cell models using CRISPR-Cas9 in autoimmune diseases.
Recently, gene therapy and especially CRISPR-Cas9 method has become a promising approach as our understanding of the immunological and molecular basis of autoimmune diseases advances. Possibilities are not limited to frequently occurring diseases; it even holds great promise to treat rare diseases. Nevertheless, many of the studies on the treatment of autoimmune diseases using CRISPR-Cas9 have been conducted in cell studies so far and, therefore, more studies in humans are needed. Additionally, several technical challenges need to be addressed, such as off-target activity, insufficient indel or low homology-directed repair efficiency, in vivo delivery of the CRISPR-Cas9 system components, and immune responses [64]. Despite these limitations, the current evidence implies a promising role of CRISPR-Cas9 in the regulation of autoimmune diseases and further studies that focus on effectiveness of CRISPR-Cas9 method to human treatment are needed.
Author Contributions
All authors made substantial contributions to all of the following: conception and design of the study, J.I.S. and J.H.K.; data acquisition, M.H.L., S.P., J.B.H., Y.P. and E.C.; analysis, K.H.L., D.H.C. and K.T.; interpretation of data, A.K., L.J. and S.P.; drafting or critical revision of the article for intellectual content, J.W.Y., J.I.S., L.S., K.T. and K.H.L.; final approval of version to be submitted, J.W.Y., J.H.K. and M.H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We express our sincere gratitude to the students at Yonsei University College of Medicine who assisted in the preparation of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Marrack, P.; Kappler, J.; Kotzin, B.L. Autoimmune disease: Why and where it occurs. Nat. Med. 2001, 7, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Benros, M.E.; Waltoft, B.L.; Nordentoft, M.; Ostergaard, S.D.; Eaton, W.W.; Krogh, J.; Mortensen, P.B. Autoimmune diseases and severe infections as risk factors for mood disorders a nationwide study. JAMA Psychiatry 2013, 70, 812–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, G.S.; Miller, F.W.; Pandey, J.P. The role of genetic factors in autoimmune disease: Implications for environmental research. Environ. Health Perspect. 1999, 107 (Suppl. S5), 693–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, S.-A.; Wang, J.; Tao, M.-H.; Leung, P.S.C. Gene therapy for autoimmune disease. Clin. Rev. Allergy Immunol. 2015, 49, 163–176. [Google Scholar] [CrossRef]
- Yin, H.; Kauffman, K.J.; Anderson, D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017, 16, 387–399. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. Crispr-mediated modular rna-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [Green Version]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with crispr-cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of crispr-cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [Green Version]
- Ding, Q.; Strong, A.; Patel, K.M.; Ng, S.L.; Gosis, B.S.; Regan, S.N.; Cowan, C.A.; Rader, D.J.; Musunuru, K. Permanent alteration of pcsk9 with in vivo crispr-cas9 genome editing. Circ. Res. 2014, 115, 488–492. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Smith, J.; Breton, C.; Clark, P.; Zhang, J.; Ying, L.; Che, Y.; Lape, J.; Bell, P.; Calcedo, R.; et al. Meganuclease targeting of pcsk9 in macaque liver leads to stable reduction in serum cholesterol. Nat. Biotechnol. 2018, 36, 717–725. [Google Scholar] [CrossRef]
- Pankowicz, F.P.; Barzi, M.; Legras, X.; Hubert, L.; Mi, T.; Tomolonis, J.A.; Ravishankar, M.; Sun, Q.; Yang, D.; Borowiak, M.; et al. Reprogramming metabolic pathways in vivo with crispr/cas9 genome editing to treat hereditary tyrosinaemia. Nat. Commun. 2016, 7, 12642. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Bell, P.; McMenamin, D.; He, Z.; White, J.; Yu, H.; Xu, C.; Morizono, H.; Musunuru, K.; et al. A dual aav system enables the cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016, 34, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, N.E.; Hall, J.K.; Odom, G.L.; Phelps, M.P.; Andrus, C.R.; Hawkins, R.D.; Hauschka, S.D.; Chamberlain, J.R.; Chamberlain, J.S. Muscle-specific crispr/cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for duchenne muscular dystrophy. Nat. Commun. 2017, 8, 14454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendell, J.R.; Rodino-Klapac, L.R. Duchenne muscular dystrophy: Crispr/cas9 treatment. Cell Res. 2016, 26, 513–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [Green Version]
- Daya, S.; Berns, K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008, 21, 583–593. [Google Scholar] [CrossRef] [Green Version]
- Tycko, J.; Myer, V.E.; Hsu, P.D. Methods for optimizing crispr-cas9 genome editing specificity. Mol. Cell 2016, 63, 355–370. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using staphylococcus aureus cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small cas9 orthologue derived from campylobacter jejuni. Nat Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Su, J.; Liu, Y.; Jin, X.; Zhong, X.M.; Mo, L.; Wang, Q.N.; Deng, H.X.; Yang, Y. In vivo pcsk9 gene editing using an all-in-one self-cleavage aav-crispr system. Mol. Ther.-Methods Clin. Dev. 2021, 20, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Interleukin-1 and tumor necrosis factor: Effector cytokines in autoimmune diseases. Semin. Immunol. 1992, 4, 133–145. [Google Scholar] [PubMed]
- Garlanda, C.; Dinarello, C.A.; Mantovani, A. The interleukin-1 family: Back to the future. Immunity 2013, 39, 1003–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, A.; Hidaka, K.; Kanda, T.; Imaeda, H.; Shioya, M.; Inatomi, O.; Bamba, S.; Kitoh, K.; Sugimoto, M.; Andoh, A. Increased expression of interleukin-36, a member of the interleukin-1 cytokine family, in inflammatory bowel disease. Inflamm. Bowel Dis. 2016, 22, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, F.; Accardo-Palumbo, A.; Alessandro, R.; Alessandri, C.; Priori, R.; Guggino, G.; Raimondo, S.; Carubbi, F.; Valesini, G.; Giacomelli, R.; et al. Interleukin-36alpha axis is modulated in patients with primary sjogren’s syndrome. Clin. Exp. Immunol. 2015, 181, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wang, B.; Ma, Z.; Sun, X.; Tang, Y.; Li, X.; Wu, X. Detection of the novel il-1 family cytokines by qah-il1f-1 assay in rheumatoid arthritis. Cell Mol. Biol. 2016, 62, 31–34. [Google Scholar]
- Swindell, W.R.; Beamer, M.A.; Sarkar, M.K.; Loftus, S.; Fullmer, J.; Xing, X.; Ward, N.L.; Tsoi, L.C.; Kahlenberg, M.J.; Liang, Y.; et al. Rna-seq analysis of il-1b and il-36 responses in epidermal keratinocytes identifies a shared myd88-dependent gene signature. Front. Immunol. 2018, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Shembade, N.; Ma, A.; Harhaj, E.W. Inhibition of nf-kappab signaling by a20 through disruption of ubiquitin enzyme complexes. Science 2010, 327, 1135–1139. [Google Scholar] [CrossRef] [Green Version]
- Stahl, E.A.; Raychaudhuri, S.; Remmers, E.F.; Xie, G.; Eyre, S.; Thomson, B.P.; Li, Y.; Kurreeman, F.A.; Zhernakova, A.; Hinks, A.; et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010, 42, 508–514. [Google Scholar] [CrossRef]
- Zhang, M.; Peng, L.L.; Wang, Y.; Wang, J.S.; Liu, J.; Liu, M.M.; Hu, J.; Song, B.; Yang, H.B. Roles of a20 in autoimmune diseases. Immunol. Res. 2016, 64, 337–344. [Google Scholar] [CrossRef]
- Bulatov, E.; Khaiboullina, S.; dos Reis, H.J.; Palotás, A.; Venkataraman, K.; Vijayalakshmi, M.; Rizvanov, A. Mookambeswaran Vijayalakshmi & Albert Rizvanov Ubiquitin-proteasome system: Promising therapeutic targets in autoimmune and neurodegenerative diseases. BioNanoScience 2016, 6, 341–344. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, H.; Schwartz, D.M.; Stoffels, M.; Park, Y.H.; Zhang, Y.; Yang, D.; Demirkaya, E.; Takeuchi, M.; Tsai, W.L.; et al. Loss-of-function mutations in tnfaip3 leading to a20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat. Genet. 2016, 48, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Huang, Y.; Ye, Z.; Wang, Y.; Tang, Z.; Lu, J.; Wu, J.; Zhou, Y.; Wang, L.; Huang, Z.; et al. Infantile onset intractable inflammatory bowel disease due to novel heterozygous mutations in tnfaip3 (a20). Inflamm. Bowel Dis. 2018, 24, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wen, F.; Tessneer, K.L.; Gaffney, P.M. Talen-mediated enhancer knockout influences tnfaip3 gene expression and mimics a molecular phenotype associated with systemic lupus erythematosus. Genes Immun. 2016, 17, 165–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Yang, S.; Yu, D.; Gao, W.; Liu, X.; Zhang, K.; Fu, X.; Bao, W.; Zhang, K.; Yu, J.; et al. Crispr/cas9 mediated knockout of an intergenic variant rs6927172 identified il-20ra as a new risk gene for multiple autoimmune diseases. Genes Immun. 2018, 20, 103–111. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Rasmussen, J.P.; Williams, L.M.; Dooley, J.L.; Farr, A.G.; Rudensky, A.Y. Regulatory t cell lineage specification by the forkhead transcription factor foxp3. Immunity 2005, 22, 329–341. [Google Scholar] [CrossRef]
- Bennett, C.L.; Christie, J.; Ramsdell, F.; Brunkow, M.E.; Ferguson, P.J.; Whitesell, L.; Kelly, T.E.; Saulsbury, F.T.; Chance, P.F.; Ochs, H.D. The immune dysregulation, polyendocrinopathy, enteropathy, x-linked syndrome (ipex) is caused by mutations of foxp3. Nat. Genet. 2001, 27, 20–21. [Google Scholar] [CrossRef]
- Jeffries, M.A. Epigenetic editing: How cutting-edge targeted epigenetic modification might provide novel avenues for autoimmune disease therapy. Clin. Immunol. 2018, 196, 49–58. [Google Scholar] [CrossRef]
- Brunger, J.M.; Zutshi, A.; Willard, V.P.; Gersbach, C.A.; Guilak, F. Genome engineering of stem cells for autonomously regulated, closed-loop delivery of biologic drugs. Stem Cell Rep. 2017, 8, 1202–1213. [Google Scholar] [CrossRef] [Green Version]
- Brunger, J.M.; Zutshi, A.; Willard, V.P.; Gersbach, C.A.; Guilak, F. Crispr/cas9 editing of murine induced pluripotent stem cells for engineering inflammation-resistant tissues. Arthritis Rheumatol. 2017, 69, 1111–1121. [Google Scholar] [CrossRef]
- Gerace, D.; Martiniello-Wilks, R.; Nassif, N.T.; Lal, S.; Steptoe, R.; Simpson, A.M. Crispr-targeted genome editing of mesenchymal stem cell-derived therapies for type 1 diabetes: A path to clinical success? Stem Cell Res. 2017, 8, 62. [Google Scholar] [CrossRef] [Green Version]
- Gimenez, C.A.; Ielpi, M.; Mutto, A.; Grosembacher, L.; Argibay, P.; Pereyra-Bonnet, F. Crispr-on system for the activation of the endogenous human ins gene. Gene 2016, 23, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; McGovern, A.; Martin, P.; Duffus, K.; Ge, X.; Zarrineh, P.; Morris, A.P.; Adamson, A.; Fraser, P.; Rattray, M.; et al. Analysis of chromatin organization and gene expression in t cells identifies functional genes for rheumatoid arthritis. Nat. Commun. 2020, 11, 4402. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Jin, Z.; Liu, X.; Gao, W.; Gao, X.; Zhang, S.; Ma, S.; Yu, J.; Wang, S. Manipulation of an ra-associated intergenic snp by crispr/cas9 system alters expression of multiple genes at the tnfaip3 locus, poster presentation abstracts. Int. J. Rheum. Dis. 2016, 19 (Suppl. S2), 89. [Google Scholar] [CrossRef] [Green Version]
- Jing, W.; Zhang, X.; Sun, W.; Hou, X.; Yao, Z.; Zhu, Y. Crispr/cas9-mediated genome editing of mirna-155 inhibits proinflammatory cytokine production by raw264.7 cells. Biomed. Res. Int. 2015, 2015, 326042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardinale, C.J.; March, M.E.; Lin, X.; Liu, Y.; Spruce, L.A.; Bradfield, J.P.; Wei, Z.; Seeholzer, S.H.; Grant, S.F.A.; Hakonarson, H. Regulation of janus kinase 2 by an inflammatory bowel disease causal non-coding single nucleotide polymorphism. J. Crohns Colitis 2020, 14, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, N.; Den Low, E.N.; Yahaya, A.; Ali, R.A.R. Characterisation of sgk2 gene using crispr/cas9 gene editing tool in ulcerative colitis. J. Gastroenterol. Hepatol. 2019, 34 (Suppl. S3), 274. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Kuemmerle, J. P119 the role of ptpn2 snp in the pathogenesis of fibrosis in crohn’s disease. Inflamm. Bowel Dis. 2019, 25, S57–S58. [Google Scholar] [CrossRef]
- Matthews, S.M.; Eshelman, M.A.; Berg, A.S.; Koltun, W.A.; Yochum, G.S. The crohn’s disease associated snp rs6651252 impacts myc gene expression in human colonic epithelial cells. PLoS ONE 2019, 14, e0212850. [Google Scholar] [CrossRef]
- Pai, Y.-C.; Shih, D.Q.; Targan, S.R.; Turner, J.; Wei, S.C.; Yu, L.C.H. Su1097 tl1a-mediated bacterial transcytosis by a mlck splicing variant contributes to inflammatory bowel disease. Gastroenterology 2020, 158, S-508. [Google Scholar] [CrossRef]
- Friedrich, M.; Ganther, J.; Breiderhoff, T.; Rosenthal, R.; Glauben, R.; Siegmund, B. P063 hdac as versatile regulators of the intestinal epithelial barrier in inflammatory bowel disease. J. Crohns Colitis 2017, 11, S109. [Google Scholar] [CrossRef] [Green Version]
- Eftychi, C.; Schwarzer, R.; Vlantis, K.; Wachsmuth, L.; Basic, M.; Wagle, P.; Neurath, M.F.; Becker, C.; Bleich, A.; Pasparakis, M. Temporally distinct functions of the cytokines il-12 and il-23 drive chronic colon inflammation in response to intestinal barrier impairment. Immunity 2019, 51, 367–380.e4. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.D.; Sun, M.M.; Wu, W.; Ma, C.Y.; Zhang, C.; He, C.; Li, J.X.; Cong, Y.Z.; Zhang, D.K.; Liu, Z.J. Microrna-125a suppresses intestinal mucosal inflammation through targeting ets-1 in patients with inflammatory bowel diseases. J. Autoimmun. 2019, 101, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Odqvist, L.; Jevnikar, Z.; Riise, R.; Oberg, L.; Rhedin, M.; Leonard, D.; Yrlid, L.; Jackson, S.; Mattsson, J.; Nanda, S.; et al. Genetic variations in a20 dub domain provide a genetic link to citrullination and neutrophil extracellular traps in systemic lupus erythematosus. Ann. Rheum. Dis. 2019, 78, 1363–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, V.M.; Koelsch, K.A.; Kurien, B.T.; Harley, I.T.W.; Wren, J.D.; Harley, J.B.; Scofield, R.H. Characterization of cxorf21 provides molecular insight into female-bias immune response in sle pathogenesis. Front. Immunol. 2019, 10, 2160. [Google Scholar] [CrossRef]
- Gregory, S.G.; Schmidt, S.; Seth, P.; Oksenberg, J.R.; Hart, J.; Prokop, A.; Caillier, S.J.; Ban, M.; Goris, A.; Barcellos, L.F.; et al. Interleukin 7 receptor alpha chain (il7r) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007, 39, 1083–1091. [Google Scholar] [CrossRef]
- Galarza-Munoz, G.; Briggs, F.B.S.; Evsyukova, I.; Schott-Lerner, G.; Kennedy, E.M.; Nyanhete, T.; Wang, L.; Bergamaschi, L.; Widen, S.G.; Tomaras, G.D.; et al. Human epistatic interaction controls il7r splicing and increases multiple sclerosis risk. Cell 2017, 169, 72–84.e13. [Google Scholar] [CrossRef] [Green Version]
- Maier, L.M.; Lowe, C.E.; Cooper, J.; Downes, K.; Anderson, D.E.; Severson, C.; Clark, P.M.; Healy, B.; Walker, N.; Aubin, C.; et al. Il2ra genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009, 5, e1000322. [Google Scholar] [CrossRef]
- Gregory, A.P.; Dendrou, C.A.; Attfield, K.E.; Haghikia, A.; Xifara, D.K.; Butter, F.; Poschmann, G.; Kaur, G.; Lambert, L.; Leach, O.A.; et al. Tnf receptor 1 genetic risk mirrors outcome of anti-tnf therapy in multiple sclerosis. Nature 2012, 488, 508–511. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Wang, J.; Zhang, L.; Bian, W.; Lin, M.; Xu, X.; Zhou, X. Lck rs10914542-g allele associates with type 1 diabetes in children via t cell hyporesponsiveness. Pediatr. Res. 2019, 86, 311–315. [Google Scholar] [CrossRef]
- Ratiu, J.J.; Racine, J.J.; Hasham, M.G.; Wang, Q.; Branca, J.A.; Chapman, H.D.; Zhu, J.; Donghia, N.; Philip, V.; Schott, W.H.; et al. Genetic and small molecule disruption of the aid/rad51 axis similarly protects nonobese diabetic mice from type 1 diabetes through expansion of regulatory b lymphocytes. J. Immunol. 2017, 198, 4255–4267. [Google Scholar] [CrossRef] [Green Version]
- Arakawa, A.; Vollmer, S.; Arakawa, Y.; He, M.; Reeves, E.; James, E.; Prinz, J. Erap1 Regulates the Risk for Psoriasis through Affecting Immunogenicity of Melanocytes and the Psoriatic Autoantigen Adamts-Like Protein 5. In Proceedings of the 46th Annual Meeting of the Arbeitsgemeinschaft Dermatologische Forschung (ADF), Munich, Germany, 13–16 March 2019; Wiley: Munich, Germany, 2019; pp. E66–E67. [Google Scholar]
- Roth-Carter, Q.R.; Godsel, L.; Koetsier, J.L.; Broussard, J.A.; Burks, H.E.; Fitz, G.; Huffine, A.L.; Amagai, S.; Lloyd, S.; Kweon, J.; et al. 225 desmoglein 1 deficiency in knockout mice impairs epidermal barrier formation and results in a psoriasis-like gene signature in e18. 5 embryos. J. Investig. Dermatol. 2020, 140, S26. [Google Scholar] [CrossRef]
- Jouanin, A.; Schaart, J.G.; Boyd, L.A.; Cockram, J.; Leigh, F.J.; Bates, R.; Wallington, E.J.; Visser, R.G.F.; Smulders, M.J.M. Outlook for coeliac disease patients: Towards bread wheat with hypoimmunogenic gluten by gene editing of alpha- and gamma-gliadin gene families. BMC Plant Biol. 2019, 19, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Xu, J.; Ge, S.; Lai, L. Crispr/cas: Advances, limitations, and applications for precision cancer research. Front. Med. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).