Aberrant Methylation of 20 miRNA Genes Specifically Involved in Various Steps of Ovarian Carcinoma Spread: From Primary Tumors to Peritoneal Macroscopic Metastases
Abstract
1. Introduction
2. Results
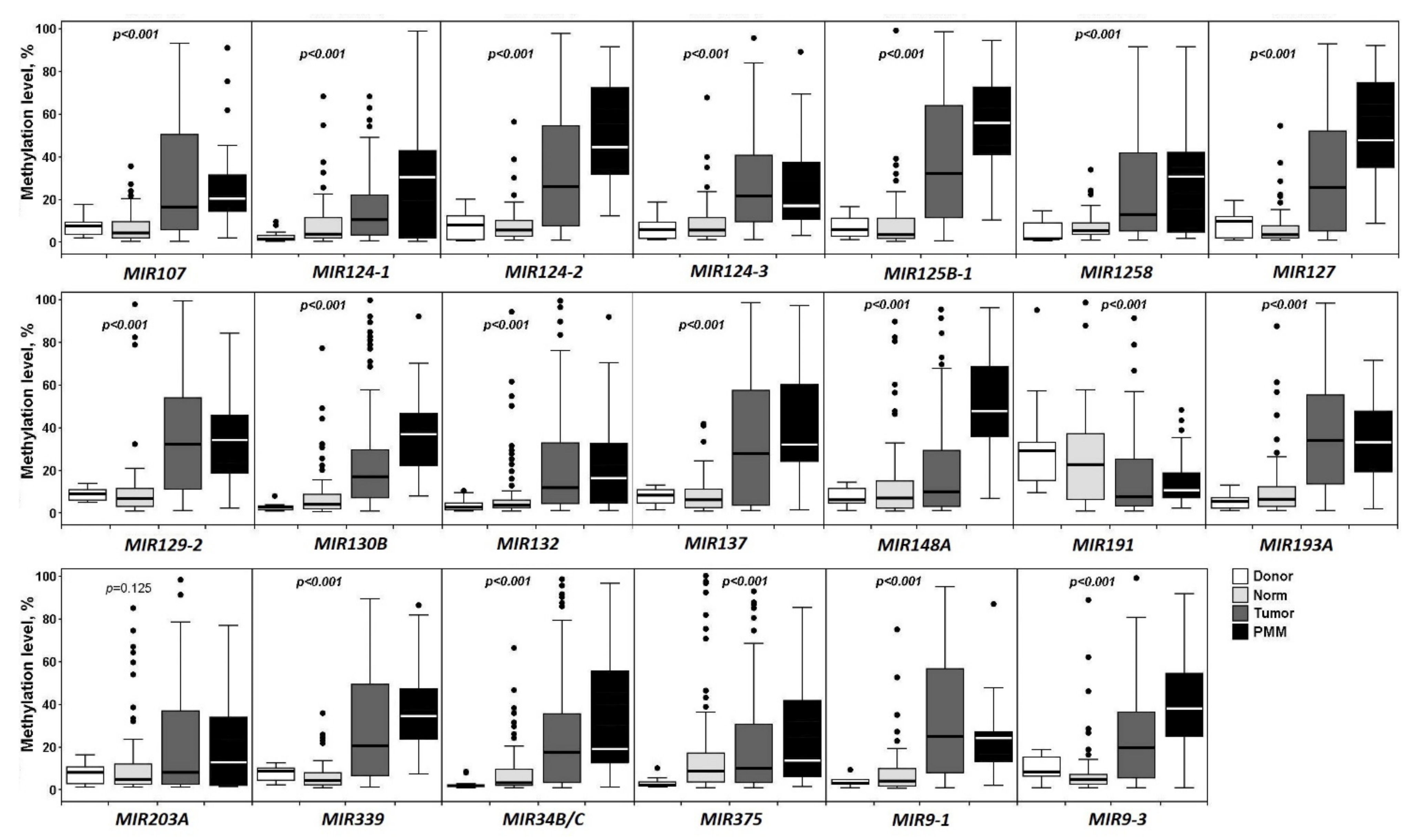
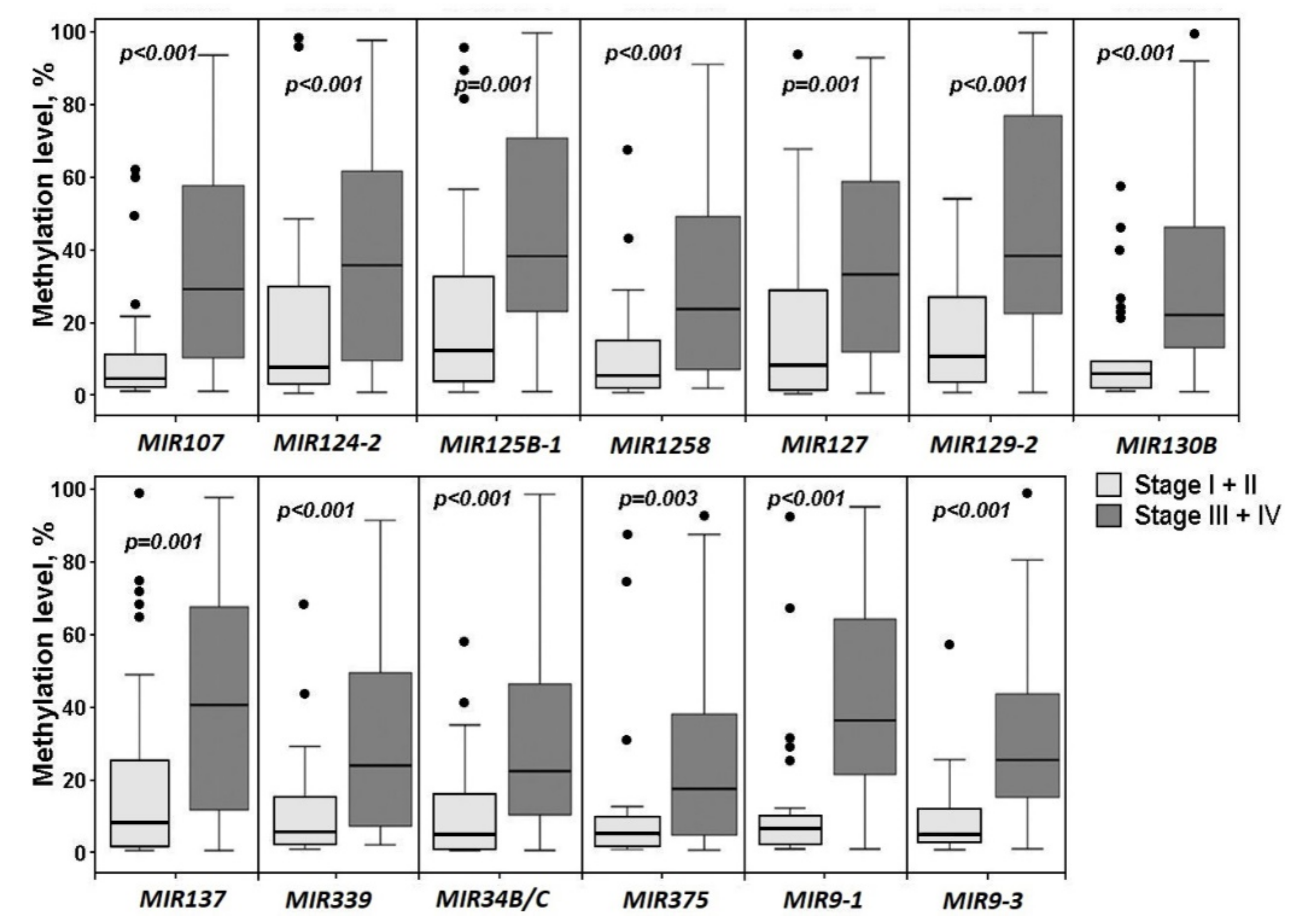
2.1. Aberrant Methylation of 20 miRNA Genes in the Development of Primary Tumors and Metastasis in Ovarian Cancer
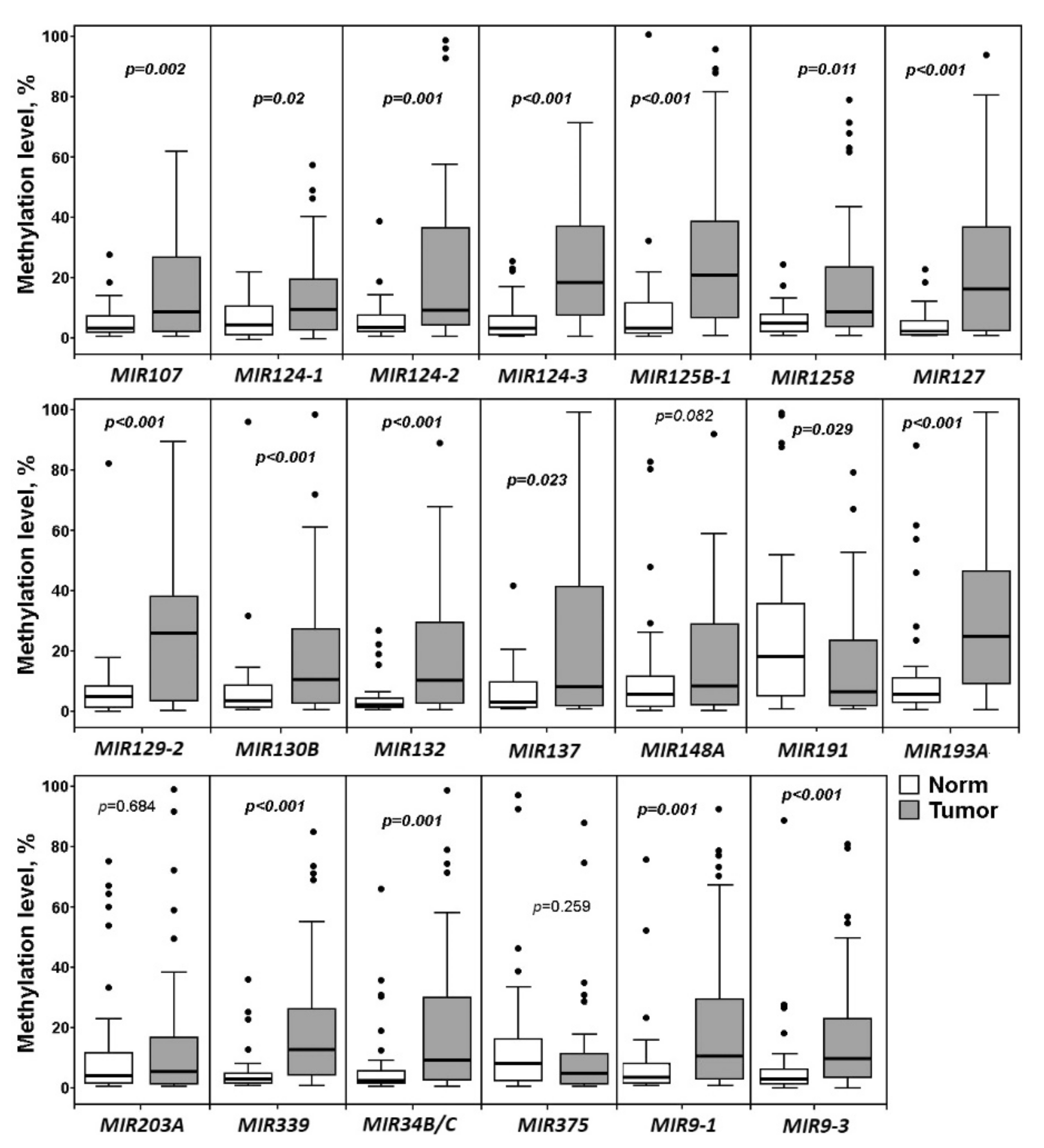
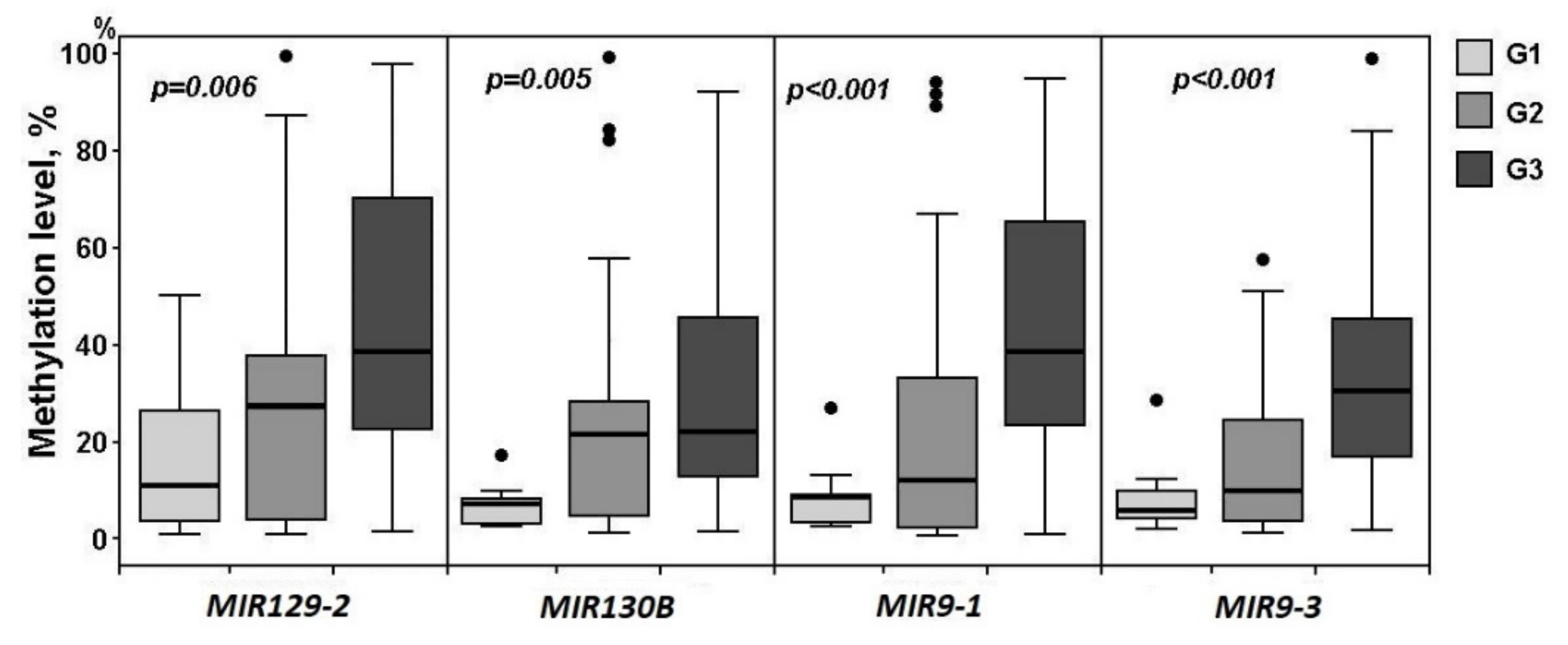
2.2. Relationship of Methylation of Examined miRNA Genes with the Clinical Stage and Histological Grade of Ovarian Cancer
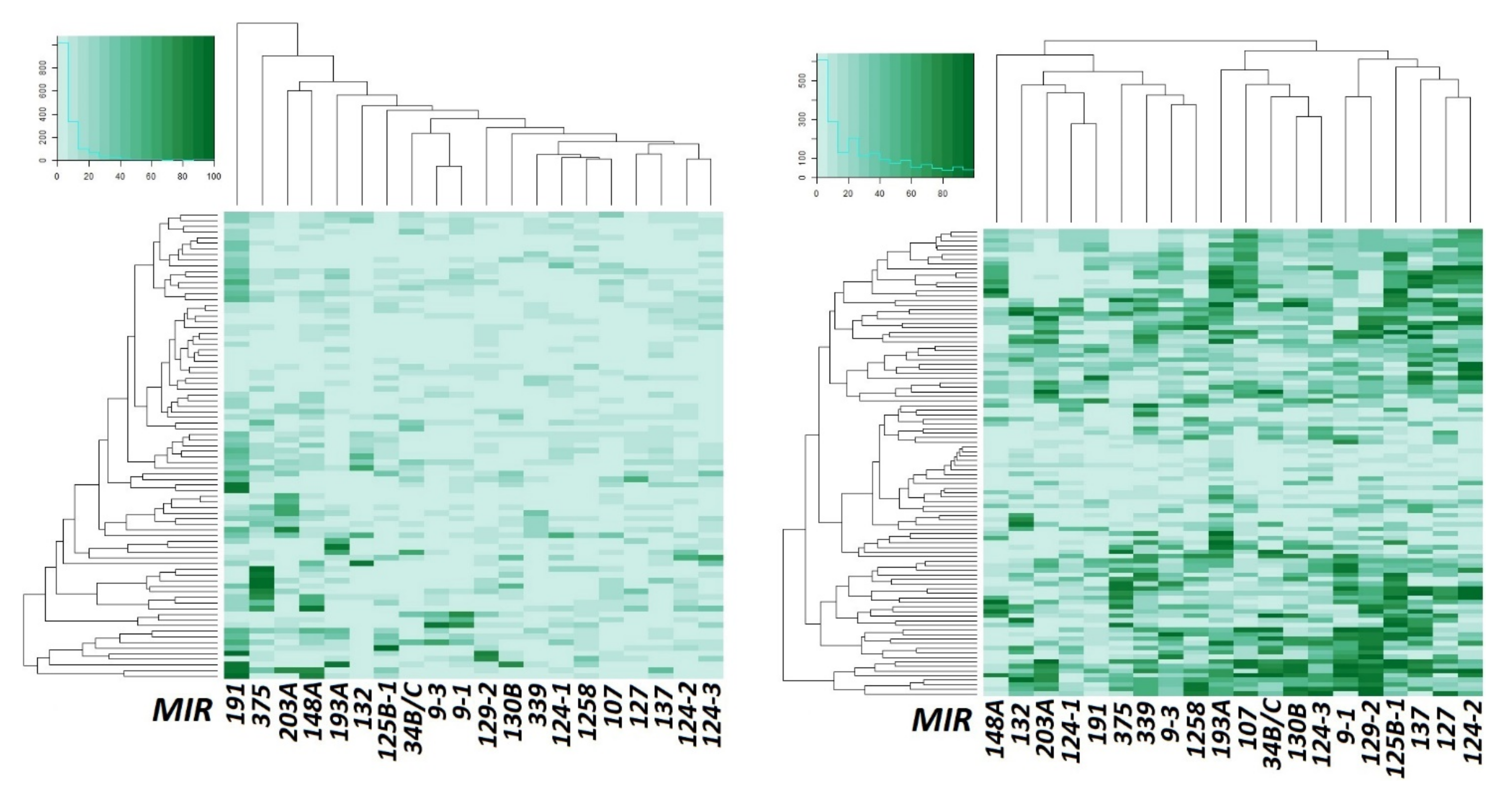
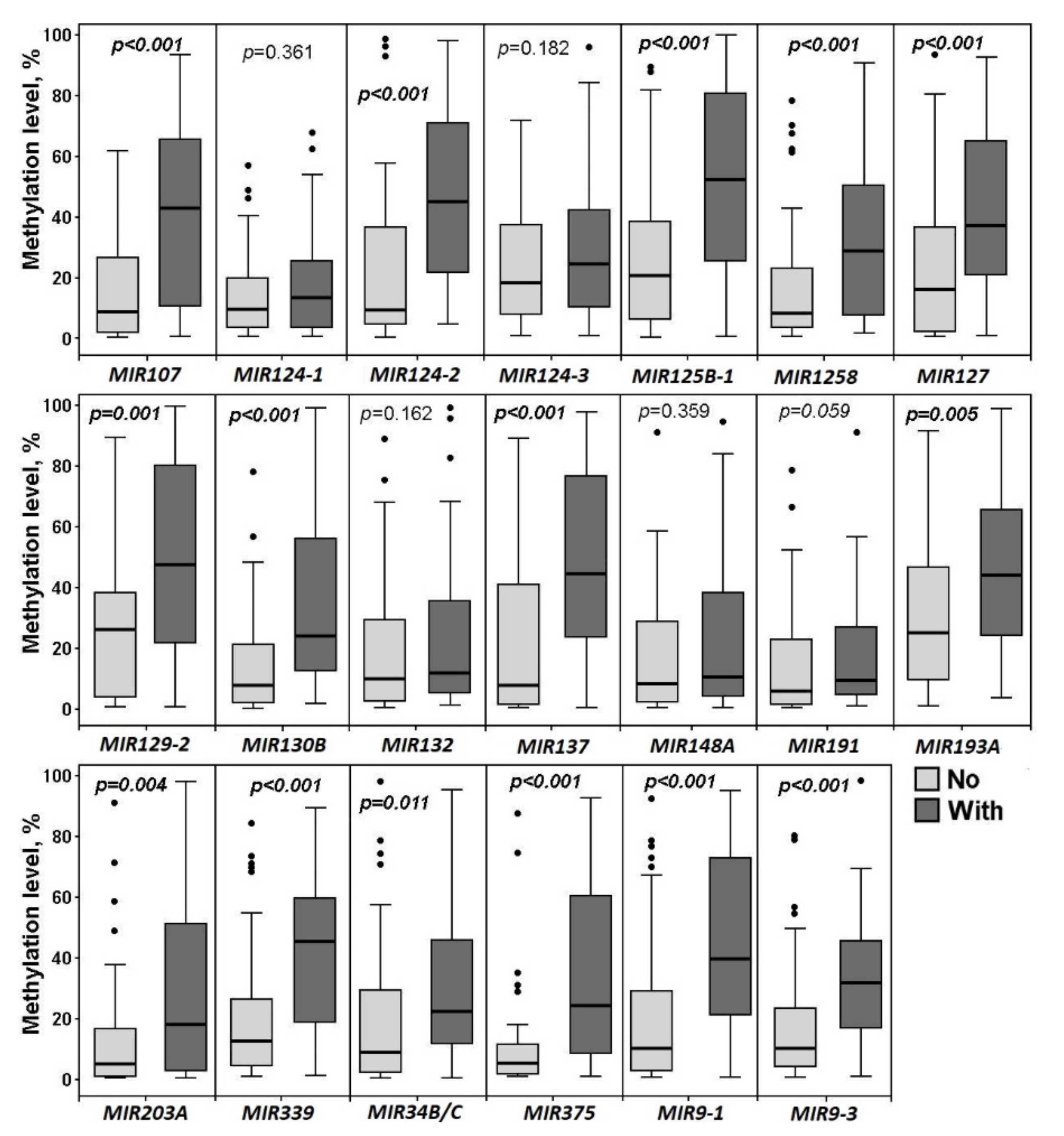
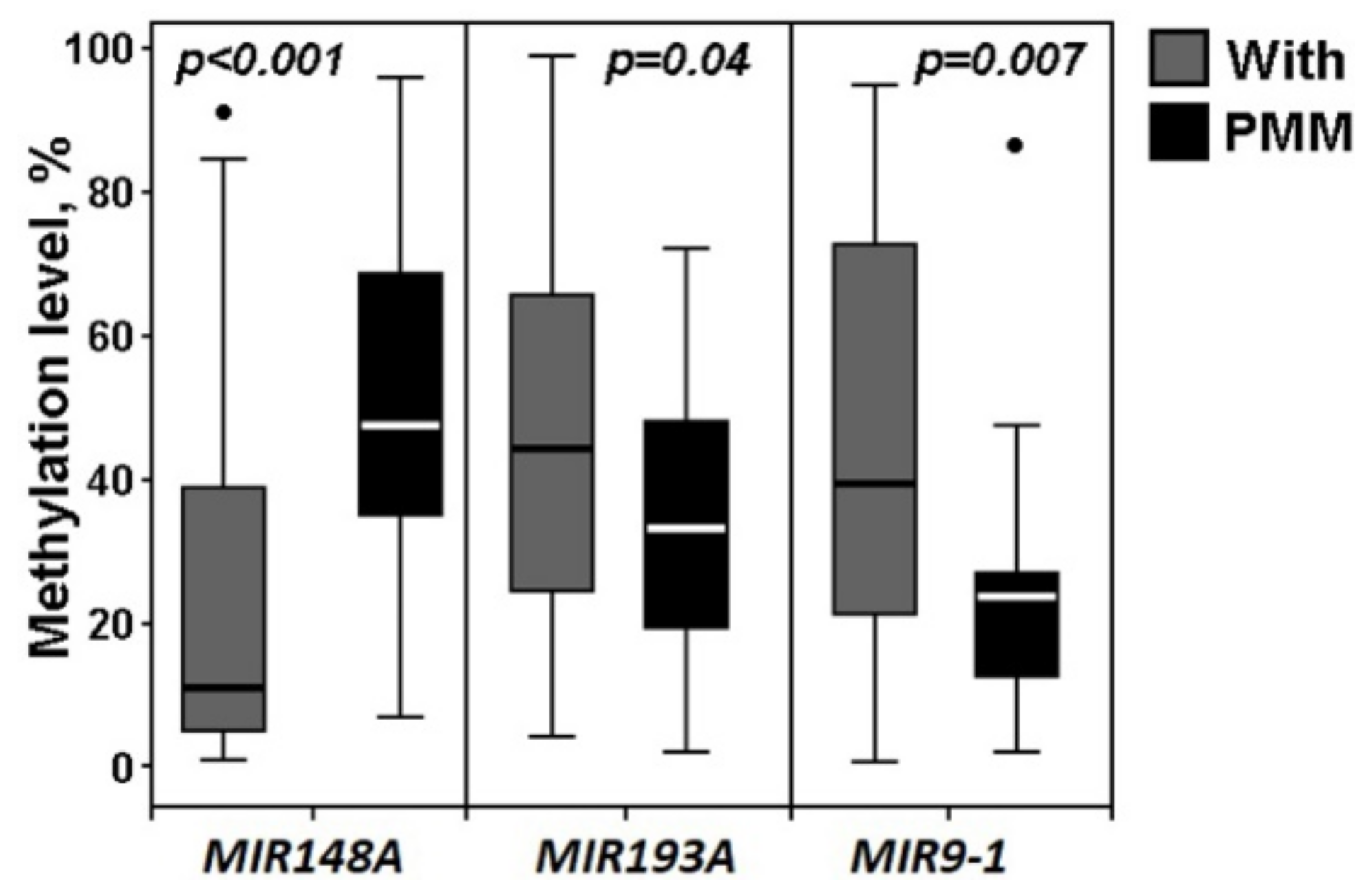
2.3. Hypermethylated miRNA Genes Specifically Involved in the Formation of Macroscopic Metastases in the Peritoneum of Ovarian Cancer Patients
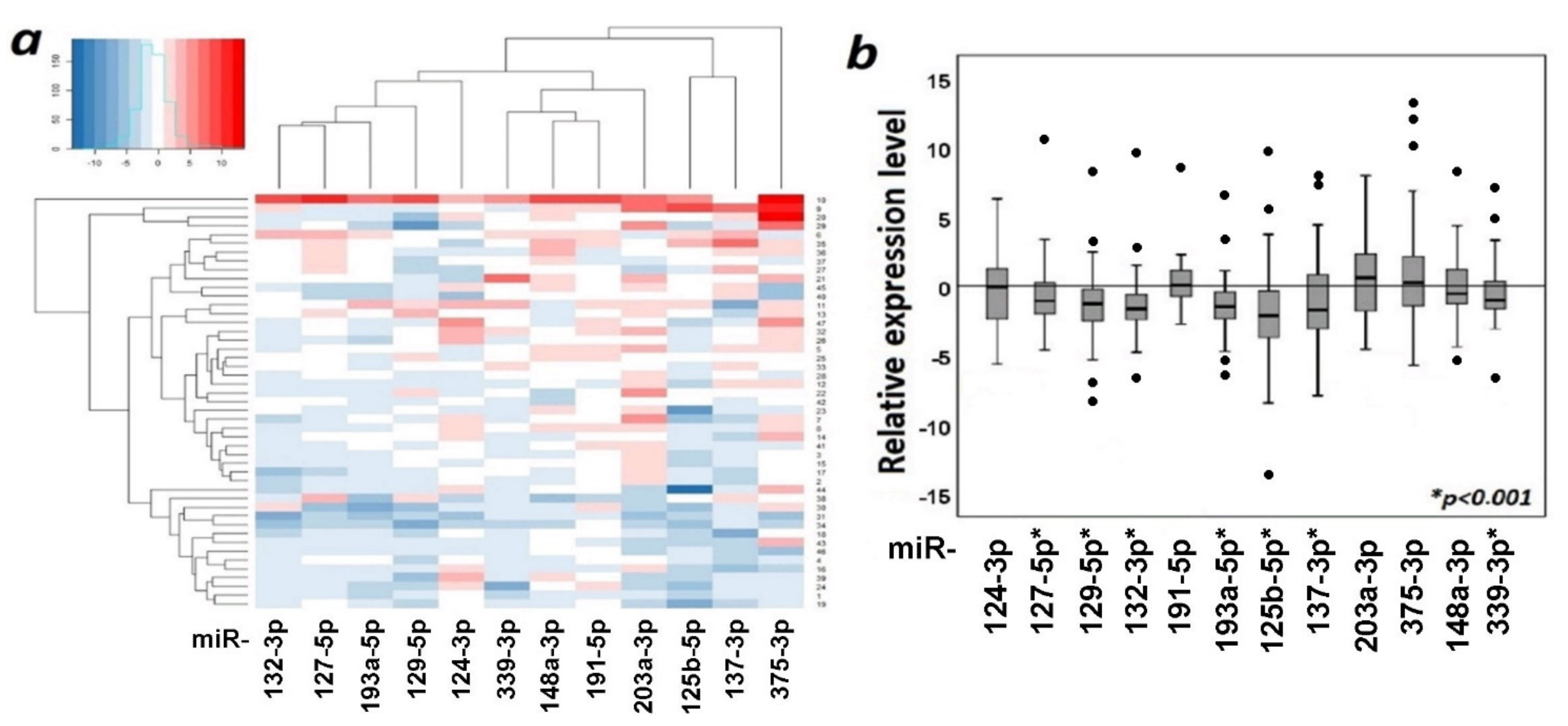
2.4. The Functional Significance of Methylation of Examined miRNA Genes in the Regulation of Their Expression in Ovarian Cancer
2.5. Comparison of Expression Levels of 12 miRNAs in Primary Tumors of Ovarian Cancer Patients without and with Metastases and in Macroscopic Peritoneal Metastases
2.6. Reverse Relationship between Expression Levels of miRNA-203a-3p and ZEB1 and ZEB2 Genes in Ovarian Cancer
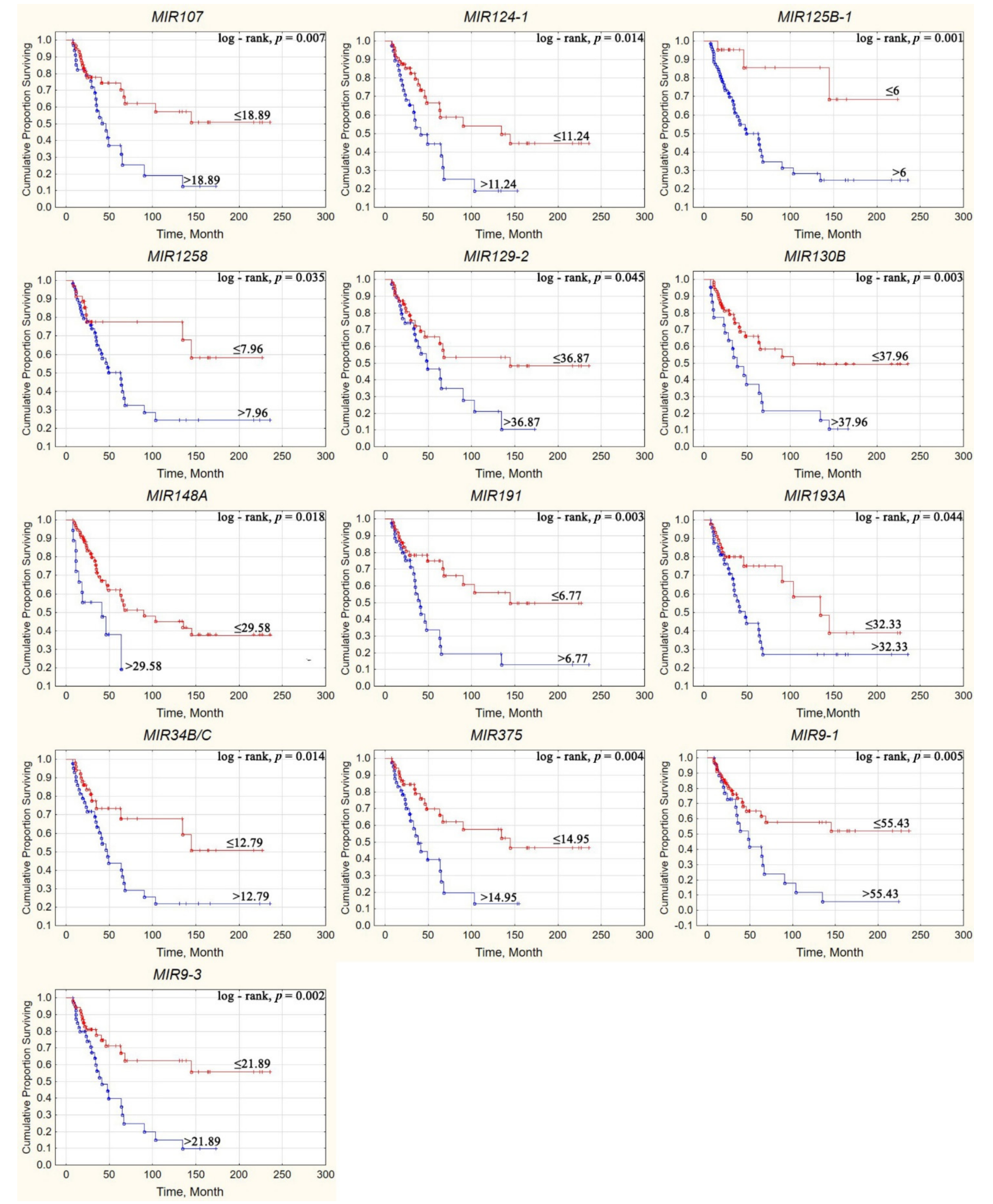
2.7. Effect of Aberrant Methylation of Examined miRNA Genes on the Survival of Patients with Ovarian Cancer
3. Discussion
4. Materials and Methods
4.1. Tissue Samples
4.2. Bioinformatics
4.3. DNA and Total RNA Isolation and Reverse Transcription
4.4. Quantitative PCR (qPCR) for Expression Analysis
4.5. Quantitative Methylation-Specific PCR (qMSP)
4.6. Overall Survival Markers
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vogell, A.; Evans, M.L. Cancer Screening in Women. Obstet. Gynecol. Clin. N. Am. 2019, 46, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Fridman, M.V.; Kushlinskii, N.E. Molecular Mechanisms of Ovarian Carcinoma Metastasis: Key Genes and Regulatory MicroRNAs. Biochemistry 2017, 82, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Fridman, M.V.; Moscovtsev, A.A.; Filippova, E.A.; Dmitriev, A.A.; Kushlinskii, N.E. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. Int. J. Mol. Sci. 2020, 21, 8855. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.; Agarwal, R.; Kaye, S.B. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006, 7, 925–934. [Google Scholar] [CrossRef]
- Cohen, M.; Petignat, P. The bright side of ascites in ovarian cancer. Cell Cycle 2014, 13, 2319. [Google Scholar] [CrossRef] [PubMed]
- van Baal, J.; van Noorden, C.J.F.; Nieuwland, R.; Van de Vijver, K.K.; Sturk, A.; van Driel, W.J.; Kenter, G.G.; Lok, C.A.R. Development of Peritoneal Carcinomatosis in Epithelial Ovarian Cancer: A Review. J. Histochem. Cytochem. 2018, 66, 67–83. [Google Scholar] [CrossRef]
- Barbolina, M.V. Molecular Mechanisms Regulating Organ-Specific Metastases in Epithelial Ovarian Carcinoma. Cancers 2018, 10, 444. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Taheri, M. miRNA profile in ovarian cancer. Exp. Mol. Pathol. 2020, 113, 104381. [Google Scholar] [CrossRef]
- Mahdian-Shakib, A.; Dorostkar, R.; Tat, M.; Hashemzadeh, M.S.; Saidi, N. Differential role of microRNAs in prognosis, diagnosis, and therapy of ovarian cancer. Biomed. Pharmacother. 2016, 84, 592–600. [Google Scholar] [CrossRef]
- Nguyen, V.H.L.; Yue, C.; Du, K.Y.; Salem, M.; O’Brien, J.; Peng, C. The Role of microRNAs in Epithelial Ovarian Cancer Metastasis. Int. J. Mol. Sci. 2020, 21, 7093. [Google Scholar] [CrossRef] [PubMed]
- Piletic, K.; Kunej, T. MicroRNA epigenetic signatures in human disease. Arch. Toxicol. 2016, 90, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Pronina, I.V.; Loginov, V.I.; Burdennyy, A.M.; Fridman, M.V.; Senchenko, V.N.; Kazubskaya, T.P.; Kushlinskii, N.E.; Dmitriev, A.A.; Braga, E.A. DNA methylation contributes to deregulation of 12 cancer-associated microRNAs and breast cancer progression. Gene 2017, 604, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Loginov, V.I.; Pronina, I.V.; Burdennyy, A.M.; Filippova, E.A.; Kazubskaya, T.P.; Kushlinsky, D.N.; Utkin, D.O.; Khodyrev, D.S.; Kushlinskii, N.E.; Dmitriev, A.A.; et al. Novel miRNA genes deregulated by aberrant methylation in ovarian carcinoma are involved in metastasis. Gene 2018, 662, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Guo, Y.; Chen, Z.; Wang, Y.; Yang, C.; Dudas, A.; Du, Z.; Liu, W.; Zou, Y.; Szabo, E.; et al. miR-203 Functions as a Tumor Suppressor by Inhibiting Epithelial to Mesenchymal Transition in Ovarian Cancer. J. Cancer Sci. Ther. 2015, 7, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Gao, W.; Chan, J.Y. Transcription regulation of E-cadherin by zinc finger E-box binding homeobox proteins in solid tumors. BioMed Res. Int. 2014, 2014, 921564. [Google Scholar] [CrossRef]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Loret, N.; Denys, H.; Tummers, P.; Berx, G. The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers 2019, 11, 838. [Google Scholar] [CrossRef]
- Chen, D.; Wang, J.; Zhang, Y.; Chen, J.; Yang, C.; Cao, W.; Zhang, H.; Liu, Y.; Dou, J. Effect of down-regulated transcriptional repressor ZEB1 on the epithelial-mesenchymal transition of ovarian cancer cells. Int. J. Gynecol. Cancer 2013, 23, 1357–1366. [Google Scholar] [CrossRef]
- Prislei, S.; Martinelli, E.; Zannoni, G.F.; Petrillo, M.; Filippetti, F.; Mariani, M.; Mozzetti, S.; Raspaglio, G.; Scambia, G.; Ferlini, C. Role and prognostic significance of the epithelial-mesenchymal transition factor ZEB2 in ovarian cancer. Oncotarget 2015, 6, 18966–18979. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Liu, H.; Zhang, M.; Huang, Z.; Zhou, H.; Zhu, Y.; Tao, Y.; Xie, N.; Liu, X.; Hou, J.; et al. Prognostic value of epithelial-mesenchymal transition-inducing transcription factors in head and neck squamous cell carcinoma: A meta-analysis. Head Neck 2020, 42, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Ahmadiankia, N.; Khosravi, A. Significance of epithelial-to-mesenchymal transition inducing transcription factors in predicting distance metastasis and survival in patients with colorectal cancer: A systematic review and meta-analysis. J. Res. Med. Sci. 2020, 25, 60. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.; Prasad, R.; Katiyar, S.K. Therapeutic intervention of silymarin on the migration of non-small cell lung cancer cells is associated with the axis of multiple molecular targets including class 1 HDACs, ZEB1 expression, and restoration of miR-203 and E-cadherin expression. Am. J. Cancer Res. 2016, 6, 1287–1301. [Google Scholar] [PubMed]
- Jiang, Y.; Jin, S.; Tan, S.; Shen, Q.; Xue, Y. MiR-203 acts as a radiosensitizer of gastric cancer cells by directly targeting ZEB1. OncoTargets Ther. 2019, 12, 6093–6104. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Pascual, A.; Hale, J.S.; Kordowski, A.; Pugh, J.; Silver, D.J.; Bayik, D.; Roversi, G.; Alban, T.J.; Rao, S.; Chen, R.; et al. ADAMDEC1 Maintains a Growth Factor Signaling Loop in Cancer Stem Cells. Cancer Discov. 2019, 9, 1574–1589. [Google Scholar] [CrossRef]
- Gu, Y.; Zhu, Z.; Pei, H.; Xu, D.; Jiang, Y.; Zhang, L.; Xiao, L. Long non-coding RNA NNT-AS1 promotes cholangiocarcinoma cells proliferation and epithelial-to-mesenchymal transition through down-regulating miR-203. Aging 2020, 12, 2333–2346. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, J.; Wu, Q.; Liu, Y.; Ma, C. HOTAIR Contributes to Stemness Acquisition of Cervical Cancer through Regulating miR-203 Interaction with ZEB1 on Epithelial-Mesenchymal Transition. J. Oncol. 2021, 2021, 4190764. [Google Scholar] [CrossRef]
- Saini, S.; Majid, S.; Yamamura, S.; Tabatabai, L.; Suh, S.O.; Shahryari, V.; Chen, Y.; Deng, G.; Tanaka, Y.; Dahiya, R. Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clin. Cancer Res. 2011, 17, 5287–5298. [Google Scholar] [CrossRef]
- Duan, X.; Fu, Z.; Gao, L.; Zhou, J.; Deng, X.; Luo, X.; Fang, W.; Luo, R. Direct interaction between miR-203 and ZEB2 suppresses epithelial-mesenchymal transition signaling and reduces lung adenocarcinoma chemoresistance. Acta Biochim. Biophys. Sin. 2016, 48, 1042–1049. [Google Scholar] [CrossRef]
- Jiang, Q.; Zhou, Y.; Yang, H.; Li, L.; Deng, X.; Cheng, C.; Xie, Y.; Luo, X.; Fang, W.; Liu, Z. A directly negative interaction of miR-203 and ZEB2 modulates tumor stemness and chemotherapy resistance in nasopharyngeal carcinoma. Oncotarget 2016, 7, 67288–67301. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Qiao, F.; Wu, H.; Cui, H.; Li, Y.; Zheng, Y.; Zhou, M.; Fan, H. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018, 9, 1158. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wei, Z.; Wu, K.; Dai, W.; Zhang, C.; Peng, J.; He, Y. Long noncoding RNA B3GALT5-AS1 suppresses colon cancer liver metastasis via repressing microRNA-203. Aging 2018, 10, 3662–3682. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhong, Y.; Li, L. miR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC). J. Transl. Med. 2020, 18, 69. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. How does multistep tumorigenesis really proceed? Cancer Discov. 2015, 5, 22–24. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Williams, E.D.; Gao, D.; Redfern, A.; Thompson, E.W. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer 2019, 19, 716–732. [Google Scholar] [CrossRef]
- Davidson, B.; Trope, C.G.; Reich, R. Epithelial-mesenchymal transition in ovarian carcinoma. Front. Oncol. 2012, 2, 33. [Google Scholar] [CrossRef]
- Klymenko, Y.; Kim, O.; Stack, M.S. Complex Determinants of Epithelial: Mesenchymal Phenotypic Plasticity in Ovarian Cancer. Cancers 2017, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Skrypek, N.; Goossens, S.; De Smedt, E.; Vandamme, N.; Berx, G. Epithelial-to-Mesenchymal Transition: Epigenetic Reprogramming Driving Cellular Plasticity. Trends Genet. 2017, 33, 943–959. [Google Scholar] [CrossRef] [PubMed]
- Burdennyy, A.M.; Filippova, E.A.; Ivanova, N.A.; Lukina, S.S.; Pronina, I.V.; Loginov, V.I.; Fridman, M.V.; Kazubskaya, T.P.; Utkin, D.O.; Braga, E.A.; et al. Hypermethylation of Genes in New Long Noncoding RNA in Ovarian Tumors and Metastases: A Dual Effect. Bull. Exp. Biol. Med. 2021, 171, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef]
- Pal, A.; Barrett, T.F.; Paolini, R.; Parikh, A.; Puram, S.V. Partial EMT in head and neck cancer biology: A spectrum instead of a switch. Oncogene 2021, 40, 5049–5065. [Google Scholar] [CrossRef]
- Li, X.; Pan, Q.; Wan, X.; Mao, Y.; Lu, W.; Xie, X.; Cheng, X. Methylation-associated Has-miR-9 deregulation in paclitaxel-resistant epithelial ovarian carcinoma. BMC Cancer 2015, 15, 509. [Google Scholar] [CrossRef]
- Yang, C.; Cai, J.; Wang, Q.; Tang, H.; Cao, J.; Wu, L.; Wang, Z. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol. Oncol. 2012, 124, 325–334. [Google Scholar] [CrossRef]
- Chen, C.; Hu, Y.; Li, L. NRP1 is targeted by miR-130a and miR-130b, and is associated with multidrug resistance in epithelial ovarian cancer based on integrated gene network analysis. Mol. Med. Rep. 2016, 13, 188–196. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, X.; Sun, S.; Lu, C.; Wang, L. Serum miR-125b levels associated with epithelial ovarian cancer (EOC) development and treatment responses. Bioengineered 2020, 11, 311–317. [Google Scholar] [CrossRef]
- Parayath, N.N.; Gandham, S.K.; Leslie, F.; Amiji, M.M. Improved anti-tumor efficacy of paclitaxel in combination with MicroRNA-125b-based tumor-associated macrophage repolarization in epithelial ovarian cancer. Cancer Lett. 2019, 461, 1–9. [Google Scholar] [CrossRef]
- Tang, Z.; Fang, Y.; Du, R. MicroRNA-107 induces cell cycle arrests by directly targeting cyclin E1 in ovarian cancer. Biochem. Biophys. Res. Commun. 2019, 512, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yan, L.; Chi, Y.; Sun, Y.; Yang, X. Long non-coding RNA AFAP1-AS1 facilitates ovarian cancer progression by regulating the miR-107/PDK4 axis. J. Ovarian Res. 2021, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Loginov, V.I.; Burdennyy, A.M.; Filippova, E.A.; Pronina, I.V.; Kazubskaya, T.P.; Kushlinsky, D.N.; Ermilova, V.D.; Rykov, S.V.; Khodyrev, D.S.; Braga, E.A. Hypermethylation of miR-107, miR-130b, miR-203a, miR-1258 Genes Associated with Ovarian Cancer Development and Metastasis. Mol. Biol. 2018, 52, 693–700. [Google Scholar] [CrossRef]
- Shi, J.; Chen, P.; Sun, J.; Song, Y.; Ma, B.; Gao, P.; Chen, X.; Wang, Z. MicroRNA-1258: An invasion and metastasis regulator that targets heparanase in gastric cancer. Oncol. Lett. 2017, 13, 3739–3745. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, H.; Jiang, S.; Guo, L.; Li, X. MicroRNA-1258, regulated by c-Myb, inhibits growth and epithelial-to-mesenchymal transition phenotype via targeting SP1 in oral squamous cell carcinoma. J. Cell. Mol. Med. 2019, 23, 2813–2821. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Watari, H.; Hanley, S.J.; Konno, Y.; Ihira, K.; Yamada, T.; Kudo, M.; Yue, J.; Sakuragi, N. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 132. [Google Scholar] [CrossRef]
- Yang, S.; Yang, R.; Lin, R.; Si, L. MicroRNA-375 inhibits the growth, drug sensitivity and metastasis of human ovarian cancer cells by targeting PAX2. JBUON 2019, 24, 2341–2346. [Google Scholar]
- Gong, L.; Wang, C.; Gao, Y.; Wang, J. Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed. Pharmacother. Biomed. Pharmacother. 2016, 83, 58–63. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, Y.; Jasper, J.; Lykken, E.; Alexander, P.B.; Markowitz, G.J.; McDonnell, D.P.; Li, Q.J.; Wang, X.F. MiR-148a functions to suppress metastasis and serves as a prognostic indicator in triple-negative breast cancer. Oncotarget 2016, 7, 20381–20394. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours, 8th ed.; Brierley, J.D., Gospodarowicz, M.K., Wittekind, C., Eds.; Wiley-Blackwell: Oxford, UK, 2017; p. 272. [Google Scholar]
- Kurman, R.J.; Carcangiu, M.L.; Herrington, C.S.; Young, R.H. WHO Classification of Tumours of Female Reproductive Organs, 4th ed.; Kurman, R.J., Carcangiu, M.L., Herrington, C.S., Young, R.H., Eds.; IARC Press: Lyon, France, 2014; p. 307. [Google Scholar]
- Pronina, I.V.; Loginov, V.I.; Khodyrev, D.S.; Kazubskaya, T.P.; Braga, E.A. RASSF1A expression level in primary epithelial tumors of various locations. Mol. Biol. 2012, 46, 236–243. [Google Scholar] [CrossRef]
- You, J.; Zhao, Q.; Fan, X.; Wang, J. SOX5 promotes cell invasion and metastasis via activation of Twist-mediated epithelial-mesenchymal transition in gastric cancer. OncoTargets Ther. 2019, 12, 2465–2476. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, G.S.; Kudryavtseva, A.V.; Snezhkina, A.V.; Lakunina, V.A.; Beniaminov, A.D.; Melnikova, N.V.; Dmitriev, A.A. Pan-Cancer Analysis of TCGA Data Revealed Promising Reference Genes for qPCR Normalization. Front. Genet. 2019, 10, 97. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, G.S.; Oparina, N.Y.; Dmitriev, A.A.; Kudryavtseva, A.V.; Anedchenko, E.A.; Kondrat’eva, T.T.; Zabarovsky, E.R.; Senchenko, V.N. RPN1, a new reference gene for quantitative data normalization in lung and kidney cancer. Mol. Biol. 2011, 45, 211–220. [Google Scholar] [CrossRef]
- Dreijerink, K.; Braga, E.; Kuzmin, I.; Geil, L.; Duh, F.M.; Angeloni, D.; Zbar, B.; Lerman, M.I.; Stanbridge, E.J.; Minna, J.D.; et al. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc. Natl. Acad. Sci. USA 2001, 98, 7504–7509. [Google Scholar] [CrossRef]
- Hattermann, K.; Mehdorn, H.M.; Mentlein, R.; Schultka, S.; Held-Feindt, J. A methylation-specific and SYBR-green-based quantitative polymerase chain reaction technique for O6-methylguanine DNA methyltransferase promoter methylation analysis. Anal. Biochem. 2008, 377, 62–71. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Biziota, E.; Koukaki, T.; Karamitrousis, E.; Nena, E.; Tsamardinos, I.; Kolios, G.; Lianidou, E.; et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2019, 38, 3387–3401. [Google Scholar] [CrossRef]
- van Hoesel, A.Q.; Sato, Y.; Elashoff, D.A.; Turner, R.R.; Giuliano, A.E.; Shamonki, J.M.; Kuppen, P.J.; van de Velde, C.J.; Hoon, D.S. Assessment of DNA methylation status in early stages of breast cancer development. Br. J. Cancer 2013, 108, 2033–2038. [Google Scholar] [CrossRef]
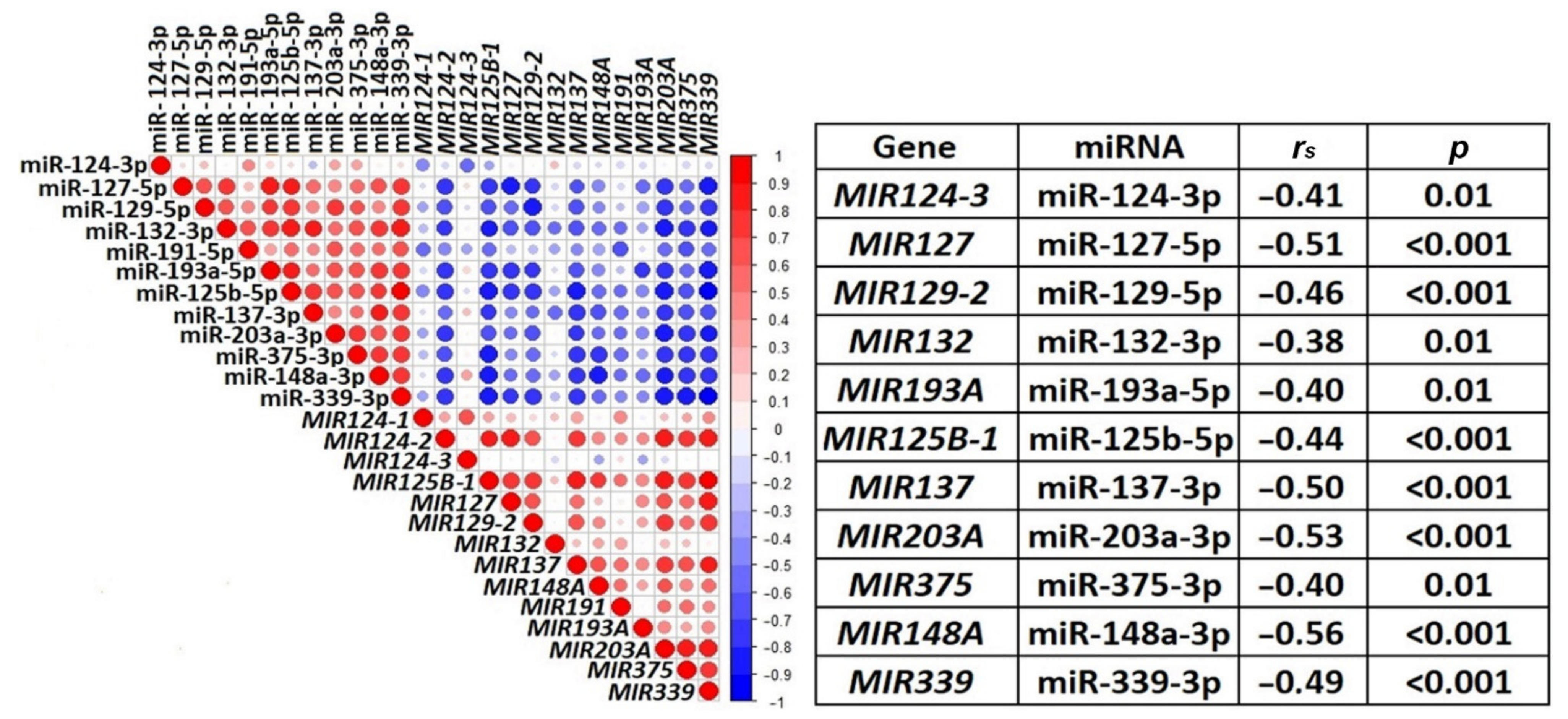
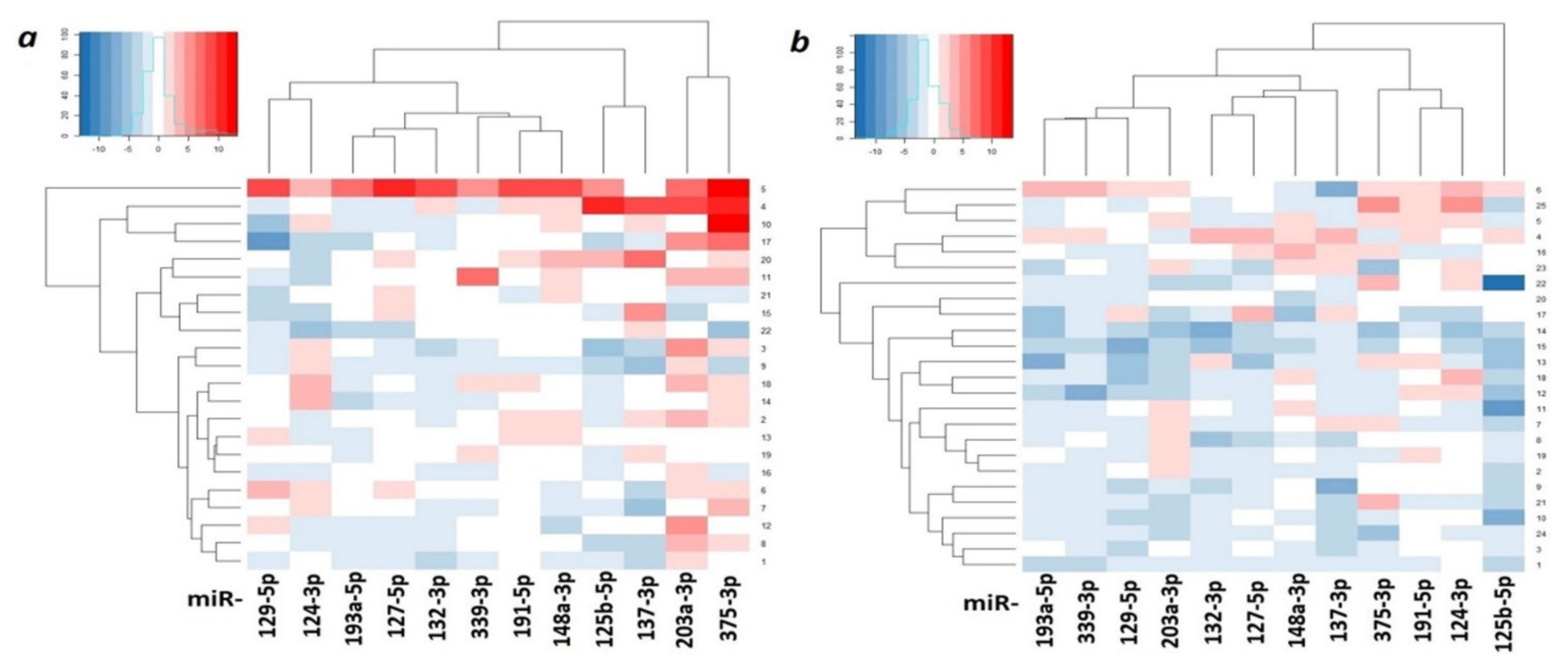
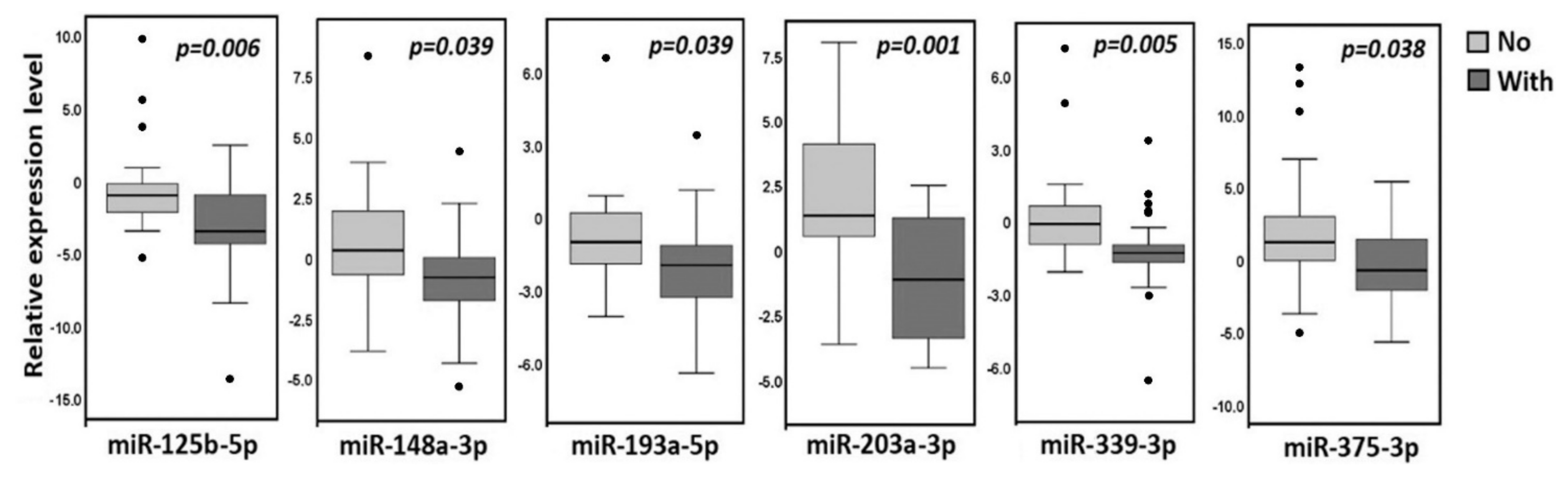
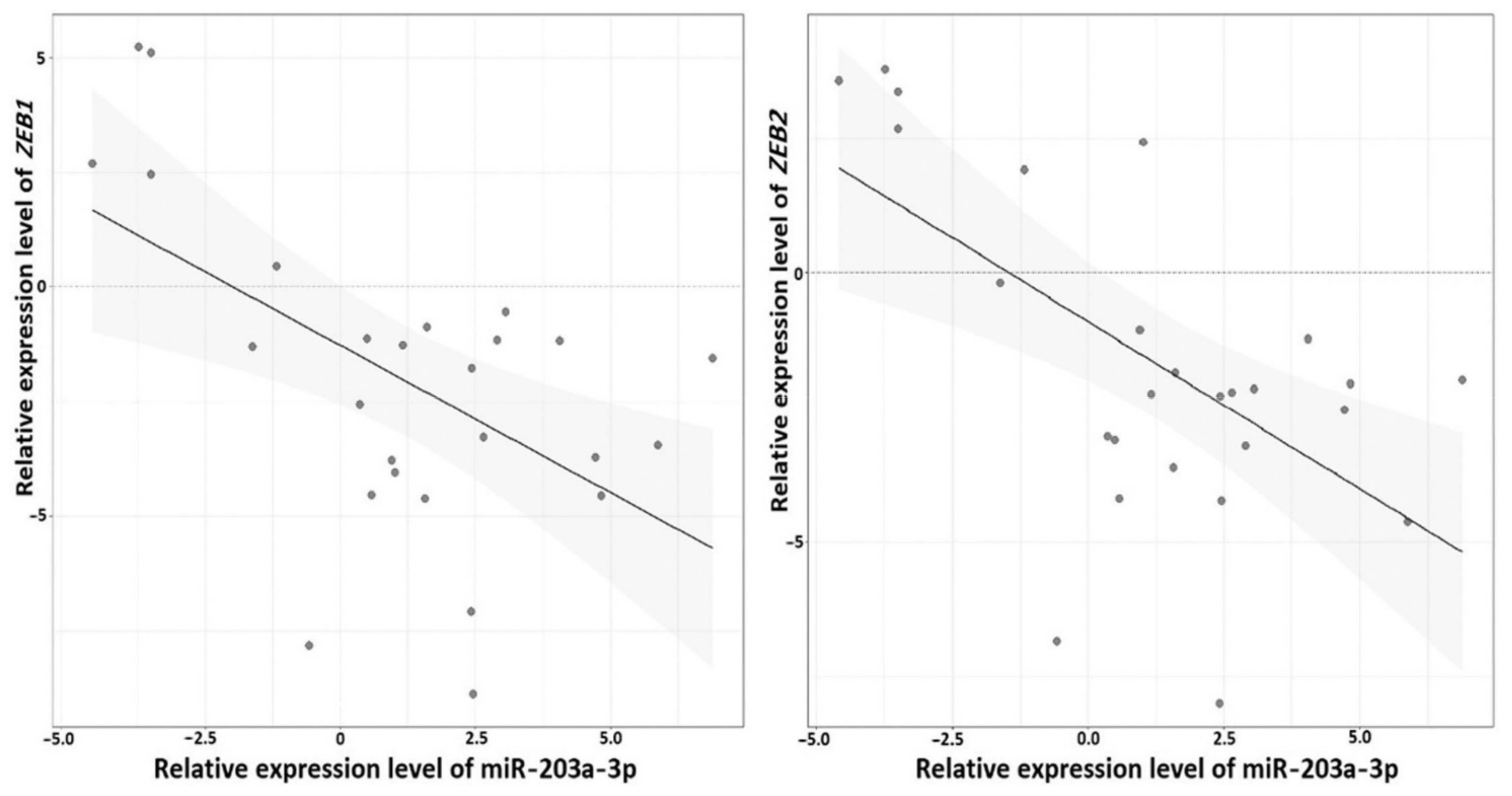
| miRNA | Tissue Samples | 1st Quartile | Median | 3rd Quartile |
|---|---|---|---|---|
| miR-124-3p | primary tumor | –2.46 | 0.54 | 3.28 |
| PMM | –2.60 | –0.21 | 2.12 | |
| miR-127-5p | primary tumor | –3.16 | –2.07 | –0.24 |
| PMM | –5.70 | –2.56 | –0.97 | |
| miR-129-5p | primary tumor | –2.04 | –1.76 | –1.33 |
| PMM | –3.46 | –2.37 | –1.98 | |
| miR-132-3p | primary tumor | –1.79 | –1.64 | –0.57 |
| PMM | –5.54 | –3.40 | –0.16 | |
| miR-191-5p | primary tumor | –0.78 | 0.40 | 0.99 |
| PMM | –1.40 | –0.29 | 0.03 | |
| miR-193a-5p | primary tumor | –3.01 | –2.21 | –1.61 |
| PMM | –4.06 | –2.35 | –1.98 | |
| miR-125b-5p | primary tumor | –4.33 | –2.25 | –0.61 |
| PMM | –4.53 | –1.93 | –0.81 | |
| miR-137-3p | primary tumor | –3.51 | –2.21 | 0.74 |
| PMM | –4.28 | –3.11 | –0.56 | |
| miR-203a-3p | primary tumor | –3.43 | –0.90 | 1.17 |
| PMM | –3.43 | –1.21 | –1.10 | |
| miR-375-3p | primary tumor | –5.03 | –1.26 | 3.25 |
| PMM | –3.94 | –2.35 | –0.61 | |
| miR-148a-3p | primary tumor | –0.94 | –0.70 | 0.42 |
| PMM | –2.91 | –1.45 | –0.38 | |
| miR-339-3p | primary tumor | –1.35 | –1.21 | 0.43 |
| PMM | –1.74 | –1.11 | –0.95 |
| Gene | AUC | 95% CI | Cutoff | The Significance Level (Area = 0.5) | Sn | Sp |
|---|---|---|---|---|---|---|
| MIR107 | 0.66 | 0.56–0.76 | >18.89 | 0.004 | 53.7 | 75.9 |
| MIR124-1 | 0.59 | 0.49–0.69 | >11.24 | 0.124 | 53.7 | 68.5 |
| MIR124-2 | 0.56 | 0.45–0.66 | >7.19 | 0.345 | 78.1 | 44.4 |
| MIR124-3 | 0.62 | 0.51–0.72 | >34.97 | 0.043 | 41.5 | 79.6 |
| MIR125B-1 | 0.64 | 0.53–0.73 | >6.00 | 0.017 | 92.7 | 33.3 |
| MIR1258 | 0.61 | 0.51–0.71 | >7.96 | 0.051 | 78.1 | 48.2 |
| MIR127 | 0.50 | 0.40–0.61 | >5.29 | 0.959 | 73.2 | 37.0 |
| MIR129-2 | 0.55 | 0.45–0.66 | >36.87 | 0.385 | 53.7 | 66.7 |
| MIR130B | 0.68 | 0.57–0.77 | >37.96 | 0.003 | 43.9 | 90.7 |
| MIR132 | 0.55 | 0.45–0.66 | ≤11.25 | 0.374 | 65.9 | 53.7 |
| MIR137 | 0.50 | 0.40–0.61 | >0.34 | 0.991 | 85.4 | 3.7 |
| MIR148A | 0.52 | 0.41–0.62 | >29.58 | 0.763 | 26.8 | 87.0 |
| MIR191 | 0.58 | 0.47–0.68 | >6.77 | 0.199 | 61.0 | 61.1 |
| MIR193A | 0.62 | 0.51–0.71 | >32.33 | 0.052 | 65.9 | 59.3 |
| MIR203A | 0.54 | 0.43–0.64 | >32.26 | 0.560 | 36.6 | 74.1 |
| MIR339 | 0.57 | 0.47–0.68 | >43.76 | 0.231 | 45.0 | 75.0 |
| MIR34B/C | 0.67 | 0.56–0.76 | >12.79 | 0.003 | 65.9 | 68.5 |
| MIR375 | 0.61 | 0.50–0.71 | >14.95 | 0.064 | 56.1 | 64.8 |
| MIR9-1 | 0.67 | 0.56–0.76 | >55.43 | 0.004 | 46.3 | 85.2 |
| MIR9-3 | 0.62 | 0.52–0.72 | >21.89 | 0.046 | 61.0 | 70.4 |
| Gene, MIR | Frequency of Methylation Level Higher/Lower Cutoff Value for Living Patients | Frequency of Methylation Level Higher/Lower Cutoff Value for Deceased Patients | Fisher’s Exact Test, p | Cox Regr., p | Odds Ratio/ 95% CI | Relative Risk/ 95% CI |
|---|---|---|---|---|---|---|
| 107 | 13/41 | 22/19 | 0.005 | 0.008 | 3.65/1.52–8.76 | 2.23/1.28–3.87 |
| 124-1 | 17/37 | 22/19 | 0.037 | 0.004 | 2.52/1.09–5.84 | 1.70/1.05–2.77 |
| 124-2 | 30/24 | 32/9 | 0.030 | 0.134 | 2.84/1.14–7.09 | 1.40/1.05–1.87 |
| 124-3 | 11/43 | 17/24 | 0.040 | 0.038 | 2.77/1.12–6.87 | 2.04/1.07–3.86 |
| 125B-1 | 36/18 | 38/3 | 0.003 | 0.006 | 6.33/1.72–23.3 | 1.39/1.13–1.71 |
| 1258 | 28/26 | 32/9 | 0.010 | 0.306 | 3.30/1.33–8.22 | 1.51/1.11–2.04 |
| 127 | 34/20 | 30/11 | 0.378 | 0.178 | 1.60/0.66–3.89 | 1.16/0.88–1.53 |
| 129-2 | 18/36 | 22/19 | 0.060 | 0.124 | 2.32/1.00–5.34 | 1.61/1.00–2.58 |
| 130B | 5/49 | 18/23 | 0.0002 | 0.003 | 7.67/2.53–23.2 | 4.74/1.92–11.71 |
| 132 | 25/29 | 27/14 | 0.065 | 0.645 | 2.24/0.97–5.17 | 1.42/0.99–2.04 |
| 137 | 52/2 | 35/6 | 0.072 | 0.528 | 0.22/0.04–1.18 | 0.89/0.77–1.02 |
| 148A | 7/47 | 11/30 | 0.115 | 0.020 | 2.46/0.86–7.05 | 2.07/0.88–4.87 |
| 191 | 21/33 | 25/16 | 0.040 | 0.037 | 2.46/1.07–5.65 | 1.57/1.04–2.37 |
| 193A | 22/32 | 27/14 | 0.022 | 0.063 | 2.81/1.21–6.52 | 1.62/1.09–2.39 |
| 203A | 14/40 | 15/26 | 0.273 | 0.416 | 1.65/0.68–3.97 | 1.41/0.77–2.58 |
| 339 | 15/39 | 19/22 | 0.084 | 0.188 | 2.25/0.95–5.28 | 1.67/0.97–2.87 |
| 34B/C | 17/37 | 27/14 | 0.002 | 0.141 | 4.20/1.77–9.96 | 2.09/1.33–3.28 |
| 375 | 19/35 | 23/18 | 0.060 | 0.077 | 2.35/1.02–5.41 | 1.59/1.01–2.51 |
| 9-1 | 8/46 | 19/22 | 0.001 | 0.017 | 4.97/1.88–13.1 | 3.13/1.52–6.42 |
| 9-3 | 16/38 | 25/16 | 0.003 | 0.025 | 3.71/1.57–8.75 | 2.06/1.28–3.32 |
| Clinical and Histological Characteristics | N = 102 | With PMM N = 30 | |
|---|---|---|---|
| Histological type | Borderline serous adenocarcinoma | 6 | 0 |
| High-grade serous adenocarcinoma | 73 | 25 | |
| Low-grade serous adenocarcinoma | 7 | 0 | |
| Endometrioid adenocarcinoma | 9 | 4 | |
| Clear cell adenocarcinoma | 2 | 0 | |
| Mucinous adenocarcinoma | 2 | 1 | |
| Mixed epithelial tumors | 2 | 0 | |
| Undifferentiated carcinoma | 1 | 0 | |
| Stage | I | 12 | 1 |
| II | 17 | 4 | |
| III | 67 | 24 | |
| IV | 6 | 1 | |
| Grade | G1 | 9 | 1 |
| G2 | 27 | 11 | |
| G3 | 60 | 18 | |
| Grade Unknown | 6 | 0 | |
| Size | T1 | 12 | 1 |
| T2 | 18 | 4 | |
| T3 | 72 | 25 | |
| Peritoneal metastases | Absent | 72 | 0 |
| Present | 30 | 30 | |
| Lymph node metastases | N0 | 81 | 30 |
| N1 | 21 | 0 | |
| Distant metastases | M0 | 96 | 29 |
| M1 | 6 | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loginov, V.I.; Pronina, I.V.; Filippova, E.A.; Burdennyy, A.M.; Lukina, S.S.; Kazubskaya, T.P.; Uroshlev, L.A.; Fridman, M.V.; Brovkina, O.I.; Apanovich, N.V.; et al. Aberrant Methylation of 20 miRNA Genes Specifically Involved in Various Steps of Ovarian Carcinoma Spread: From Primary Tumors to Peritoneal Macroscopic Metastases. Int. J. Mol. Sci. 2022, 23, 1300. https://doi.org/10.3390/ijms23031300
Loginov VI, Pronina IV, Filippova EA, Burdennyy AM, Lukina SS, Kazubskaya TP, Uroshlev LA, Fridman MV, Brovkina OI, Apanovich NV, et al. Aberrant Methylation of 20 miRNA Genes Specifically Involved in Various Steps of Ovarian Carcinoma Spread: From Primary Tumors to Peritoneal Macroscopic Metastases. International Journal of Molecular Sciences. 2022; 23(3):1300. https://doi.org/10.3390/ijms23031300
Chicago/Turabian StyleLoginov, Vitaly I., Irina V. Pronina, Elena A. Filippova, Alexey M. Burdennyy, Svetlana S. Lukina, Tatiana P. Kazubskaya, Leonid A. Uroshlev, Marina V. Fridman, Olga I. Brovkina, Natalya V. Apanovich, and et al. 2022. "Aberrant Methylation of 20 miRNA Genes Specifically Involved in Various Steps of Ovarian Carcinoma Spread: From Primary Tumors to Peritoneal Macroscopic Metastases" International Journal of Molecular Sciences 23, no. 3: 1300. https://doi.org/10.3390/ijms23031300
APA StyleLoginov, V. I., Pronina, I. V., Filippova, E. A., Burdennyy, A. M., Lukina, S. S., Kazubskaya, T. P., Uroshlev, L. A., Fridman, M. V., Brovkina, O. I., Apanovich, N. V., Karpukhin, A. V., Stilidi, I. S., Kushlinskii, N. E., Dmitriev, A. A., & Braga, E. A. (2022). Aberrant Methylation of 20 miRNA Genes Specifically Involved in Various Steps of Ovarian Carcinoma Spread: From Primary Tumors to Peritoneal Macroscopic Metastases. International Journal of Molecular Sciences, 23(3), 1300. https://doi.org/10.3390/ijms23031300







