Functional Characterization of an In-Frame Deletion in the Basic Domain of the Retinal Transcription Factor ATOH7
Abstract
1. Introduction
2. Results
2.1. The NCRNA-Associated In-Frame Deletion R41_R48del Removes Evolutionary Conserved Amino Acids in the Basic Domain of ATOH7
2.2. The Mutant ATOH7:R41_R48del Protein Is Misslocalized Due to the Loss of a Predicted NLS within the Basic Domain
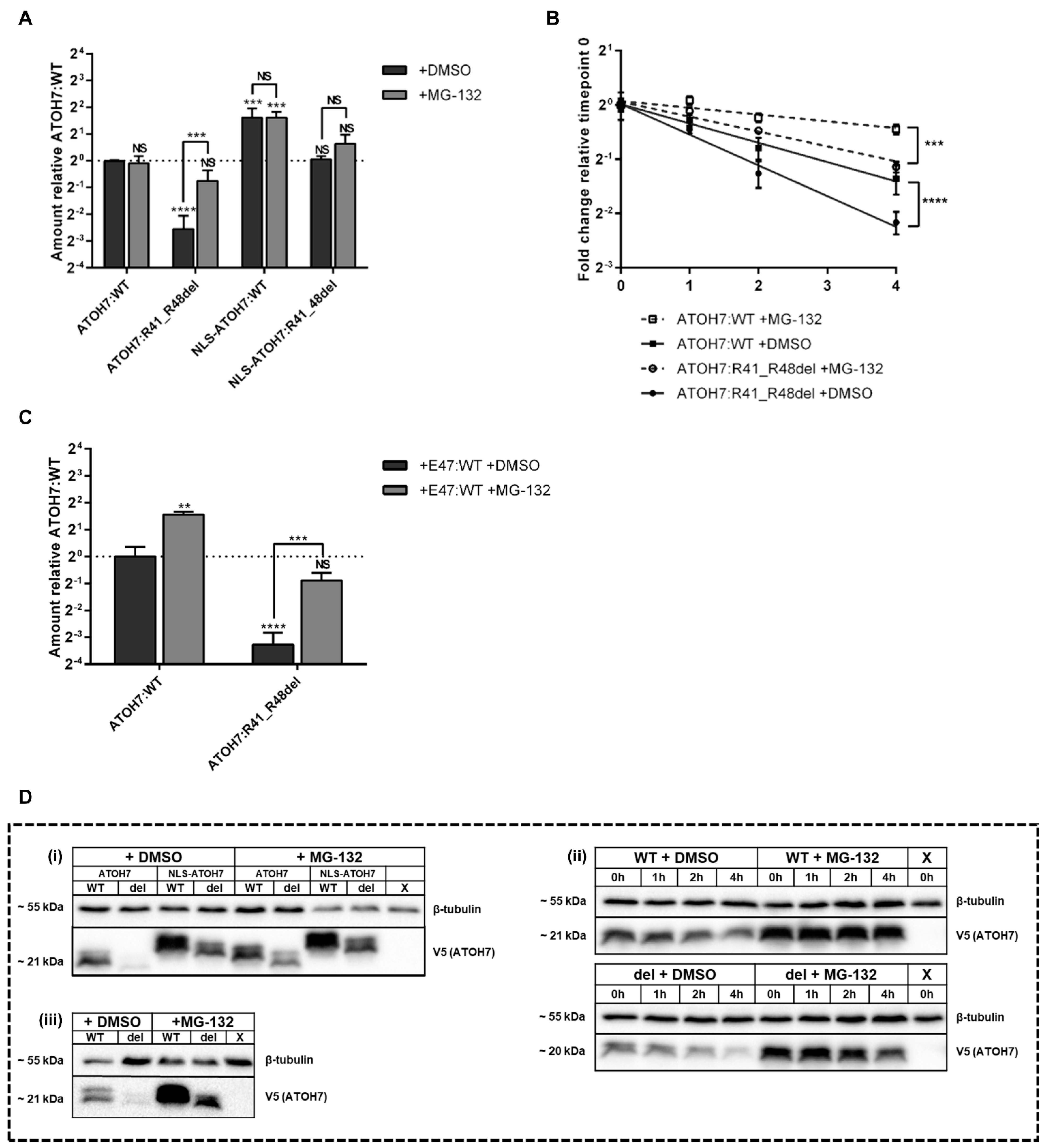
2.3. Decreased Amount of the Mutant ATOH7:R41_R48del Protein Is the Result of an Increased Proteasomal Degradation
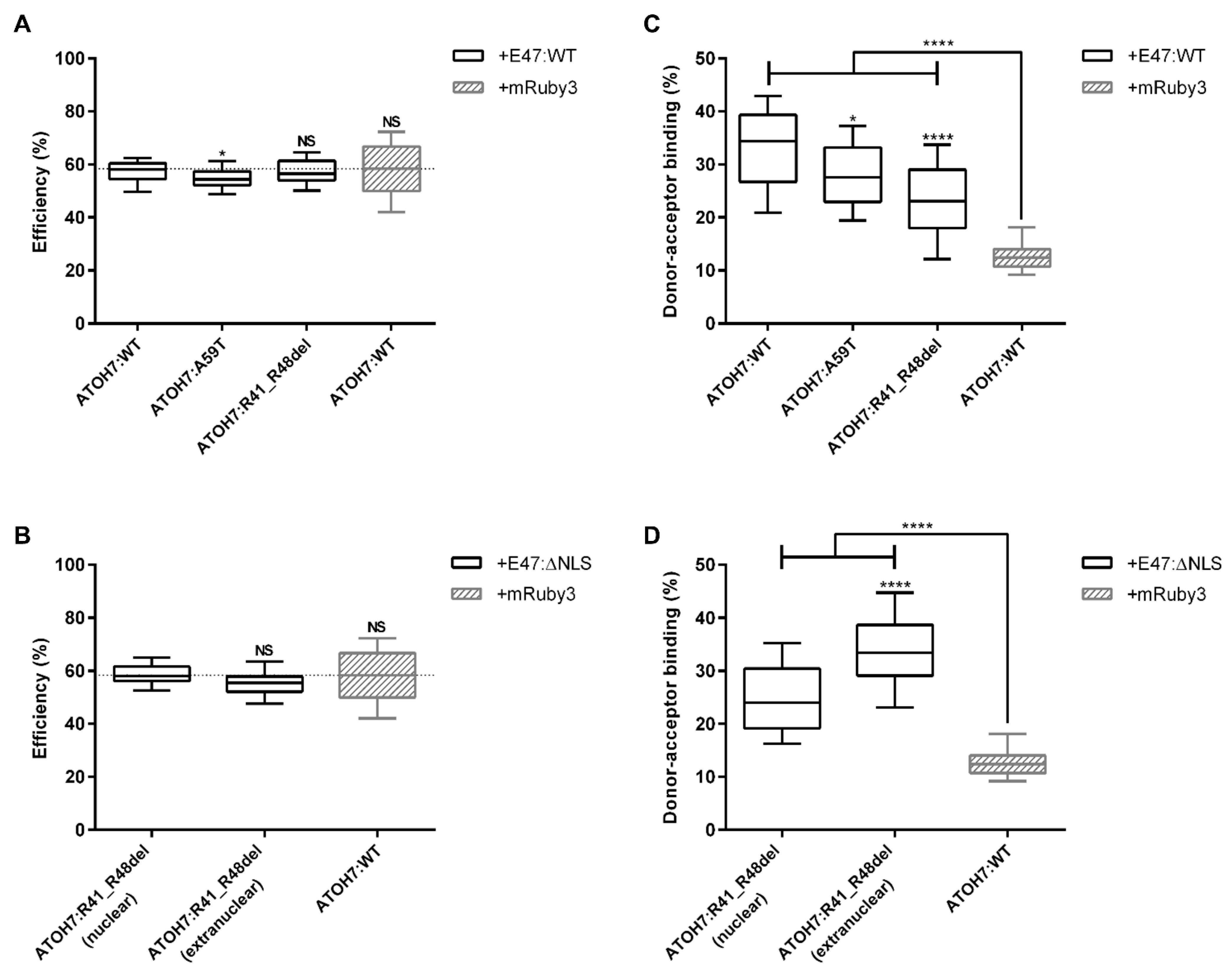
2.4. Reduced Nuclear Dimerization of Mutant ATOH7:R41_R48del Protein and Extranuclear Protein Interaction with E47 in Living Cells
2.4.1. FLIM–FRET Confirms Dimerization of ATOH7 and E47 in Living Cells
2.4.2. Establishing a Background Threshold for Significant Protein Interaction
2.4.3. Measurements of Protein Interactions between ATOH7 and E47
2.4.4. The Reduced Dimerization of Mutant ATOH7:R41_R48del Protein with E47 Is Not a Result of Altered Protein Amounts
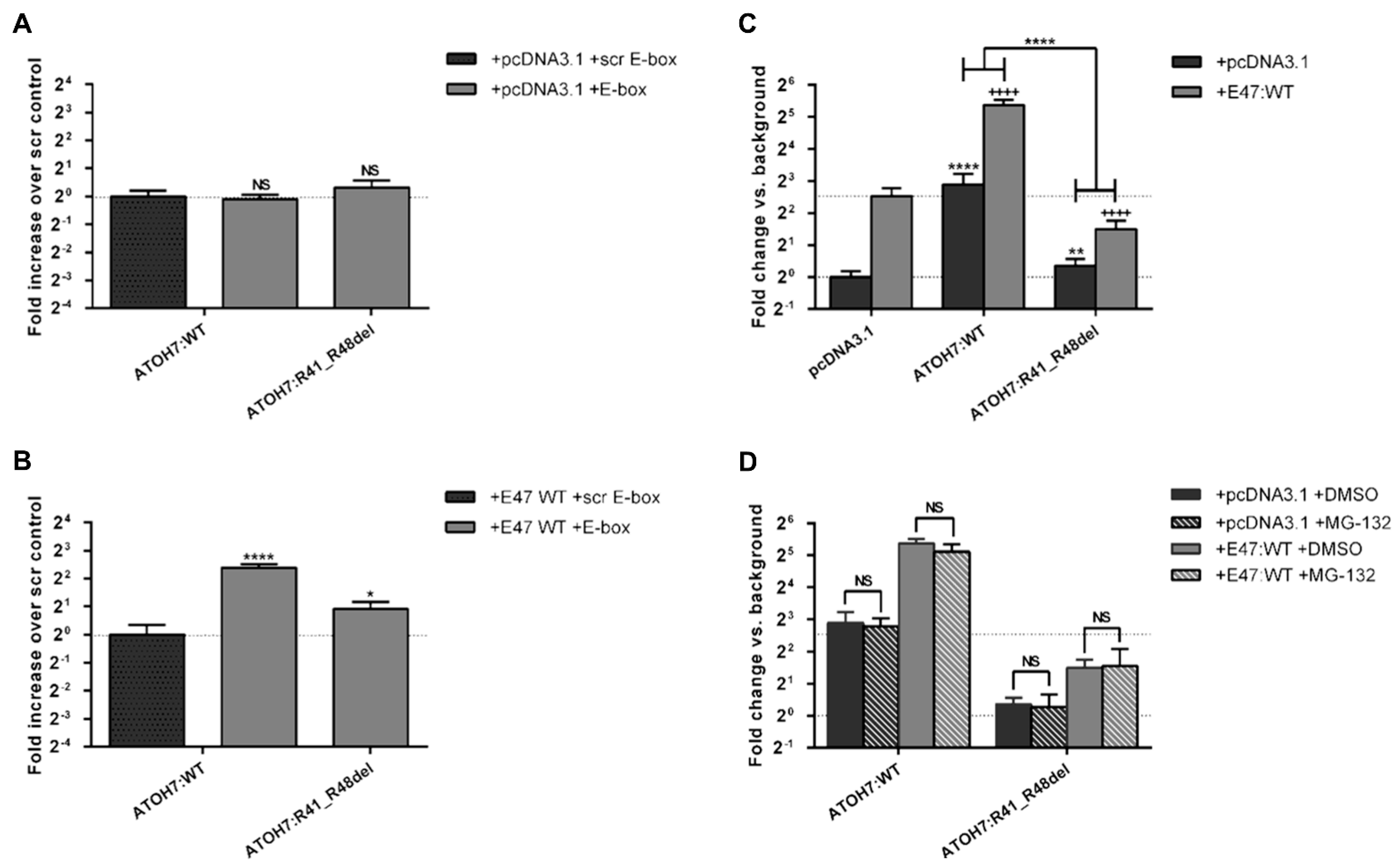
2.5. The Mutant ATOH7:R41_R48del Protein Shows Reduced DNA-Binding, Reduced Transcriptional Activation and Inhibition of E47 Mediated Transcription
3. Discussion
3.1. Nuclear Localization of ATOH7 Is Directed by an NLS Contained within the Basic Domain and Can Also Be Stimulated by Synergistic Nuclear Import with E47
3.2. Basic Amino Acid Residues in the ATOH7 Basic Domain May Be Involved in Regulating Protein Turnover through Proteasomal Degradation
3.3. The ATOH7 Basic Domain Plays a Role in Protein Dimerization
3.4. Mutation of the ATOH7 Basic Domain Reduces Transcriptional Activation and Inhibits the Ability of Heterodimers to Regulate Transcription
3.5. The Basic Domain Mediates Various Functional Properties of the ATOH7 Protein
3.6. Conclusions
4. Materials and Methods
4.1. Bioinformatics Analysis
4.2. Protein Modeling
4.3. Expression Vectors
4.4. Cell Culture and Protein Assays
4.5. Confocal Fluorescence Microscopy
4.6. FLIM–FRET
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murre, C.; Page-McCaw, P.; Vaessin, H.; Caudy, M.; Jan, L.; Jan, Y.N.; Cabrera, C.V.; Buskin, J.N.; Hauschka, S.D.; Lassar, A.B.; et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 1989, 58, 537–544. [Google Scholar] [CrossRef]
- Bertrand, N.; Castro, D.; Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002, 3, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Dennis, D.J.; Han, S.; Schuurmans, C. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 2019, 1705, 48–65. [Google Scholar] [CrossRef]
- Longo, A.; Guanga, G.P.; Rose, R. Crystal Structure of E47−NeuroD1/Beta2 bHLH Domain−DNA Complex: Heterodimer Selectivity and DNA Recognition. Biochemistry 2008, 47, 218–229. [Google Scholar] [CrossRef]
- Garrell, J.; Campuzano, S. The helix-loop-helix domain: A common motif for bristles, muscles and sex. BioEssays 1991, 13, 493–498. [Google Scholar] [CrossRef]
- Curtis, D.J.; Salmon, J.M.; Pimanda, J.E. Concise Review: Blood Relatives: Formation and regulation of hematopoietic stem cells by the basic helix-loop-helix transcription factors stem cell leukemia and lymphoblastic leukemia-derived sequence 1. Stem Cells 2012, 30, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Berkes, C.A.; Tapscott, S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef]
- Tomita, K.; Nakanishi, S.; Guillemot, F.; Kageyama, R.; Nishikawa, S.-I. Mash1 promotes neuronal differentiation in the retina. Genes Cells 1996, 1, 765–774. [Google Scholar] [CrossRef]
- Brown, N.; Kanekar, S.; Vetter, M.; Tucker, P.; Gemza, D.; Glaser, T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 1998, 125, 4821–4833. [Google Scholar] [CrossRef] [PubMed]
- Morrow, E.; Furukawa, T.; Lee, J.; Cepko, C. NeuroD regulates multiple functions in the developing neural retina in rodent. Development 1999, 126, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Perron, M.; Opdecamp, K.; Butler, K.; Harris, W.A.; Bellefroid, E.J. X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc. Natl. Acad. Sci. USA 1999, 96, 14996–15001. [Google Scholar] [CrossRef]
- Wang, J.-C.; Harris, W. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev. Biol. 2005, 285, 101–115. [Google Scholar] [CrossRef]
- Hernandez, J.; Matter-Sadzinski, L.; Skowronska-Krawczyk, D.; Chiodini, F.; Alliod, C.; Ballivet, M.; Matter, J.-M. Highly Conserved Sequences Mediate the Dynamic Interplay of Basic Helix-Loop-Helix Proteins Regulating Retinogenesis. J. Biol. Chem. 2007, 282, 37894–37905. [Google Scholar] [CrossRef]
- Brown, N.L.; Dagenais, S.L.; Chen, C.-M.; Glaser, T. Molecular characterization and mapping of ATOH7, a human atonal homolog with a predicted role in retinal ganglion cell development. Mamm. Genome 2002, 13, 95–101. [Google Scholar] [CrossRef]
- Brennan, T.J.; Chakraborty, T.; Olson, E.N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 5675–5679. [Google Scholar] [CrossRef]
- Shimizu, T.; Toumoto, A.; Ihara, K.; Shimizu, M.; Kyogoku, Y.; Ogawa, N.; Oshima, Y.; Hakoshima, T. Crystal structure of PHO4 bHLH domain-DNA complex: Flanking base recognition. EMBO J. 1997, 16, 4689–4697. [Google Scholar] [CrossRef] [PubMed]
- Kanekar, S.; Perron, M.; Dorsky, R.; Harris, W.A.; Jan, L.; Jan, Y.N.; Vetter, M.L. Xath5 Participates in a Network of bHLH Genes in the Developing Xenopus Retina. Neuron 1997, 19, 981–994. [Google Scholar] [CrossRef]
- Masai, I.; Stemple, D.L.; Okamoto, H.; Wilson, S.W. Midline Signals Regulate Retinal Neurogenesis in Zebrafish. Neuron 2000, 27, 251–263. [Google Scholar] [CrossRef]
- Liu, W.; Mo, Z.; Xiang, M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc. Natl. Acad. Sci. USA 2001, 98, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Matter-Sadzinski, L.; Matter, J.; Ong, M.; Hernandez, J.; Ballivet, M. Specification of neurotransmitter receptor identity in developing retina: The chick ATH5 promoter integrates the positive and negative effects of several bHLH proteins. Development 2001, 128, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Kim, B.S.; Ding, K.; Wang, H.; Sun, D.; Johnson, R.L.; Klein, W.H.; Gan, L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001, 15, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Patel, S.; Brzezinski, J.; Glaser, T. Math5 is required for retinal ganglion cell and optic nerve formation. Development 2001, 128, 2497–2508. [Google Scholar] [CrossRef]
- Kay, J.N.; Finger-Baier, K.C.; Roeser, T.; Staub, W.; Baier, H. Retinal Ganglion Cell Genesis Requires lakritz, a Zebrafish atonal Homolog. Neuron 2001, 30, 725–736. [Google Scholar] [CrossRef]
- Ghiasvand, N.M.; Rudolph, D.D.; Mashayekhi, M.; A Brzezinski, J.; Goldman, D.; Glaser, T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 2011, 14, 578–586. [Google Scholar] [CrossRef]
- Khan, K.; Logan, C.V.; McKibbin, M.; Sheridan, E.; Elçioglu, N.H.; Yenice, O.; Parry, D.A.; Fernandez-Fuentes, N.; Abdelhamed, Z.I.; Al-Maskari, A.; et al. Next generation sequencing identifies mutations in Atonal homolog 7 (ATOH7) in families with global eye developmental defects. Hum. Mol. Genet. 2011, 21, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Prasov, L.; Masud, T.; Khaliq, S.; Mehdi, S.Q.; Abid, A.; Oliver, E.R.; Silva, E.; Lewanda, A.; Brodsky, M.C.; Borchert, M.; et al. ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum. Mol. Genet. 2012, 21, 3681–3694. [Google Scholar] [CrossRef]
- Kondo, H.; Matsushita, I.; Tahira, T.; Uchio, E.; Kusaka, S. Mutations in ATOH7 gene in patients with nonsyndromic congenital retinal nonattachment and familial exudative vitreoretinopathy. Ophthalmic Genet. 2016, 37, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Keser, V.; Khan, A.; Siddiqui, S.; Lopez, I.; Ren, H.; Qamar, R.; Nadaf, J.; Majewski, J.; Chen, R.; Koenekoop, R.K. The Genetic Causes of Nonsyndromic Congenital Retinal Detachment: A Genetic and Phenotypic Study of Pakistani Families. Investig. Opthalmol. Vis. Sci. 2017, 58, 1028–1036. [Google Scholar] [CrossRef]
- Atac, D.; Koller, S.; Hanson, J.V.M.; Feil, S.; Tiwari, A.; Bahr, A.; Baehr, L.; Magyar, I.; Kottke, R.; Gerth-Kahlert, C.; et al. Atonal homolog 7 (ATOH7) loss-of-function mutations in predominant bilateral optic nerve hypoplasia. Hum. Mol. Genet. 2019, 29, 132–148. [Google Scholar] [CrossRef]
- Macgregor, S.; Hewitt, A.W.; Hysi, P.G.; Ruddle, J.B.; Medland, S.E.; Henders, A.K.; Gordon, S.D.; Andrew, T.; McEvoy, B.; Sanfilippo, P.G.; et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum. Mol. Genet. 2010, 19, 2716–2724. [Google Scholar] [CrossRef]
- Ramdas, W.D.; van Koolwijk, L.M.; Lemij, H.G.; Pasutto, F.; Cree, A.J.; Thorleifsson, G.; Janssen, S.F.; Jacoline, T.B.; Amin, N.; Rivadeneira, F.; et al. Common genetic variants associated with open-angle glaucoma. Hum. Mol. Genet. 2011, 20, 2464–2471. [Google Scholar] [CrossRef]
- Ba, A.N.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 1–11. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Szmacinski, H.; Nowaczyk, K.; Johnson, M.L. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. USA 1992, 89, 1271–1275. [Google Scholar] [CrossRef]
- Bastiaens, P.I. Fluorescence lifetime imaging microscopy: Spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999, 9, 48–52. [Google Scholar] [CrossRef]
- Rajoria, S.; Zhao, L.; Intes, X.; Barroso, M. FLIM-FRET for Cancer Applications. Curr. Mol. Imaging 2015, 3, 144–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Groszmann, M.; Paicu, T.; Smyth, D.R. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. Plant J. 2008, 55, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Sawadogo, M. Functional domains of the transcription factor USF2: Atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol. Cell. Biol. 1996, 16, 1367–1375. [Google Scholar] [CrossRef]
- Li, C.; Wu, R.-C.; Amazit, L.; Tsai, S.Y.; Tsai, M.-J.; O’Malley, B.W. Specific Amino Acid Residues in the Basic Helix-Loop-Helix Domain of SRC-3 Are Essential for Its Nuclear Localization and Proteasome-Dependent Turnover. Mol. Cell. Biol. 2007, 27, 1296–1308. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yeung, P.L.; Zhang, A.; Chen, J.D. Nuclear localization of coactivator RAC3 is mediated by a bipartite NLS and importin α3. Biochem. Biophys. Res. Commun. 2006, 348, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, R.; Yasuhara, N.; Oe, S.; Nagai, M.; Yoneda, Y. Synergistic nuclear import of NeuroD1 and its partner transcription factor, E47, via heterodimerization. Exp. Cell Res. 2009, 315, 1639–1652. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.S.; Blobel, G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 1993, 365, 661–663. [Google Scholar] [CrossRef]
- Melchior, F.; Paschal, B.; Evans, J.; Gerace, L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol. 1993, 123, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Massari, M.E.; Murre, C. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Cheng, P.-F.; Lassar, A.B.; Weintraub, H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 1990, 60, 733–746. [Google Scholar] [CrossRef]
- Wang, L.-H.; Baker, N.E. E Proteins and ID Proteins: Helix-Loop-Helix Partners in Development and Disease. Dev. Cell 2015, 35, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Skowronska-Krawczyk, D.; Matter-Sadzinski, L.; Ballivet, M.; Matter, J.-M. The Basic Domain of ATH5 Mediates Neuron-Specific Promoter Activity during Retina Development. Mol. Cell. Biol. 2005, 25, 10029–10039. [Google Scholar] [CrossRef][Green Version]
- Higgins, D.G.; Thompson, J.D.; Gibson, T.J. Using CLUSTAL for multiple sequence alignments. In Methods in Enzymology; Elsevier BV: Amsterdam, The Netherlands, 1996; Volume 266, pp. 383–402. [Google Scholar]
- Fernandez-Fuentes, N.; Rai, B.K.; Madrid-Aliste, C.J.; Fajardo, J.E.; Fiser, A. Comparative protein structure modeling by combining multiple templates and optimizing sequence-to-structure alignments. Bioinformatics 2007, 23, 2558–2565. [Google Scholar] [CrossRef]
- Fernandez-Fuentes, N.; Madrid-Aliste, C.J.; Rai, B.K.; Fajardo, J.E.; Fiser, A. M4T: A comparative protein structure modeling server. Nucleic Acids Res. 2007, 35, W363–W368. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef]
- Molina, R.S.; Tran, T.M.; Campbell, R.E.; Lambert, G.G.; Salih, A.; Shaner, N.C.; Hughes, T.E.; Drobizhev, M. Blue-Shifted Green Fluorescent Protein Homologues Are Brighter than Enhanced Green Fluorescent Protein under Two-Photon Excitation. J. Phys. Chem. Lett. 2017, 8, 2548–2554. [Google Scholar] [CrossRef]
- Bajar, B.T.; Wang, E.S.; Lam, A.J.; Kim, B.B.; Jacobs, C.L.; Howe, E.S.; Davidson, M.W.; Lin, M.Z.; Chu, J. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 2016, 6, 20889. [Google Scholar] [CrossRef] [PubMed]



| Identifier | DNA/Protein Variant | Region | Phenotype(s) | Zygosity | Segregation | Transcription Activity | Published |
|---|---|---|---|---|---|---|---|
| CM1213359 1 | c.139G>A; p.(Ala47Thr) | Basic | ONH | HTZ | – | Reduced | 2010 [26,30] |
| C0027672 2 | c.193A>G; p.(Arg65Gly) | Helix 1 | ONH | HTZ | – | Normal | 2010 [26,30] |
| CG115298 1 | c.-22208_-15686del | Enhancer | NCRNA, ONH | HMZ | Yes | – | 2011 [24] |
| rs1900004 3 | c.-9021C>T | 5′-UTR | POAG | – | – | – | 2011 [31] |
| CM121097 1 | c.53delC; p.(Pro18ArgfsTer69) | Coding * | NCRNA, ONH | HMZ | Yes | – | 2012 [25] |
| CM121094 1 | c.146A>T; p.(Glu49Val) | Basic | NCRNA, ONH | HMZ | Yes | – | 2012 [25] |
| CM126904 1 | c.136A>C; p.(His46Asn) | Basic | PHPVAR | HMZ | Yes | LOF | 2012 [26] |
| CG1613845 1 | c.121_144del; p.(Arg41_Arg48del) | Basic | NCRNA EVR | HMZ HTZ | Yes No | – | 2016 [27] |
| CM1613857 1 | c.400G>T; p.(Glu134 *) | Helix 2 | EVR | HTZ | No | – | 2017 [27] |
| CM172971 1 | c.125G>C; p.(Arg42Pro) | Basic | NCRNA | HMZ | Yes | – | 2017 [28] |
| CM204439 1 CM204440 1 | c.175G>A; p.(Ala59Thr) c.176C>T; p.(Ala59Val) | Helix 1 | ONH, FVH | Compound HTZ | Yes | LOF | 2020 [29] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atac, D.; Mohn, L.; Feil, S.; Maggi, K.; Haenni, D.; Seebauer, B.; Koller, S.; Berger, W. Functional Characterization of an In-Frame Deletion in the Basic Domain of the Retinal Transcription Factor ATOH7. Int. J. Mol. Sci. 2022, 23, 1053. https://doi.org/10.3390/ijms23031053
Atac D, Mohn L, Feil S, Maggi K, Haenni D, Seebauer B, Koller S, Berger W. Functional Characterization of an In-Frame Deletion in the Basic Domain of the Retinal Transcription Factor ATOH7. International Journal of Molecular Sciences. 2022; 23(3):1053. https://doi.org/10.3390/ijms23031053
Chicago/Turabian StyleAtac, David, Lucas Mohn, Silke Feil, Kevin Maggi, Dominik Haenni, Britta Seebauer, Samuel Koller, and Wolfgang Berger. 2022. "Functional Characterization of an In-Frame Deletion in the Basic Domain of the Retinal Transcription Factor ATOH7" International Journal of Molecular Sciences 23, no. 3: 1053. https://doi.org/10.3390/ijms23031053
APA StyleAtac, D., Mohn, L., Feil, S., Maggi, K., Haenni, D., Seebauer, B., Koller, S., & Berger, W. (2022). Functional Characterization of an In-Frame Deletion in the Basic Domain of the Retinal Transcription Factor ATOH7. International Journal of Molecular Sciences, 23(3), 1053. https://doi.org/10.3390/ijms23031053







