Expression Profile of Selected Genes Involved in Storage Lipid Synthesis in a Model Oleaginous Yeast Species Yarrowia lipolytica
Abstract
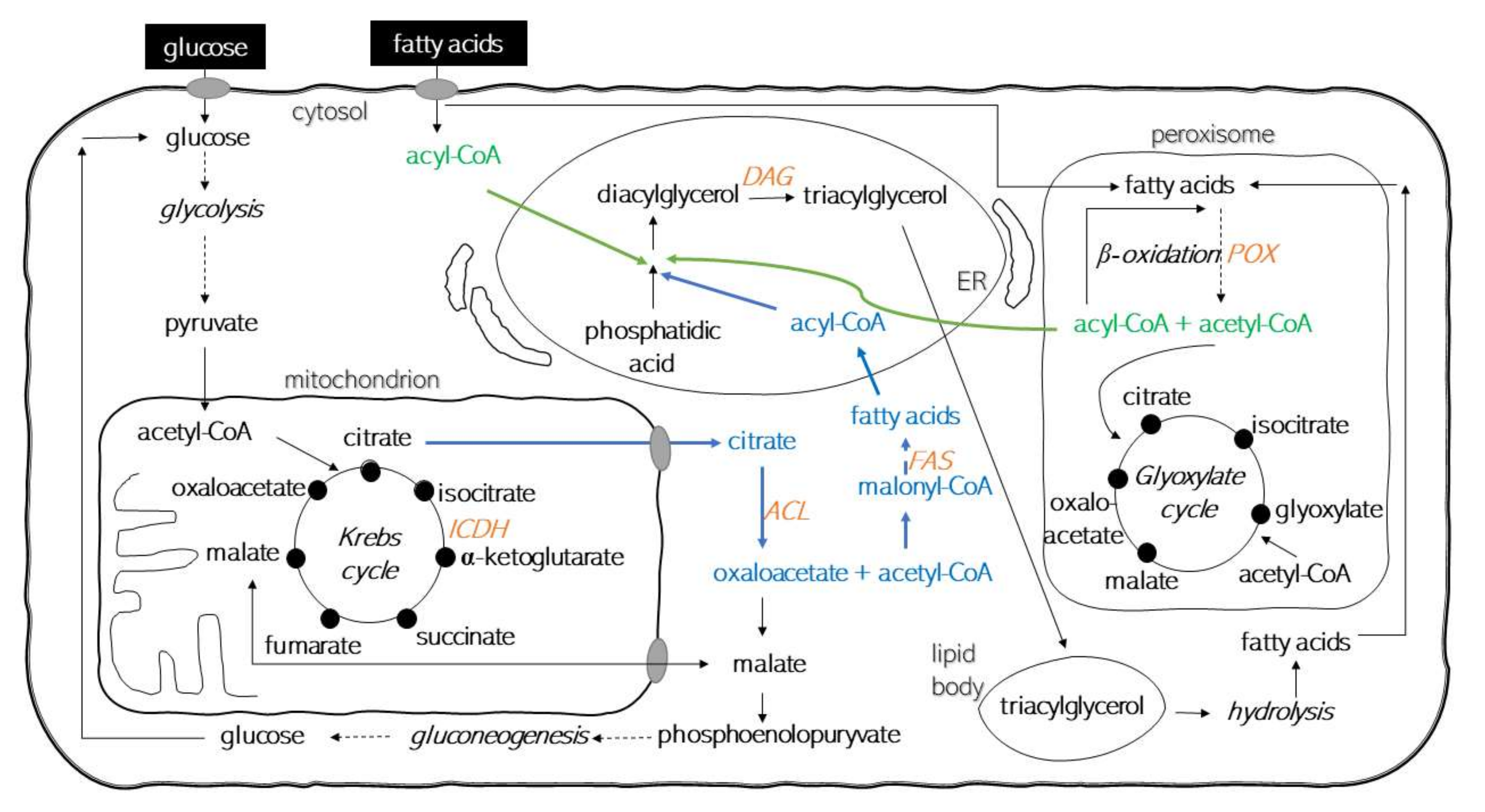
1. Introduction
2. Results
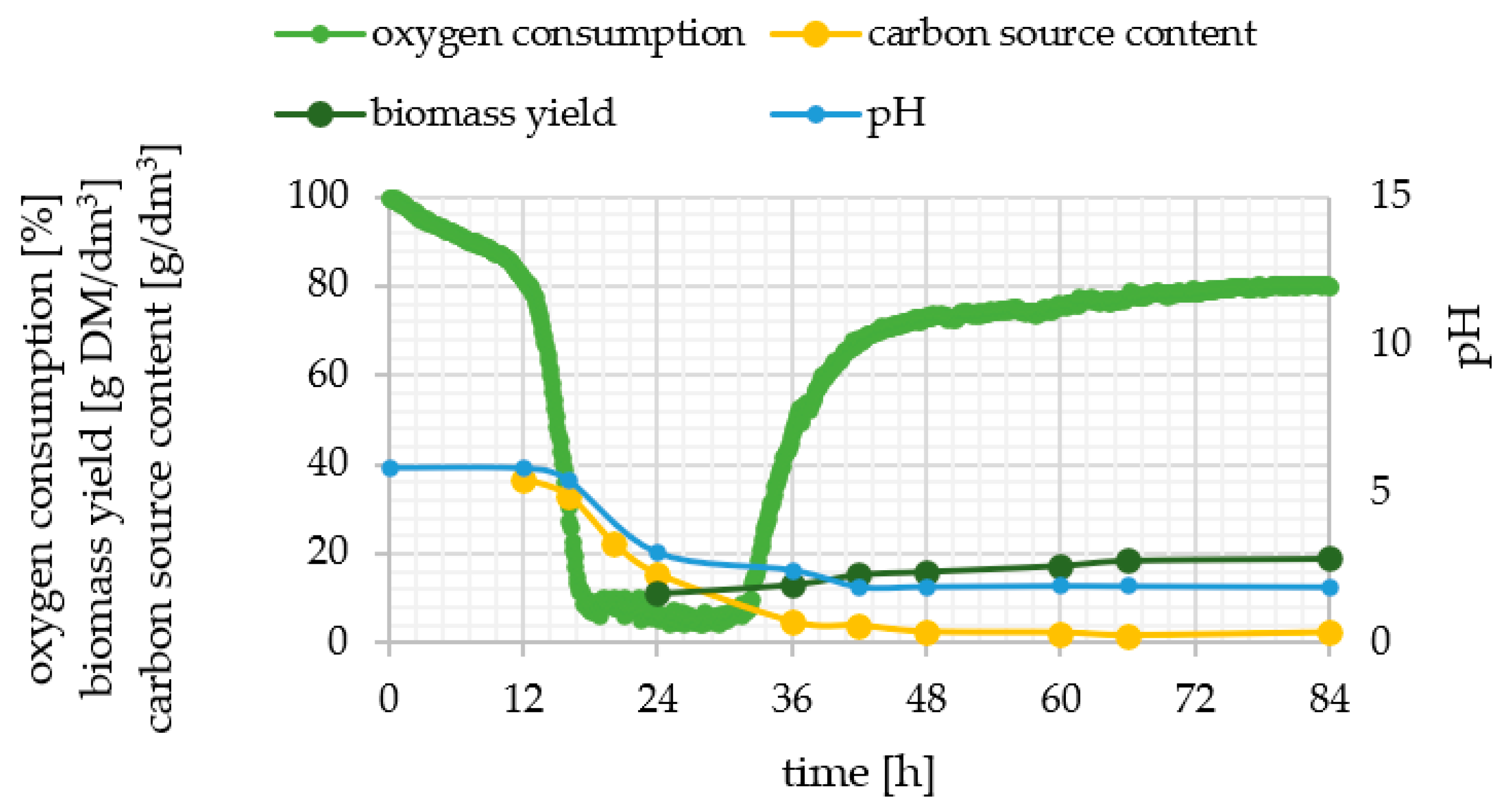
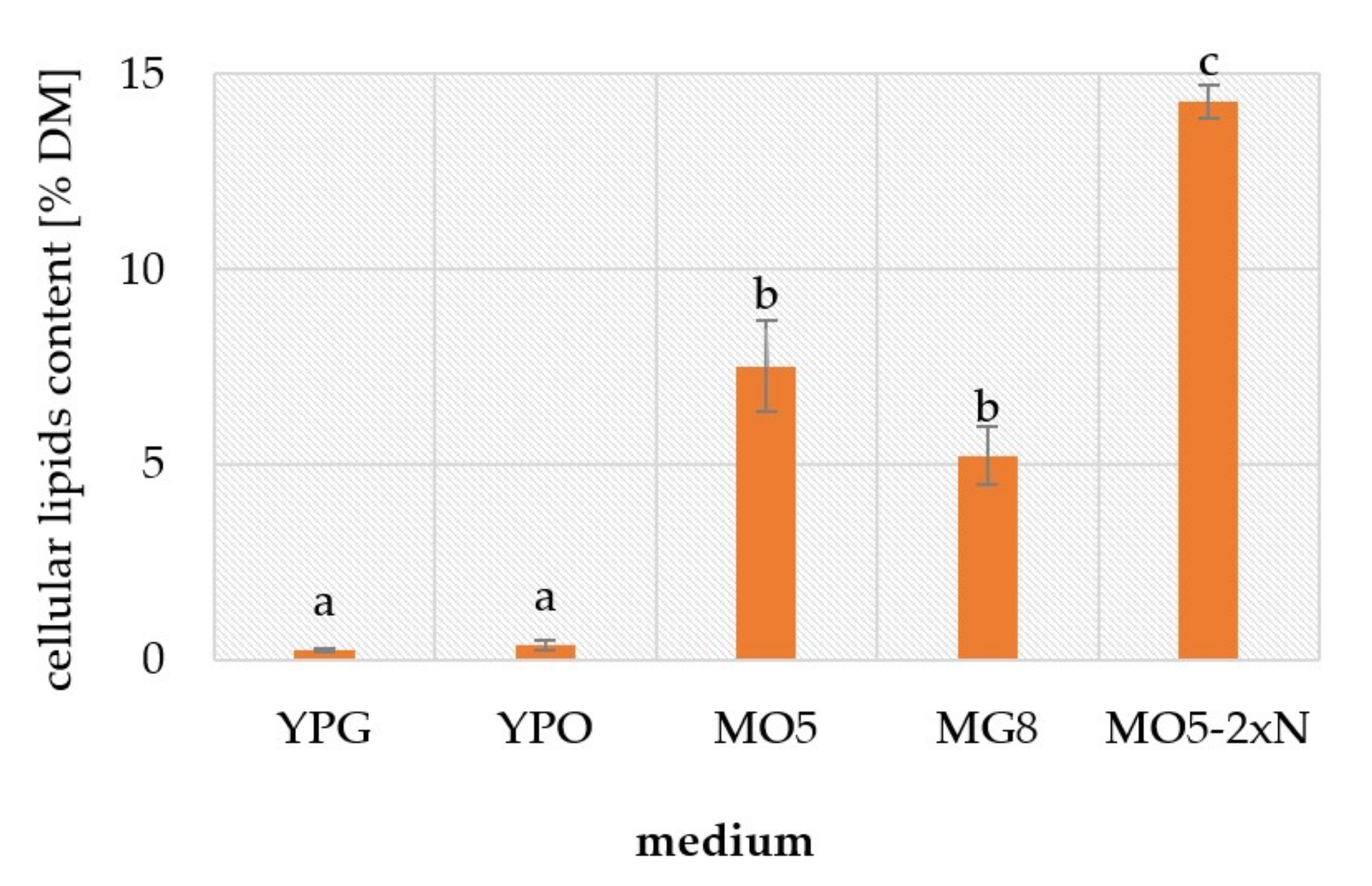
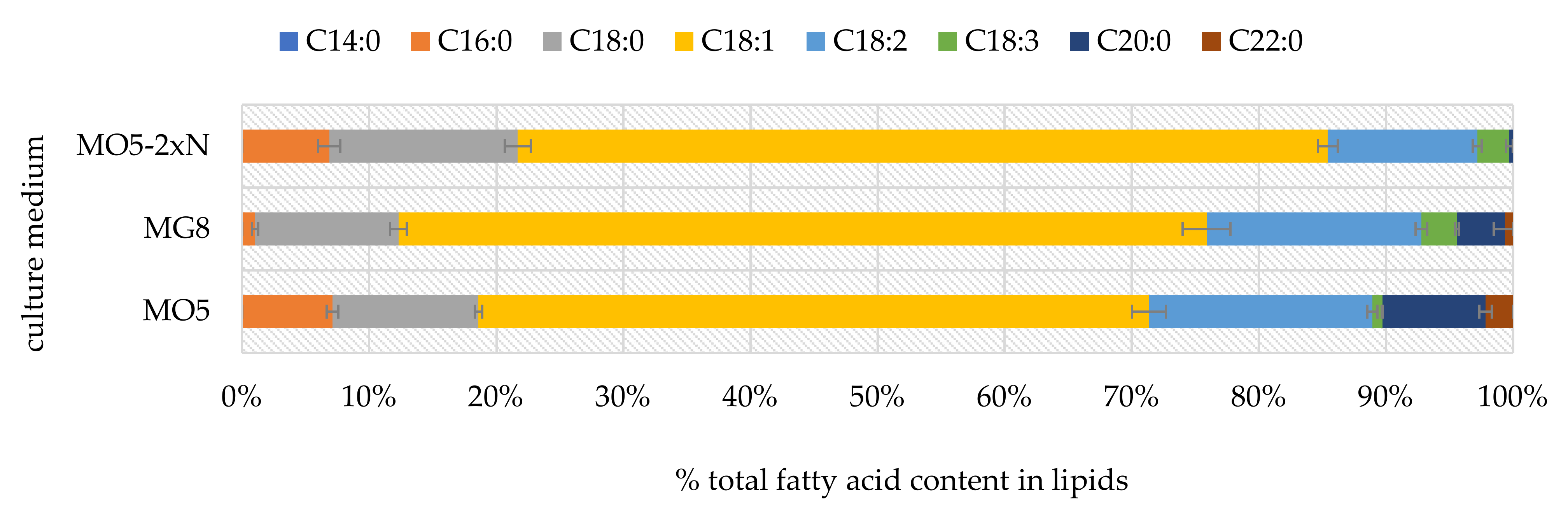
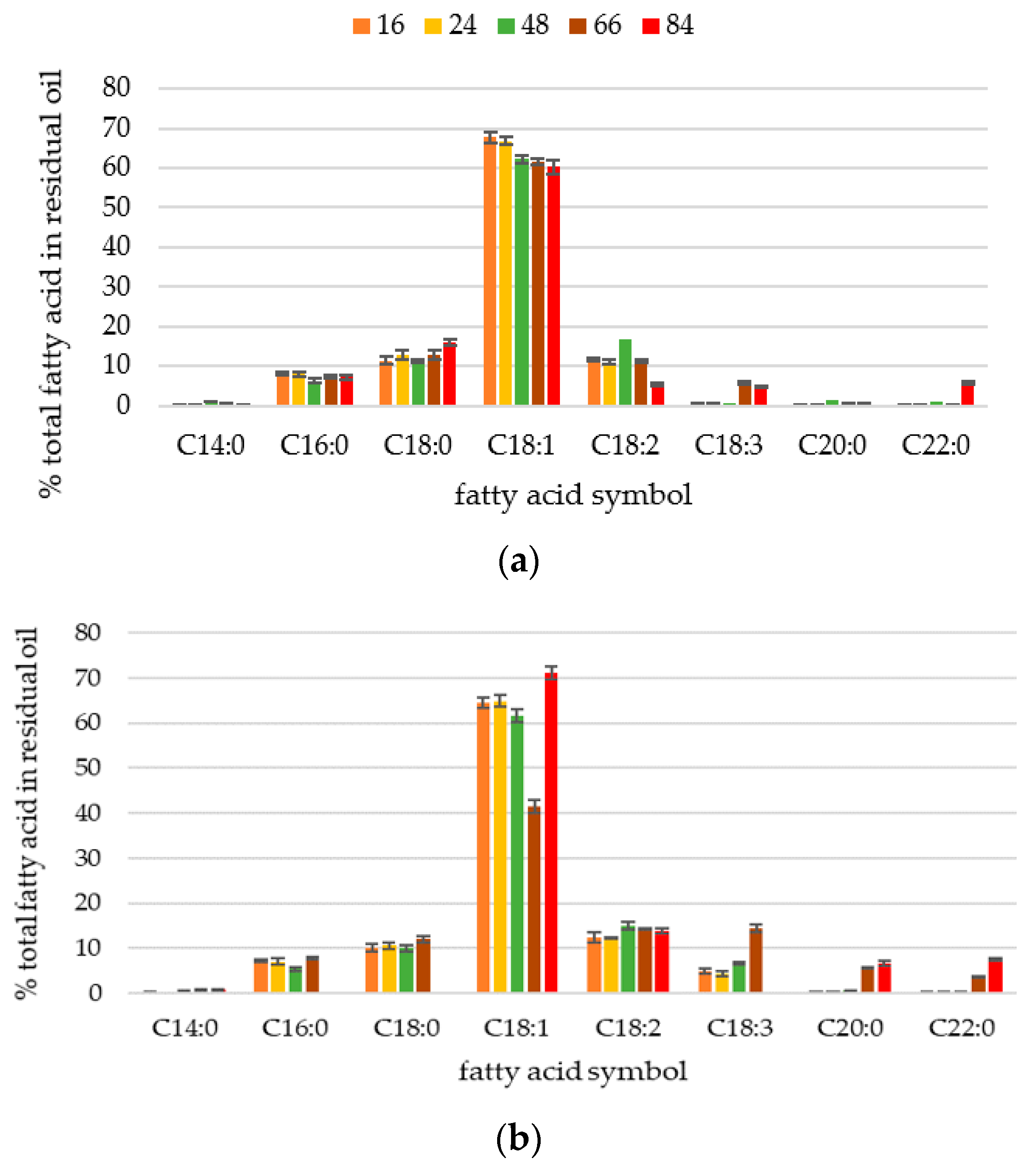
2.1. Phenotypic Observations during Batch Cultures
2.2. Transcription Levels of Selected Genes Involved in Storage Lipid Synthesis
3. Discussion
4. Materials and Methods
4.1. Yeast Strain and Culture Conditions
4.2. General Analytical Techniques
4.3. Lipase Activity Assay
4.4. Quantification of Genes Expression
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Papanikolau, S.; Aggelis, G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Czabany, T.; Athenstaedt, K.; Daum, G. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 2007, 1771, 299–309. [Google Scholar] [CrossRef]
- Fickers, P.; Marty, A.; Nicaud, J.-M. The lipases from Yarrowia lipolytica: Genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol. Adv. 2011, 29, 632–644. [Google Scholar] [CrossRef]
- Wynn, J.P.; Ratledge, C. Microbial production of oils and fats. In Food Biotechnology, Second Edition Food Science and Technology; Shetty, K., Paliyath, G., Pometto, A., Levin, R.E., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 443–472. [Google Scholar]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.L.; Molina-Jouve, C.; Nicaud, J.M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef]
- Dulermo, R.; Gamboa-Melendez, H.; Ledesma-Amaro, R.; Thevenieau, F.; Nicaud, J.-M. Unraveling fatty acid transport and activation mechanisms in Yarrowia lipolytica. Biochim. Biophys. Acta 2015, 1851, 1202–1217. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Nicaud, J.-M. Yarrowia lipolytica as a biotechnological chasis to produce usual and unusual fatty acids. Progr. Lipid Res. 2016, 61, 40–50. [Google Scholar] [CrossRef]
- Abghari, A.; Chen, S. Yarrowia lipolytica as an oleaginous cell factory platform for production of fatty acid-based biofuel and bioproducts. Frontiers Energy Res. 2014, 2, 21. [Google Scholar] [CrossRef]
- Probst, K.V.; Vadlani, P.V. Production of single cell oil from Lipomyces starkeyi ATCC 56304 using biorefinery by-products. Bioresour. Technol. 2015, 198, 268–275. [Google Scholar] [CrossRef]
- Dulermo, T.; Lazar, Z.; Dulermo, R.; Rakicka, M.; Haddouche, R.; Nicaud, J.-M. Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acids synthesis. Biochim. Biophys. Acta 2015, 1851, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Fabiszewska, A.; Misiukiewicz-Stępień, P.; Paplińska-Goryca, M.; Zieniuk, B.; Białecka-Florjańczyk, E. An Insight into Storage Lipid Synthesis by Yarrowia lipolytica Yeast Relating to Lipid and Sugar Substrates Metabolism. Biomolecules 2019, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Fabiszewska, A.U.; Zieniuk, B.; Kozłowska, M.; Mazurczak-Zieniuk, P.M.; Wołoszynowska, M.; Misiukiewicz-Stępień, P.; Nowak, D. Studies on Upgradation of Waste Fish Oil to Lipid-Rich Yeast Biomass in Yarrowia lipolytica Batch Cultures. Foods 2021, 10, 436. [Google Scholar] [CrossRef]
- Ochsenrether, K.; Gluck, C.; Stressler, T.; Fischer, L.; Syldatk, C. Production strategies and applications of microbial single cell oils. Front. Microbiol. 2016, 7, 1539–1565. [Google Scholar] [CrossRef]
- Tai, M.; Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013, 15, 1–9. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Chevalot, I.; Komaitis, M.; Aggelis, G.; Marc, I. Kinetic profile of the cellular lipid composition in an oleaginous Yarrowia lipolytica capable of producing a cocoa-butter substitute from industrial fats. Antonie Van Leeuwenhoek 2001, 80, 215–224. [Google Scholar] [CrossRef]
- Sarantou, S.; Stoforos, N.G.; Kalantzi, O.; Papanikolaou, S. Biotechnological valorization of biodiesel-derived glycerol: Trials with the non-conventional yeasts Yarrowia lipolytica and Rhodosporidium sp. Carb. Res. Convers. 2021, 4, 61–75. [Google Scholar] [CrossRef]
- Saygun, A.; Sahin-Yesilcubuk, N.; Aran, N. Effects of different oil sources and residues on biomass and metabolite production by Yarrowia lipolytica YB 423-12. J. Am. Oil Chem. Soc. 2014, 91, 1521–1530. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Muniglia, L.; Chevalot, I.; Aggelis, G.; Marc, I. Yarrowia lipolytica as a potential producer of citric acid from raw glycerol. J. Appl. Microbiol. 2002, 92, 737–744. [Google Scholar] [CrossRef]
- Pathan, E.K.; Ghormade, V.; Deshpande, M.V. Selection of reference genes for quantitative real-time RT-PCR assays in different morphological forms of dimorphic zygomycetous fungus Benjaminiella poitrasii. PLoS ONE 2017, 12, e0179454. [Google Scholar] [CrossRef]
- Rzechonek, D.A.; Neuvéglise, C.; Devillers, H.; Rymowicz, W.; Mirończuk, A.M. EUF1—A newly identified gene involved in erythritol utilization in Yarrowia lipolytica. Sci. Rep. 2017, 7, 12507. [Google Scholar] [CrossRef]
- Mansour, S.; Bailly, J.; Landaud, S.; Monnet, C.; Sarthou, A.S.; Cocaign-Bousquet, M.; Leroy, S.; Irlinger, F.; Bonnarme, P. Investigation of associations of Yarrowia lipolytica, Staphylococcus xylosus, and Lactococcus lactis in culture as a first step in microbial interaction analysis. Appl. Environ. Microbiol. 2009, 75, 6422–6430. [Google Scholar] [CrossRef] [PubMed]
- Feddersen, S.; Neergaard, T.B.; Knudsen, J.; Faergeman, N.J. Transcriptional regulation of phospholipid biosynthesis is linked to fatty acid metabolism by an acyl-CoA-binding-protein-dependent mechanism in Saccharomyces cerevisiae. Biochem. J. 2007, 407, 219–230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Papanikolaou, S.; Galiotou-Panayotou, M.; Chevalot, I.; Komaitis, M.; Marc, I.; Aggelis, G. Influence of glucose and saturated free-fatty acid mixtures on citric acid and lipid production by Yarrowia lipolytica. Curr. Microbiol. 2006, 52, 134–142. [Google Scholar] [CrossRef]
- Athenstaedt, K. YALI0E32769g (DGA1) and YALI0E16797g (LRO1) encode major triacylglycerol synthases of the oleaginous yeast Yarrowia lipolytica. Biochim. Biophys. Acta. 2011, 1811, 587–596. [Google Scholar] [CrossRef]
- Beopoulos, A.; Haddouche, R.; Kabran, P.; Dulermo, T.; Chardot, T.; Nicaud, J.-M. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 2012, 93, 1523–1537. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A.; Lewin, T.M.; Van Horn, C.G.; Gonzalez-Baró, M.R. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 2002, 132, 2123–2126. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Song, H.; Wang, Z.; Ding, Y. Expression of POX2 gene and disruption of POX3 genes in the industrial Yarrowia lipolytica on the γ-decalactone production. Microbiol. Res. 2012, 167, 246–252. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, L.; Stryer, L. Biochemistry, 5th ed.; W. H. Freeman: New York, NY, USA, 2002. [Google Scholar]
- Borkowska, M.; Białas, W.; Celińska, E. A new set of reference genes for comparative gene expression analyses in Yarrowia lipolytica. FEMS Yeast Res. 2020, 20, foaa059. [Google Scholar] [CrossRef]
- Ratledge, C.; Cohen, Z. Microbial and algal oils: Do they have a future for biodiesel or as commodity oils? Lipid Technol. 2008, 20, 155–160. [Google Scholar] [CrossRef]
- Kapturowska, A.; Stolarzewicz, I.; Krzyczkowska, J.; Białecka-Florjańczyk, E. Studies on lipolytic activity of sonicated enzymes from Yarrowia lipolytica. Ultrason. Sonochem. 2012, 19, 86–191. [Google Scholar] [CrossRef] [PubMed]
- Fojan, P.; Jonson, P.H.; Petersen, M.T.N.; Petersen, S.B. What distinguishes an esterase from a lipase: A novel structural approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef]
- Chomczyński, P.; Sacchi, N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitive PCR and the 2−ΔΔCT. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Culture Medium | Carbon Source [g/dm3] | Inorganic Nitrogen Source (NH4)2SO4 [g/dm3] | Medium Composition [g/dm3] |
|---|---|---|---|
| YPG | glucose, 20.0 | - | yeast extract, 10.0; peptone, 20.0 |
| YPO | olive oil, 20.0 | - | |
| MO5 | olive oil, 50.0 | 2.5 | KH2PO4, 7.0; Na2HPO4, 2.5; FeSO4 x H2O, 0.16; CaCl2, 0.15; MnCl2 x 4H2O, 0.08; ZnSO4, 0.02; yeast extract, 2.0; peptone, 1.0 |
| MO5x2N | olive oil, 50.0 | 5.0 | |
| MG8 | Glucose, 80.0 | 2.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabiszewska, A.; Paplińska-Goryca, M.; Misiukiewicz-Stępień, P.; Wołoszynowska, M.; Nowak, D.; Zieniuk, B. Expression Profile of Selected Genes Involved in Storage Lipid Synthesis in a Model Oleaginous Yeast Species Yarrowia lipolytica. Int. J. Mol. Sci. 2022, 23, 1041. https://doi.org/10.3390/ijms23031041
Fabiszewska A, Paplińska-Goryca M, Misiukiewicz-Stępień P, Wołoszynowska M, Nowak D, Zieniuk B. Expression Profile of Selected Genes Involved in Storage Lipid Synthesis in a Model Oleaginous Yeast Species Yarrowia lipolytica. International Journal of Molecular Sciences. 2022; 23(3):1041. https://doi.org/10.3390/ijms23031041
Chicago/Turabian StyleFabiszewska, Agata, Magdalena Paplińska-Goryca, Paulina Misiukiewicz-Stępień, Małgorzata Wołoszynowska, Dorota Nowak, and Bartłomiej Zieniuk. 2022. "Expression Profile of Selected Genes Involved in Storage Lipid Synthesis in a Model Oleaginous Yeast Species Yarrowia lipolytica" International Journal of Molecular Sciences 23, no. 3: 1041. https://doi.org/10.3390/ijms23031041
APA StyleFabiszewska, A., Paplińska-Goryca, M., Misiukiewicz-Stępień, P., Wołoszynowska, M., Nowak, D., & Zieniuk, B. (2022). Expression Profile of Selected Genes Involved in Storage Lipid Synthesis in a Model Oleaginous Yeast Species Yarrowia lipolytica. International Journal of Molecular Sciences, 23(3), 1041. https://doi.org/10.3390/ijms23031041








