Secondary Structures of the Transmembrane Domain of SARS-CoV-2 Spike Protein in Detergent Micelles
Abstract
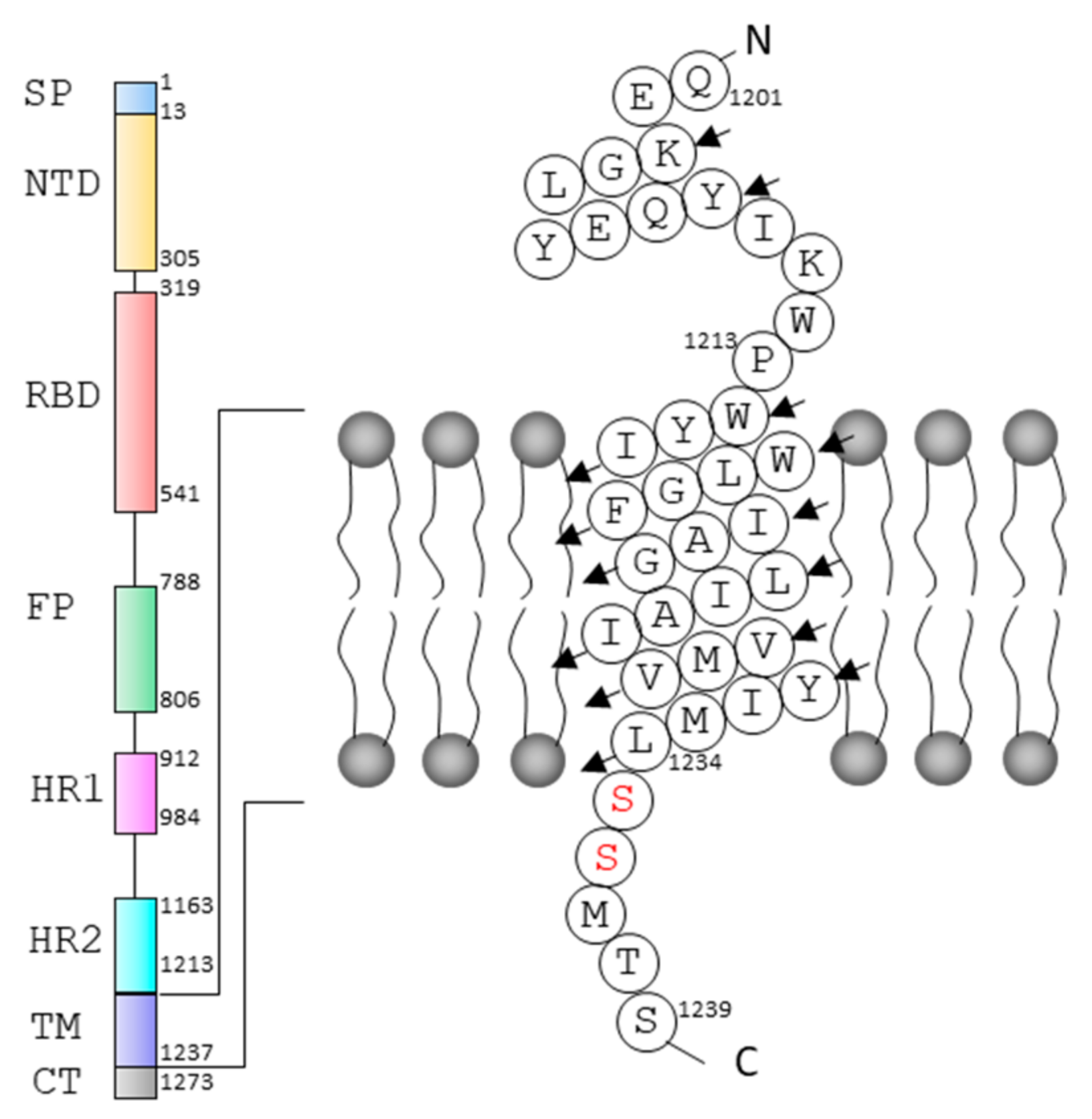
:1. Introduction
2. Results
2.1. Solution NMR Spectrum of S-TM in Micelles
2.2. Secondary Structures of S-TM
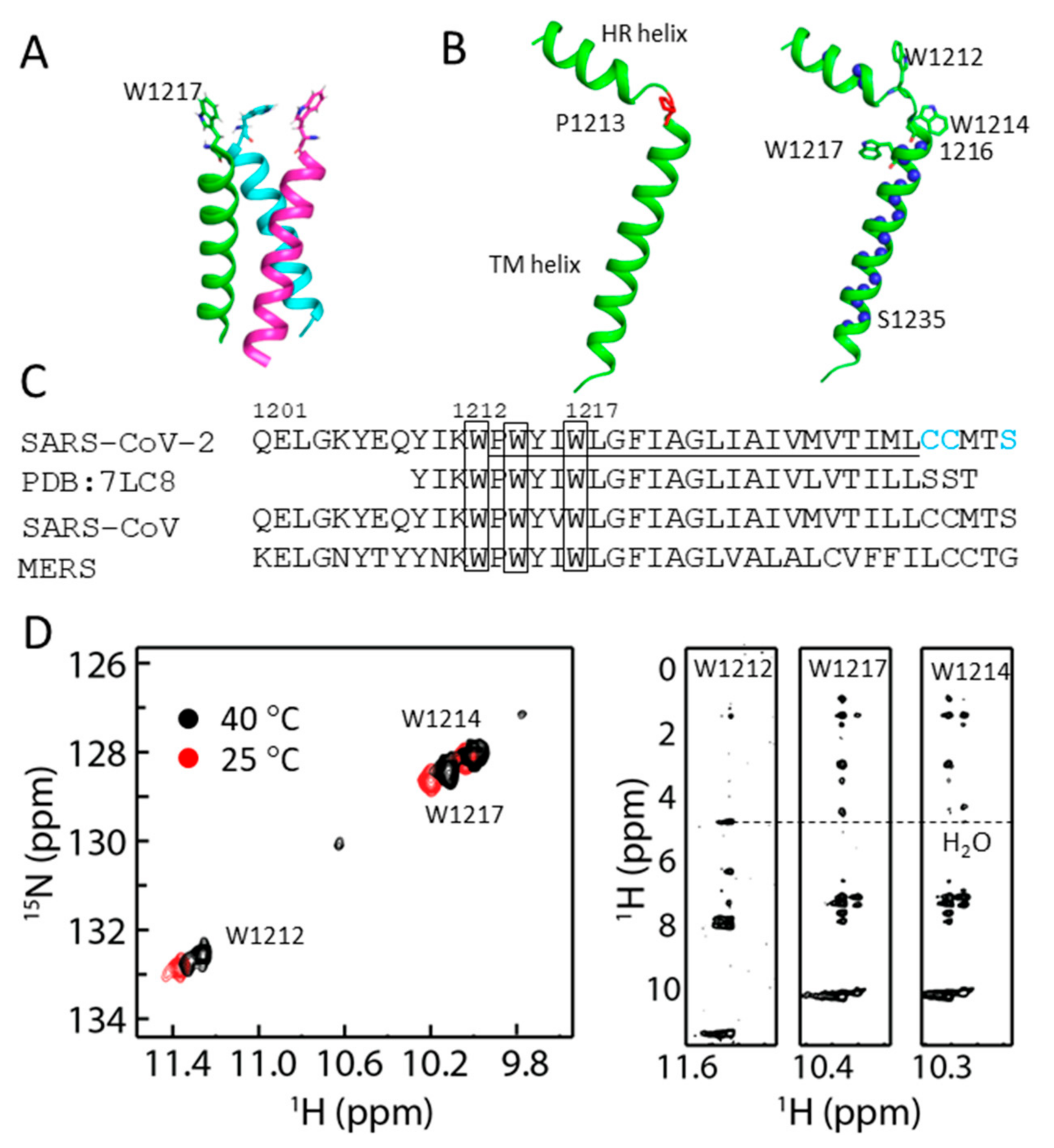
2.3. Structural Model of S-TM
2.4. Conformational Analysis of Residues in S-TM
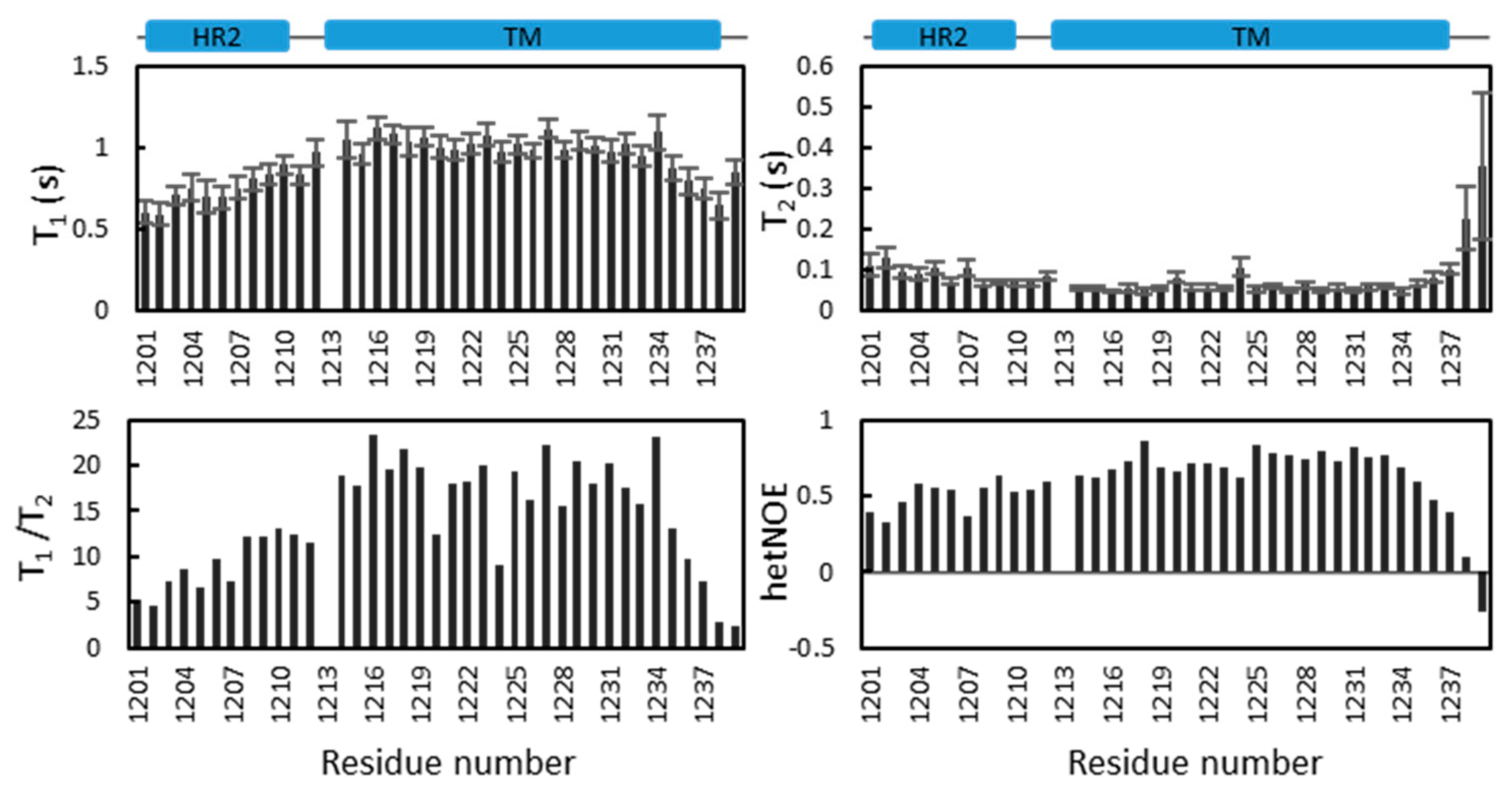
2.5. Dynamics of S-TM
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Resonance Assignment
4.3. Collection of the 1H-15N HSQC Spectrum in D2O
4.4. Relaxation Analysis
4.5. Cross-Linking Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [Green Version]
- Kaul, D. An overview of coronaviruses including the SARS-2 coronavirus—Molecular biology, epidemiology and clinical implications. Curr. Med. Res. Pract. 2020, 10, 54–64. [Google Scholar]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Lipsitch, M.; Cohen, T.; Cooper, B.; Robins, J.M.; Ma, S.; James, L.; Gopalakrishna, G.; Chew, S.K.; Tan, C.C.; Samore, M.H.; et al. Transmission dynamics and control of severe acute respiratory syndrome. Science 2003, 300, 1966–1970. [Google Scholar] [CrossRef] [Green Version]
- Sharif-Yakan, A.; Kanj, S.S. Emergence of MERS-CoV in the Middle East: Origins, transmission, treatment, and perspectives. PLoS Pathog. 2014, 10, e1004457. [Google Scholar]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar]
- Huang, Y.; Yang, C.; Xu, X.-F.; Xu, W.; Liu, S.-W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.-J.; Yu, P.-Y.; Chang, Y.-C.; Hsu, S.-T.D. D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation. J. Biol. Chem. 2021, 297, 101238. [Google Scholar]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Liu, L.; Du, L.; Zhang, C.; Jiang, S.; Li, T.; He, Y. Potent and persistent antibody responses against the receptor-binding domain of SARS-CoV spike protein in recovered patients. Virol. J. 2010, 7, 299. [Google Scholar]
- Hussain, A.; Hasan, A.; Nejadi Babadaei, M.M.; Bloukh, S.H.; Chowdhury, M.E.H.; Sharifi, M.; Haghighat, S.; Falahati, M. Targeting SARS-CoV2 spike protein receptor binding domain by therapeutic antibodies. Biomed. Pharmacother. 2020, 130, 110559. [Google Scholar]
- Lu, Y.; Neo, T.L.; Liu, D.X.; Tam, J.P. Importance of SARS-CoV spike protein Trp-rich region in viral infectivity. Biochem. Biophys. Res. Commun. 2008, 371, 356–360. [Google Scholar]
- Broer, R.; Boson, B.; Spaan, W.; Cosset, F.-L.; Corver, J. Important role for the transmembrane domain of severe acute respiratory syndrome Coronavirus spike protein during entry. J. Virol. 2006, 80, 1302–1310. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Chou, J.J. A Trimeric hydrophobic zipper mediates the intramembrane assembly of SARS-CoV-2 spike. J. Am. Chem. Soc. 2021, 143, 8543–8546. [Google Scholar]
- Van Horn, W.D.; Kim, H.-J.; Ellis, C.D.; Hadziselimovic, A.; Sulistijo, E.S.; Karra, M.D.; Tian, C.; Sönnichsen, F.D.; Sanders, C.R. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science 2009, 324, 1726–1729. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Brüschweiler, S.; Zhao, L.; Chou, J.J. Reply to ‘Concerns with yeast mitochondrial ADP/ATP carrier’s integrity in DPC’ and ‘Dynamics and interactions of AAC3 in DPC are not functionally relevant’. Nat. Struct. Mol. Biol. 2018, 25, 749–750. [Google Scholar]
- Li, Q.; Ng, H.Q.; Kang, C. Secondary structure and topology of the transmembrane domain of Syndecan-2 in detergent micelles. FEBS Lett. 2019, 593, 554–561. [Google Scholar] [CrossRef]
- Li, Y.; Lee, M.Y.; Loh, Y.R.; Kang, C. Secondary structure and membrane topology of dengue virus NS4A protein in micelles. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 442–450. [Google Scholar] [CrossRef]
- Li, Q.; Wong, Y.L.; Huang, Q.; Kang, C. Structural insight into the transmembrane domain and the juxtamembrane region of the erythropoietin receptor in micelles. Biophys. J. 2014, 107, 2325–2336. [Google Scholar] [CrossRef] [Green Version]
- Tifrea, D.F.; Pal, S.; Popot, J.-L.; Cocco, M.J.; de la Maza, L.M. Increased immunoaccessibility of MOMP epitopes in a vaccine formulated with amphipols may account for the very robust protection elicited against a vaginal challenge with Chlamydia muridarum. J. Immunol. 2014, 192, 5201–5213. [Google Scholar]
- de la Torre, J.G.; Huertas, M.; Carrasco, B. HYDRONMR: Prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000, 147, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Vinogradova, O.; Badola, P.; Czerski, L.; Sonnichsen, F.; Sanders, C. Escherichia coli diacylglycerol kinase: A case study in the application of solution NMR methods to an integral membrane protein. Biophys. J. 1997, 72, 2688–2701. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, M.; Webb, S.; Chushak, Y.; Krabacher, R.; Liu, Y.; Swami, N.; Harbaugh, S.; Chávez, J. A high-throughput pipeline for design and selection of peptides targeting the SARS-Cov-2 Spike protein. Sci. Rep. 2021, 11, 21768. [Google Scholar] [CrossRef]
- de Jesus, A.J.; Allen, T.W. The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Biochim. Biophys. Acta (BBA)-Biomembr. 2013, 1828, 864–876. [Google Scholar]
- Azad, T.; Singaravelu, R.; Crupi, M.J.F.; Jamieson, T.; Dave, J.; Brown, E.E.F.; Rezaei, R.; Taha, Z.; Boulton, S.; Martin, N.T.; et al. Implications for SARS-CoV-2 vaccine design: Fusion of Spike Glycoprotein Transmembrane Domain To Receptor-Binding Domain Induces Trimerization. Membranes 2020, 10, 215. [Google Scholar]
- Chipot, C.; Dehez, F.; Schnell, J.R.; Zitzmann, N.; Pebay-Peyroula, E.; Catoire, L.J.; Miroux, B.; Kunji, E.; Veglia, G.; Cross, T.A.; et al. Perturbations of native membrane protein structure in Alkyl Phosphocholine detergents: A critical assessment of NMR and biophysical studies. Chem. Rev. 2018, 118, 3559–3607. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Vanoye, C.G.; Welch, R.C.; Van Horn, W.D.; Sanders, C.R. Functional delivery of a membrane protein into oocyte membranes using Bicelles. Biochemistry 2010, 49, 653–655. [Google Scholar] [CrossRef] [Green Version]
- Coey, A.T.; Sahu, I.D.; Gunasekera, T.S.; Troxel, K.R.; Hawn, J.M.; Swartz, M.S.; Wickenheiser, M.R.; Reid, R.-J.; Welch, R.C.; Vanoye, C.G.; et al. Reconstitution of KCNE1 into lipid bilayers: Comparing the structural, dynamic, and activity differences in micelle and vesicle environments. Biochemistry 2011, 50, 10851–10859. [Google Scholar] [CrossRef] [Green Version]
- Yeo, K.J.; Kim, H.-Y.; Kim, Y.P.; Hwang, E.; Kim, M.H.; Cheong, C.; Choe, S.; Jeon, Y.H. Rapid exploration of the folding topology of helical membrane proteins using paramagnetic perturbation. Protein Sci. 2010, 19, 2409–2417. [Google Scholar] [CrossRef] [Green Version]
- Lakomek, N.-A.; Kaufman, J.D.; Stahl, S.J.; Wingfield, P.T. HIV-1 Envelope protein gp41: An NMR study of dodecyl phosphocholine embedded gp41 reveals a dynamic prefusion intermediate conformation. Structure 2014, 22, 1311–1321. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Wong, Y.L.; Kang, C. Solution structure of the transmembrane domain of the insulin receptor in detergent micelles. Biochim. Biophys. Acta (BBA)-Biomembr. 2014, 1838, 1313–1321. [Google Scholar] [CrossRef] [Green Version]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Johnson, B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. In Protein NMR Techniques; Springer: Berlin/Heidelberg, Germany, 2004; Volume 278, pp. 313–352. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR Structure Calculation With CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef]
- Li, Y.; Ng, H.Q.; Li, Q.; Kang, C. Structure of the Cyclic Nucleotide-Binding Homology Domain of the hERG Channel and Its Insight into Type 2 Long QT Syndrome. Sci. Rep. 2016, 6, 23712. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Wong, Y.L.; Lee, M.Y.; Li, Y.; Kang, C. Solution structure of the transmembrane domain of the mouse erythropoietin receptor in detergent micelles. Sci. Rep. 2015, 5, 13586. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Ng, H.Q.; Yoon, H.S.; Kang, C. Solution structure of the cyclic-nucleotide binding homology domain of a KCNH channel. J. Struct. Biol. 2014, 186, 68–74. [Google Scholar] [CrossRef]
- Kay, L.E.; Torchia, D.A.; Bax, A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 1989, 28, 8972–8979. [Google Scholar]
- Li, Y.; Wong, Y.L.; Lee, M.Y.; Li, Q.; Wang, Q.Y.; Lescar, J.; Shi, P.Y.; Kang, C. Secondary Structure and membrane topology of the full-length dengue virus NS4B in micelles. Angew. Chem. Int. Ed. Engl. 2016, 55, 12068–12072. [Google Scholar]
- Li, Y.; Li, Q.; Wong, Y.L.; Liew, L.S.Y.; Kang, C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 2244–2252. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kim, Y.M.; Zou, J.; Wang, Q.Y.; Gayen, S.; Wong, Y.L.; Lee le, T.; Xie, X.; Huang, Q.; Lescar, J.; et al. Secondary structure and membrane topology of dengue virus NS4B N-terminal 125 amino acids. Biochim. Biophys Acta 2015, 1848, 3150–3157. [Google Scholar]
- Gayen, S.; Li, Q.; Chen, A.S.; Nguyen, T.H.T.; Huang, Q.; Hill, J.; Kang, C. An NMR study of the N-terminal domain of wild-type hERG and a T65P trafficking deficient hERG mutant. Proteins Struct. Funct. Bioinform. 2011, 79, 2557–2565. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Huang, Q.; Kang, C. Secondary Structures of the Transmembrane Domain of SARS-CoV-2 Spike Protein in Detergent Micelles. Int. J. Mol. Sci. 2022, 23, 1040. https://doi.org/10.3390/ijms23031040
Li Q, Huang Q, Kang C. Secondary Structures of the Transmembrane Domain of SARS-CoV-2 Spike Protein in Detergent Micelles. International Journal of Molecular Sciences. 2022; 23(3):1040. https://doi.org/10.3390/ijms23031040
Chicago/Turabian StyleLi, Qingxin, Qiwei Huang, and Congbao Kang. 2022. "Secondary Structures of the Transmembrane Domain of SARS-CoV-2 Spike Protein in Detergent Micelles" International Journal of Molecular Sciences 23, no. 3: 1040. https://doi.org/10.3390/ijms23031040
APA StyleLi, Q., Huang, Q., & Kang, C. (2022). Secondary Structures of the Transmembrane Domain of SARS-CoV-2 Spike Protein in Detergent Micelles. International Journal of Molecular Sciences, 23(3), 1040. https://doi.org/10.3390/ijms23031040





