Role of the Intermediate Filament Protein Peripherin in Health and Disease
Abstract
:1. Introduction
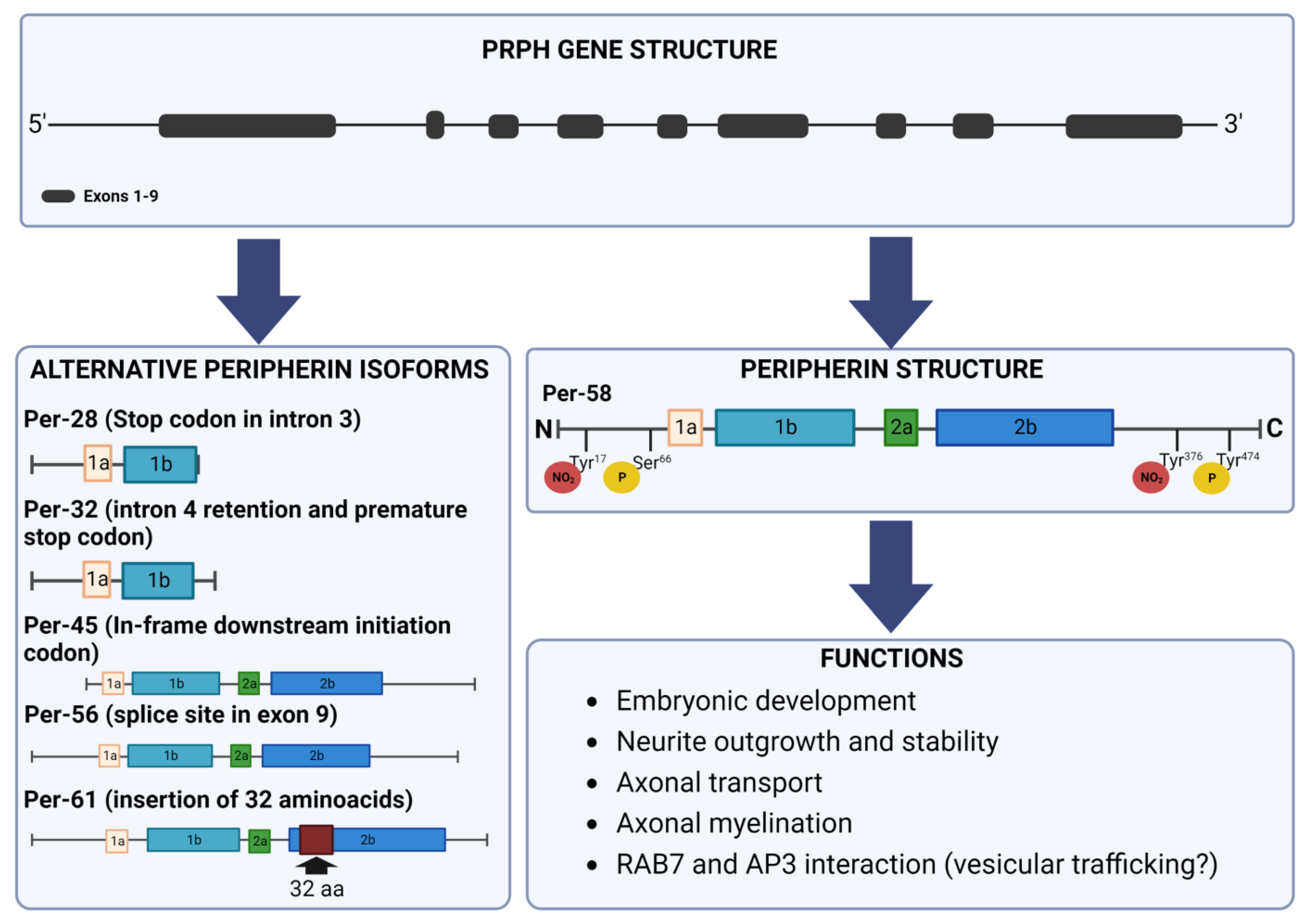
2. Peripherin Gene
3. Peripherin Expression
4. Peripherin Isoforms
5. Post-Translational Modifications
6. Peripherin Functions
7. Peripherin as a Marker of Different Neuronal Populations
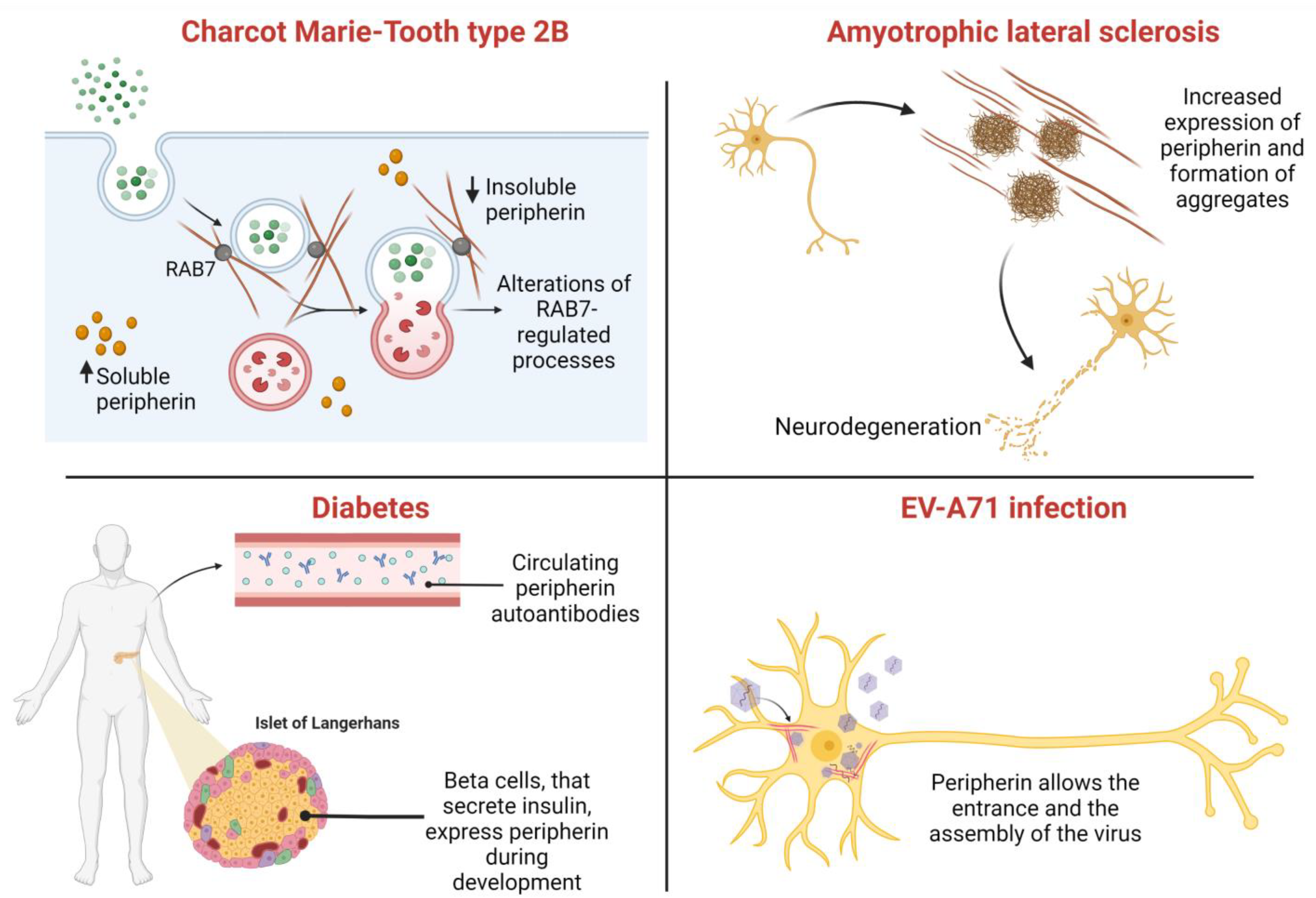
8. Role of Peripherin in Diseases
8.1. Amyotrophic Lateral Sclerosis (ALS)
8.2. Charcot–Marie–Tooth Type 2B
8.3. Other Disorders
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hohmann, T.; Dehghani, F. The Cytoskeleton—A Complex Interacting Meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margiotta, A.; Bucci, C. Role of Intermediate Filaments in Vesicular Traffic. Cells 2016, 5, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, E.; Weber, K. Intermediate filaments: Structure, dynamics, function, and disease. Annu. Rev. Biochem. 1994, 63, 345–382. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Strelkov, S.V.; Burkhard, P.; Aebi, U. Intermediate filaments: Primary determinants of cell architecture and plasticity. J. Clin. Investig. 2009, 119, 1772–1783. [Google Scholar] [CrossRef] [Green Version]
- Parry, D.A.; Strelkov, S.V.; Burkhard, P.; Aebi, U.; Herrmann, H. Towards a molecular description of intermediate filament structure and assembly. Exp. Cell Res. 2007, 313, 2204–2216. [Google Scholar] [CrossRef]
- Geisler, N.; Weber, K. The amino acid sequence of chicken muscle desmin provides a common structural model for intermediate filaments proteins. EMBO J. 1982, 1, 1649–1656. [Google Scholar] [CrossRef]
- Quax-Jeuken, Y.E.; Quax, W.J.; Bloemendal, H. Primary and secondary structure of hamster vimentin predicted from the nucleotide sequence. Proc. Natl. Acad. Sci. USA 1983, 80, 3548–3552. [Google Scholar] [CrossRef] [Green Version]
- Kornreich, M.; Avinery, R.; Malka-Gibor, E.; Laser-Azogui, A.; Beck, R. Order and disorder in intermediate filament proteins. FEBS Lett. 2015, 589, 2464–2476. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.A.; Strelkov, S.V.; Burkhard, P.; Aebi, U.; Parry, D.A. Sequence comparisons of intermediate filament chains: Evidence of a unique functional/structural role for coiled-coil segment 1A and linker L1. J. Struct. Biol. 2002, 137, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, A.V.; Kreplak, L.; Wedig, T.; Mucke, N.; Svergun, D.I.; Herrmann, H.; Aebi, U.; Strelkov, S.V. Monitoring intermediate filament assembly by small-angle x-ray scattering reveals the molecular architecture of assembly intermediates. Proc. Natl. Acad. Sci. USA 2006, 103, 16206–16211. [Google Scholar] [CrossRef]
- Herrmann, H.; Haner, M.; Brettel, M.; Ku, N.O.; Aebi, U. Characterization of distinct early assembly units of different intermediate filament proteins. J. Mol. Biol. 1999, 286, 1403–1420. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Kreplak, L.; Aebi, U. Isolation, characterization, and in vitro assembly of intermediate filaments. Methods Cell Biol. 2004, 78, 3–24. [Google Scholar] [CrossRef]
- Thompson, M.A.; Ziff, E.B. Structure of the gene encoding peripherin, an NGF-regulated neuronal-specific type III intermediate filament protein. Neuron 1989, 2, 1043–1053. [Google Scholar] [CrossRef]
- Karpov, V.; Landon, F.; Djabali, K.; Gros, F.; Portier, M.M. Structure of the mouse gene encoding peripherin: A neuronal intermediate filament protein. Biol. Cell 1992, 76, 43–48. [Google Scholar] [CrossRef]
- Foley, J.; Ley, C.A.; Parysek, L.M. The structure of the human peripherin gene (PRPH) and identification of potential regulatory elements. Genomics 1994, 22, 456–461. [Google Scholar] [CrossRef]
- Desmarais, D.; Filion, M.; Lapointe, L.; Royal, A. Cell-specific transcription of the peripherin gene in neuronal cell lines involves a cis-acting element surrounding the TATA box. EMBO J. 1992, 11, 2971–2980. [Google Scholar] [CrossRef]
- Imagawa, M.; Chiu, R.; Karin, M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: Protein kinase C and cAMP. Cell 1987, 51, 251–260. [Google Scholar] [CrossRef]
- Portier, M.M.; de Nechaud, B.; Gros, F. Peripherin, a new member of the intermediate filament protein family. Dev. Neurosci. 1983, 6, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Portier, M.M.; Brachet, P.; Croizat, B.; Gros, F. Regulation of peripherin in mouse neuroblastoma and rat PC 12 pheochromocytoma cell lines. Dev. Neurosci. 1983, 6, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Parysek, L.M.; Goldman, R.D. Distribution of a novel 57 kDa intermediate filament (IF) protein in the nervous system. J. Neurosci. 1988, 8, 555–563. [Google Scholar] [CrossRef]
- Leonard, D.G.; Gorham, J.D.; Cole, P.; Greene, L.A.; Ziff, E.B. A nerve growth factor-regulated messenger RNA encodes a new intermediate filament protein. J. Cell Biol. 1988, 106, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Escurat, M.; Djabali, K.; Gumpel, M.; Gros, F.; Portier, M.M. Differential expression of two neuronal intermediate-filament proteins, peripherin and the low-molecular-mass neurofilament protein (NF-L), during the development of the rat. J. Neurosci. 1990, 10, 764–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, D.G.; Ziff, E.B.; Greene, L.A. Identification and characterization of mRNAs regulated by nerve growth factor in PC12 cells. Mol. Cell. Biol. 1987, 7, 3156–3167. [Google Scholar] [CrossRef] [PubMed]
- Parysek, L.M.; Goldman, R.D. Characterization of intermediate filaments in PC12 cells. J. Neurosci. 1987, 7, 781–791. [Google Scholar] [CrossRef]
- Lecomte, M.J.; Basseville, M.; Landon, F.; Karpov, V.; Fauquet, M. Transcriptional activation of the mouse peripherin gene by leukemia inhibitory factor: Involvement of STAT proteins. J. Neurochem. 1998, 70, 971–982. [Google Scholar] [PubMed]
- Sterneck, E.; Kaplan, D.R.; Johnson, P.F. Interleukin-6 induces expression of peripherin and cooperates with Trk receptor signaling to promote neuronal differentiation in PC12 cells. J. Neurochem. 1996, 67, 1365–1374. [Google Scholar] [CrossRef]
- Choi, D.Y.; Toledo-Aral, J.J.; Lin, H.Y.; Ischenko, I.; Medina, L.; Safo, P.; Mandel, G.; Levinson, S.R.; Halegoua, S.; Hayman, M.J. Fibroblast growth factor receptor 3 induces gene expression primarily through Ras-independent signal transduction pathways. J. Biol. Chem. 2001, 276, 5116–5122. [Google Scholar] [CrossRef] [Green Version]
- Yuan, A.; Sasaki, T.; Kumar, A.; Peterhoff, C.M.; Rao, M.V.; Liem, R.K.; Julien, J.P.; Nixon, R.A. Peripherin is a subunit of peripheral nerve neurofilaments: Implications for differential vulnerability of CNS and peripheral nervous system axons. J. Neurosci. 2012, 32, 8501–8508. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Liem, R.K. alpha-Internexin and Peripherin: Expression, Assembly, Functions, and Roles in Disease. Methods Enzymol. 2016, 568, 477–507. [Google Scholar] [CrossRef]
- Izmiryan, A.; Li, Z.; Nothias, F.; Eyer, J.; Paulin, D.; Soares, S.; Xue, Z. Inactivation of vimentin in satellite glial cells affects dorsal root ganglion intermediate filament expression and neuronal axon growth in vitro. Mol. Cell. Neurosci. 2021, 115, 103659. [Google Scholar] [CrossRef]
- Clarke, W.T.; Edwards, B.; McCullagh, K.J.; Kemp, M.W.; Moorwood, C.; Sherman, D.L.; Burgess, M.; Davies, K.E. Syncoilin modulates peripherin filament networks and is necessary for large-calibre motor neurons. J. Cell Sci. 2010, 123, 2543–2552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, N.; Desmarais, D.; Royal, A. Transcriptional activation of the neuronal peripherin-encoding gene depends on a G + C-rich element that binds Sp1 in vitro and in vivo. Gene 1995, 159, 159–165. [Google Scholar] [CrossRef]
- Chang, L.; Thompson, M.A. Activity of the distal positive element of the peripherin gene is dependent on proteins binding to an Ets-like recognition site and a novel inverted repeat site. J. Biol. Chem. 1996, 271, 6467–6475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uveges, T.E.; Shan, Y.; Kramer, B.E.; Wight, D.C.; Parysek, L.M. Intron 1 is required for cell type-specific, but not injury-responsive, peripherin gene expression. J. Neurosci. 2002, 22, 7959–7967. [Google Scholar] [CrossRef] [Green Version]
- Karpov, V.; Fauquet, M.; Lecomte, M.J.; Portier, M.M. Three different states of the chromatin structure of the mouse peripherin gene. J. Neurosci. Res. 1996, 44, 115–123. [Google Scholar] [CrossRef]
- Gross, D.S.; Garrard, W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988, 57, 159–197. [Google Scholar] [CrossRef]
- Drapkin, R.; Merino, A.; Reinberg, D. Regulation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 1993, 5, 469–476. [Google Scholar] [CrossRef]
- Escurat, M.; Djabali, K.; Huc, C.; Landon, F.; Becourt, C.; Boitard, C.; Gros, F.; Portier, M.M. Origin of the beta cells of the islets of Langerhans is further questioned by the expression of neuronal intermediate filament proteins, peripherin and NF-L, in the rat insulinoma RIN5F cell line. Dev. Neurosci. 1991, 13, 424–432. [Google Scholar] [CrossRef]
- Landon, F.; Lemonnier, M.; Benarous, R.; Huc, C.; Fiszman, M.; Gros, F.; Portier, M.M. Multiple mRNAs encode peripherin, a neuronal intermediate filament protein. EMBO J. 1989, 8, 1719–1726. [Google Scholar] [CrossRef]
- McLean, J.; Xiao, S.; Miyazaki, K.; Robertson, J. A novel peripherin isoform generated by alternative translation is required for normal filament network formation. J. Neurochem. 2008, 104, 1663–1673. [Google Scholar] [CrossRef]
- Robertson, J.; Doroudchi, M.M.; Nguyen, M.D.; Durham, H.D.; Strong, M.J.; Shaw, G.; Julien, J.P.; Mushynski, W.E. A neurotoxic peripherin splice variant in a mouse model of ALS. J. Cell Biol. 2003, 160, 939–949. [Google Scholar] [CrossRef] [Green Version]
- Xiao, S.; Tjostheim, S.; Sanelli, T.; McLean, J.R.; Horne, P.; Fan, Y.; Ravits, J.; Strong, M.J.; Robertson, J. An aggregate-inducing peripherin isoform generated through intron retention is upregulated in amyotrophic lateral sclerosis and associated with disease pathology. J. Neurosci. 2008, 28, 1833–1840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huc, C.; Escurat, M.; Djabali, K.; Derer, M.; Landon, F.; Gros, F.; Portier, M.M. Phosphorylation of peripherin, an intermediate filament protein, in mouse neuroblastoma NIE 115 cell line and in sympathetic neurons. Biochem. Biophys. Res. Commun. 1989, 160, 772–779. [Google Scholar] [CrossRef]
- Aletta, J.M.; Shelanski, M.L.; Greene, L.A. Phosphorylation of the peripherin 58-kDa neuronal intermediate filament protein. Regulation by nerve growth factor and other agents. J. Biol. Chem. 1989, 264, 4619–4627. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Namikawa, K.; Shikata, K.; Kobatake, Y.; Tachibana, T.; Kiyama, H. Identification of peripherin as a Akt substrate in neuron. J. Biol. Chem. 2007, 282, 23491–23499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelastro, J.M.; Ho, C.L.; Frappier, T.; Liem, R.K.; Greene, L.A. Peripherin is tyrosine-phosphorylated at its carboxyl-terminal tyrosine. J. Neurochem. 1998, 70, 540–549. [Google Scholar] [CrossRef] [PubMed]
- MacTaggart, B.; Kashina, A. Posttranslational modifications of the cytoskeleton. Cytoskeleton 2021, 78, 142–173. [Google Scholar] [CrossRef]
- Tedeschi, G.; Cappelletti, G.; Nonnis, S.; Taverna, F.; Negri, A.; Ronchi, C.; Ronchi, S. Tyrosine nitration is a novel post-translational modification occurring on the neural intermediate filament protein peripherin. Neurochem. Res. 2007, 32, 433–441. [Google Scholar] [CrossRef]
- Cappelletti, G.; Maggioni, M.G.; Ronchi, C.; Maci, R.; Tedeschi, G. Protein tyrosine nitration is associated with cold- and drug-resistant microtubules in neuronal-like PC12 cells. Neurosci. Lett. 2006, 401, 159–164. [Google Scholar] [CrossRef]
- Petzold, A. The 2022 Lady Estelle Wolfson lectureship on neurofilaments. J. Neurochem. 2022, 163, 179–219. [Google Scholar] [CrossRef]
- Revill, K.; Wang, T.; Lachenmayer, A.; Kojima, K.; Harrington, A.; Li, J.; Hoshida, Y.; Llovet, J.M.; Powers, S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 2013, 145, 1424–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Vlag, J.; Otte, A.P. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 1999, 23, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Lee, Y.B.; Cho, E.J.; Lee, J.H.; Yu, S.J.; Kim, Y.J.; Yoon, J.H. CKD-5, a novel pan-histone deacetylase inhibitor, synergistically enhances the efficacy of sorafenib for hepatocellular carcinoma. BMC Cancer 2020, 20, 1001. [Google Scholar] [CrossRef] [PubMed]
- Gorham, J.D.; Baker, H.; Kegler, D.; Ziff, E.B. The expression of the neuronal intermediate filament protein peripherin in the rat embryo. Brain Res. Dev. Brain Res. 1990, 57, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Troy, C.M.; Brown, K.; Greene, L.A.; Shelanski, M.L. Ontogeny of the neuronal intermediate filament protein, peripherin, in the mouse embryo. Neuroscience 1990, 36, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Undamatla, J.; Szaro, B.G. Differential expression and localization of neuronal intermediate filament proteins within newly developing neurites in dissociated cultures of Xenopus laevis embryonic spinal cord. Cell Motil. Cytoskelet. 2001, 49, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.M.; Kriz, J.; Julien, J.P. Induction of peripherin expression in subsets of brain neurons after lesion injury or cerebral ischemia. Brain Res. 2002, 946, 153–161. [Google Scholar] [CrossRef]
- Oblinger, M.M.; Wong, J.; Parysek, L.M. Axotomy-induced changes in the expression of a type III neuronal intermediate filament gene. J. Neurosci. 1989, 9, 3766–3775. [Google Scholar] [CrossRef] [Green Version]
- Troy, C.M.; Muma, N.A.; Greene, L.A.; Price, D.L.; Shelanski, M.L. Regulation of peripherin and neurofilament expression in regenerating rat motor neurons. Brain Res. 1990, 529, 232–238. [Google Scholar] [CrossRef]
- Wong, J.; Oblinger, M.M. Differential regulation of peripherin and neurofilament gene expression in regenerating rat DRG neurons. J. Neurosci. Res. 1990, 27, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Helfand, B.T.; Mendez, M.G.; Pugh, J.; Delsert, C.; Goldman, R.D. A role for intermediate filaments in determining and maintaining the shape of nerve cells. Mol. Biol. Cell 2003, 14, 5069–5081. [Google Scholar] [CrossRef] [PubMed]
- Lariviere, R.C.; Nguyen, M.D.; Ribeiro-Da-Silva, A.; Julien, J.P. Reduced number of unmyelinated sensory axons in peripherin null mice. J. Neurochem. 2002, 81, 525–532. [Google Scholar] [CrossRef]
- Huang, L.C.; Thorne, P.R.; Housley, G.D.; Montgomery, J.M. Spatiotemporal definition of neurite outgrowth, refinement and retraction in the developing mouse cochlea. Development 2007, 134, 2925–2933. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.C.; Barclay, M.; Lee, K.; Peter, S.; Housley, G.D.; Thorne, P.R.; Montgomery, J.M. Synaptic profiles during neurite extension, refinement and retraction in the developing cochlea. Neural Dev. 2012, 7, 38. [Google Scholar] [CrossRef] [Green Version]
- Elliott, K.L.; Kersigo, J.; Lee, J.H.; Jahan, I.; Pavlinkova, G.; Fritzsch, B.; Yamoah, E.N. Developmental Changes in Peripherin-eGFP Expression in Spiral Ganglion Neurons. Front. Cell. Neurosci. 2021, 15, 678113. [Google Scholar] [CrossRef]
- Cederholm, J.M.E.; Parley, K.E.; Perera, C.J.; von Jonquieres, G.; Pinyon, J.L.; Julien, J.P.; Ryugo, D.K.; Ryan, A.F.; Housley, G.D. Noise-induced hearing loss vulnerability in type III intermediate filament peripherin gene knockout mice. Front. Neurol. 2022, 13, 962227. [Google Scholar] [CrossRef]
- Kil, H.K.; Kim, K.W.; Lee, D.H.; Lee, S.M.; Lee, C.H.; Kim, S.Y. Changes in the Gene Expression Profiles of the Inferior Colliculus Following Unilateral Cochlear Ablation in Adult Rats. Biochem. Genet. 2021, 59, 731–750. [Google Scholar] [CrossRef]
- Bucci, C.; Frunzio, R.; Chiariotti, L.; Brown, A.L.; Rechler, M.M.; Bruni, C.B. A new member of the ras gene superfamily identified in a rat liver cell line. Nucleic Acids Res. 1988, 16, 9979–9993. [Google Scholar] [CrossRef] [Green Version]
- Bucci, C.; Thomsen, P.; Nicoziani, P.; McCarthy, J.; van Deurs, B. Rab7: A key to lysosome biogenesis. Mol. Biol. Cell 2000, 11, 467–480. [Google Scholar] [CrossRef]
- Harrison, R.E.; Bucci, C.; Vieira, O.V.; Schroer, T.A.; Grinstein, S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: Role of Rab7 and RILP. Mol. Cell. Biol. 2003, 23, 6494–6506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jager, S.; Bucci, C.; Tanida, I.; Ueno, T.; Kominami, E.; Saftig, P.; Eskelinen, E.L. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004, 117, 4837–4848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cogli, L.; Progida, C.; Thomas, C.L.; Spencer-Dene, B.; Donno, C.; Schiavo, G.; Bucci, C. Charcot-Marie-Tooth type 2B disease-causing RAB7A mutant proteins show altered interaction with the neuronal intermediate filament peripherin. Acta Neuropathol. 2013, 125, 257–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Styers, M.L.; Salazar, G.; Love, R.; Peden, A.A.; Kowalczyk, A.P.; Faundez, V. The endo-lysosomal sorting machinery interacts with the intermediate filament cytoskeleton. Mol. Biol. Cell 2004, 15, 5369–5382. [Google Scholar] [CrossRef] [Green Version]
- Perrot, R.; Julien, J.P. Real-time imaging reveals defects of fast axonal transport induced by disorganization of intermediate filaments. FASEB J. 2009, 23, 3213–3225. [Google Scholar] [CrossRef]
- Gentil, B.J.; McLean, J.R.; Xiao, S.; Zhao, B.; Durham, H.D.; Robertson, J. A two-hybrid screen identifies an unconventional role for the intermediate filament peripherin in regulating the subcellular distribution of the SNAP25-interacting protein, SIP30. J. Neurochem. 2014, 131, 588–601. [Google Scholar] [CrossRef]
- Hazell, A.S.; Wang, D. Identification of complexin II in astrocytes: A possible regulator of glutamate release in these cells. Biochem. Biophys. Res. Commun. 2011, 404, 228–232. [Google Scholar] [CrossRef]
- Ilardi, J.M.; Mochida, S.; Sheng, Z.H. Snapin: A SNARE-associated protein implicated in synaptic transmission. Nat. Neurosci. 1999, 2, 119–124. [Google Scholar] [CrossRef]
- Rayala, S.K.; Hollander, P.; Balasenthil, S.; Molli, P.R.; Bean, A.J.; Vadlamudi, R.K.; Wang, R.A.; Kumar, R. Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) interacts with PELP1 and activates MAPK. J. Biol. Chem. 2006, 281, 4395–4403. [Google Scholar] [CrossRef] [Green Version]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschella, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef]
- Kumar, V.; Kundu, S.; Singh, A.; Singh, S. Understanding the Role of Histone Deacetylase and their Inhibitors in Neurodegenerative Disorders: Current Targets and Future Perspective. Curr. Neuropharmacol. 2022, 20, 158–178. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhao, Q.; Chen, L.; Luo, Y.; Shen, L.; Cao, Z.; Wang, Q. UBR3 promotes inflammation and apoptosis via DUSP1/p38 pathway in the nucleus pulposus cells of patients with intervertebral disc degeneration. Hum. Cell 2022, 35, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Sidarala, V.; Zhu, J.; Levi-D’Ancona, E.; Pearson, G.L.; Reck, E.C.; Walker, E.M.; Kaufman, B.A.; Soleimanpour, S.A. Mitofusin 1 and 2 regulation of mitochondrial DNA content is a critical determinant of glucose homeostasis. Nat. Commun. 2022, 13, 2340. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.B.; Kang, K.W.; Sharma, K.; Yi, E. Distribution of Kv3 Subunits in Cochlear Afferent and Efferent Nerve Fibers Implies Distinct Role in Auditory Processing. Exp. Neurobiol. 2020, 29, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Grasset, E.; Puel, A.; Charpentier, J.; Klopp, P.; Christensen, J.E.; Lelouvier, B.; Servant, F.; Blasco-Baque, V.; Terce, F.; Burcelin, R. Gut microbiota dysbiosis of type 2 diabetic mice impairs the intestinal daily rhythms of GLP-1 sensitivity. Acta Diabetol. 2022, 59, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, K.M.; Longacre, T.A. Utility of Peripherin Versus MAP-2 and Calretinin in the Evaluation of Hirschsprung Disease. Appl. Immunohistochem. Mol. Morphol. 2016, 24, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Galazka, P.; Szylberg, L.; Bodnar, M.; Styczynski, J.; Marszalek, A. Diagnostic Algorithm in Hirschsprung’s Disease: Focus on Immunohistochemistry Markers. In Vivo 2020, 34, 1355–1359. [Google Scholar] [CrossRef]
- Kudo, M.; Wupuer, S.; Kubota, S.; Seki, K. Distribution of Large and Small Dorsal Root Ganglion Neurons in Common Marmosets. Front. Syst. Neurosci. 2021, 15, 801492. [Google Scholar] [CrossRef]
- Kim, J.H.; Yun, J.H.; Song, E.S.; Kim, S.U.; Lee, H.J.; Song, Y.S. Improvement of damaged cavernosa followed by neuron-like differentiation at injured cavernous nerve after transplantation of stem cells seeded on the PLA nanofiber in rats with cavernous nerve injury. Mol. Biol. Rep. 2021, 48, 3549–3559. [Google Scholar] [CrossRef]
- Farnoosh, G.; Mahmoudian-Sani, M.R. Effects of Growth Factors and the MicroRNA-183 Family on Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells Towards Auditory Neuron-Like Cells. Stem Cells Cloning 2020, 13, 79–89. [Google Scholar] [CrossRef]
- Mehri Ghahfarrokhi, A.; Jami, M.S.; Hashemzadeh Chaleshtori, M. Upregulation of Neuroprogenitor and Neural Markers via Enforced miR-124 and Growth Factor Treatment. Int. J. Mol. Cell. Med. 2020, 9, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Mehri-Ghahfarrokhi, A.; Pourteymourfard-Tabrizi, Z.; Farrokhi, E.; Chaleshtori, M.H.; Jami, M.S. Increased levels of miR-124 in human dental pulp stem cells alter the expression of neural markers. J. Otol. 2019, 14, 121–127. [Google Scholar] [CrossRef]
- Feng, Z.; Li, W.; Xia, Y.; Yu, H.; Li, H.; Li, K.; Mu, Y. The Peripherin Gene Regulates the Migration of Bone Marrow Mesenchymal Stem Cells in Wuzhishan Mini Pigs. Stem Cells Int. 2020, 2020, 8856388. [Google Scholar] [CrossRef] [PubMed]
- Winbo, A.; Ramanan, S.; Eugster, E.; Jovinge, S.; Skinner, J.R.; Montgomery, J.M. Functional coculture of sympathetic neurons and cardiomyocytes derived from human-induced pluripotent stem cells. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H927–H937. [Google Scholar] [CrossRef] [PubMed]
- Bleck, D.; Erdene-Byambadoo, L.; Brinks, R.; Schneider, M.; Pongratz, G. Semi-automated Model to Accurately Counting Sympathetic Nervous Fibers. Bio-Protocol 2019, 9, e3454. [Google Scholar] [CrossRef] [PubMed]
- Bleck, D.; Ma, L.; Erdene-Bymbadoo, L.; Brinks, R.; Schneider, M.; Tian, L.; Pongratz, G. Introduction and validation of a new semi-automated method to determine sympathetic fiber density in target tissues. PLoS ONE 2019, 14, e0217475. [Google Scholar] [CrossRef] [Green Version]
- Carr-Wilkinson, J.; Prathalingam, N.; Pal, D.; Moad, M.; Lee, N.; Sundaresh, A.; Forgham, H.; James, P.; Herbert, M.; Lako, M.; et al. Differentiation of Human Embryonic Stem Cells to Sympathetic Neurons: A Potential Model for Understanding Neuroblastoma Pathogenesis. Stem Cells Int. 2018, 2018, 4391641. [Google Scholar] [CrossRef] [Green Version]
- Umehara, Y.; Toyama, S.; Tominaga, M.; Matsuda, H.; Takahashi, N.; Kamata, Y.; Niyonsaba, F.; Ogawa, H.; Takamori, K. Robust induction of neural crest cells to derive peripheral sensory neurons from human induced pluripotent stem cells. Sci. Rep. 2020, 10, 4360. [Google Scholar] [CrossRef] [Green Version]
- Solis-Castro, O.O.; Boissonade, F.M.; Rivolta, M.N. Establishment and neural differentiation of neural crest-derived stem cells from human dental pulp in serum-free conditions. Stem Cells Transl. Med. 2020, 9, 1462–1476. [Google Scholar] [CrossRef]
- Bataille, A.; Leschiera, R.; L’Herondelle, K.; Pennec, J.P.; Le Goux, N.; Mignen, O.; Sakka, M.; Plee-Gautier, E.; Brun, C.; Oddos, T.; et al. In Vitro Differentiation of Human Skin-Derived Cells into Functional Sensory Neurons-Like. Cells 2020, 9, 1000. [Google Scholar] [CrossRef]
- Mounir, M.M.F.; Rashed, F.M.; Bukhary, S.M. Regeneration of Neural Networks in Immature Teeth with Non-Vital Pulp Following a Novel Regenerative Procedure. Int. J. Stem Cells 2019, 12, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Ishikawa, H. Existence of Neural Stem Cells in Mouse Spleen. Sci. World J. 2019, 2019, 6264072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbo, M.; Hays, A.P. Peripherin and neurofilament protein coexist in spinal spheroids of motor neuron disease. J. Neuropathol. Exp. Neurol. 1992, 51, 531–537. [Google Scholar] [CrossRef]
- Migheli, A.; Pezzulo, T.; Attanasio, A.; Schiffer, D. Peripherin immunoreactive structures in amyotrophic lateral sclerosis. Lab. Investig. 1993, 68, 185–191. [Google Scholar] [PubMed]
- Tu, P.H.; Raju, P.; Robinson, K.A.; Gurney, M.E.; Trojanowski, J.Q.; Lee, V.M. Transgenic mice carrying a human mutant superoxide dismutase transgene develop neuronal cytoskeletal pathology resembling human amyotrophic lateral sclerosis lesions. Proc. Natl. Acad. Sci. USA 1996, 93, 3155–3160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, S.; McLean, J.; Robertson, J. Neuronal intermediate filaments and ALS: A new look at an old question. Biochim. Biophys. Acta 2006, 1762, 1001–1012. [Google Scholar] [PubMed] [Green Version]
- Viedma-Poyatos, A.; Pajares, M.A.; Perez-Sala, D. Type III intermediate filaments as targets and effectors of electrophiles and oxidants. Redox Biol. 2020, 36, 101582. [Google Scholar] [CrossRef] [PubMed]
- Corrado, L.; Carlomagno, Y.; Falasco, L.; Mellone, S.; Godi, M.; Cova, E.; Cereda, C.; Testa, L.; Mazzini, L.; D’Alfonso, S. A novel peripherin gene (PRPH) mutation identified in one sporadic amyotrophic lateral sclerosis patient. Neurobiol. Aging 2011, 32, 552.e1–552.e6. [Google Scholar] [CrossRef]
- Gros-Louis, F.; Larivière, R.; Gowing, G.; Laurent, S.; Camu, W.; Bouchard, J.P.; Meininger, V.; Rouleau, G.A.; Julien, J.P. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J. Biol. Chem. 2004, 279, 45951–45956. [Google Scholar] [CrossRef] [Green Version]
- Leung, C.L.; He, C.Z.; Kaufmann, P.; Chin, S.S.; Naini, A.; Liem, R.K.; Mitsumoto, H.; Hays, A.P. A pathogenic peripherin gene mutation in a patient with amyotrophic lateral sclerosis. Brain Pathol. 2004, 14, 290–296. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Nguyen, M.D.; Julien, J.P. Late onset of motor neurons in mice overexpressing wild-type peripherin. J. Cell Biol. 1999, 147, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Beaulieu, J.M.; Doroudchi, M.M.; Durham, H.D.; Julien, J.P.; Mushynski, W.E. Apoptotic death of neurons exhibiting peripherin aggregates is mediated by the proinflammatory cytokine tumor necrosis factor-alpha. J. Cell Biol. 2001, 155, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.M.; Robertson, J.; Julien, J.P. Interactions between peripherin and neurofilaments in cultured cells: Disruption of peripherin assembly by the NF-M and NF-H subunits. Biochem. Cell Biol. 1999, 77, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.M.; Julien, J.P. Peripherin-mediated death of motor neurons rescued by overexpression of neurofilament NF-H proteins. J. Neurochem. 2003, 85, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.M.; Jacomy, H.; Julien, J.P. Formation of intermediate filament protein aggregates with disparate effects in two transgenic mouse models lacking the neurofilament light subunit. J. Neurosci. 2000, 20, 5321–5328. [Google Scholar] [CrossRef] [Green Version]
- Miki, Y.; Mori, F.; Seino, Y.; Tanji, K.; Yoshizawa, T.; Kijima, H.; Shoji, M.; Wakabayashi, K. Colocalization of Bunina bodies and TDP-43 inclusions in a case of sporadic amyotrophic lateral sclerosis with Lewy body-like hyaline inclusions. Neuropathology 2018, 38, 521–528. [Google Scholar] [CrossRef]
- Muresan, V.; Ladescu Muresan, Z. Shared Molecular Mechanisms in Alzheimer’s Disease and Amyotrophic Lateral Sclerosis: Neurofilament-Dependent Transport of sAPP, FUS, TDP-43 and SOD1, with Endoplasmic Reticulum-Like Tubules. Neurodegener. Dis. 2016, 16, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Oberstadt, M.; Classen, J.; Arendt, T.; Holzer, M. TDP-43 and Cytoskeletal Proteins in ALS. Mol. Neurobiol. 2018, 55, 3143–3151. [Google Scholar] [CrossRef]
- Hawley, Z.C.E.; Campos-Melo, D.; Strong, M.J. MiR-105 and miR-9 regulate the mRNA stability of neuronal intermediate filaments. Implications for the pathogenesis of amyotrophic lateral sclerosis (ALS). Brain Res. 2019, 1706, 93–100. [Google Scholar] [CrossRef]
- Sabbatini, D.; Raggi, F.; Ruggero, S.; Seguso, M.; Mandrioli, J.; Cagnin, A.; Briani, C.; Toffanin, E.; Gizzi, M.; Fortuna, A.; et al. Evaluation of peripherin in biofluids of patients with motor neuron diseases. Ann. Clin. Transl. Neurol. 2021, 8, 1750–1754. [Google Scholar] [CrossRef]
- Liang, Y.; Tong, F.; Zhang, L.; Zhu, L.; Li, W.; Huang, W.; Zhao, S.; He, G.; Zhou, Y. iTRAQ-based proteomic analysis discovers potential biomarkers of diffuse axonal injury in rats. Brain Res. Bull. 2019, 153, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Nixon, R.A. Neurofilament Proteins as Biomarkers to Monitor Neurological Diseases and the Efficacy of Therapies. Front. Neurosci. 2021, 15, 689938. [Google Scholar] [CrossRef] [PubMed]
- Saveri, P.; De Luca, M.; Nisi, V.; Pisciotta, C.; Romano, R.; Piscosquito, G.; Reilly, M.M.; Polke, J.M.; Cavallaro, T.; Fabrizi, G.M.; et al. Charcot-Marie-Tooth Type 2B: A New Phenotype Associated with a Novel RAB7A Mutation and Inhibited EGFR Degradation. Cells 2020, 9, 1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, R.; Rivellini, C.; De Luca, M.; Tonlorenzi, R.; Beli, R.; Manganelli, F.; Nolano, M.; Santoro, L.; Eskelinen, E.L.; Previtali, S.C.; et al. Alteration of the late endocytic pathway in Charcot-Marie-Tooth type 2B disease. Cell. Mol. Life Sci. 2020, 78, 351–372. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Guerra, F.; Hu, M.; Pope, A.; Sung, K.; Yang, W.; Jetha, S.; Shoff, T.A.; Gunatilake, T.; Dahlkamp, O.; et al. Mitochondria dysfunction in Charcot Marie Tooth 2B Peripheral Sensory Neuropathy. Commun. Biol. 2022, 5, 717. [Google Scholar] [CrossRef]
- Bjornsdottir, G.; Ivarsdottir, E.V.; Bjarnadottir, K.; Benonisdottir, S.; Gylfadottir, S.S.; Arnadottir, G.A.; Benediktsson, R.; Halldorsson, G.H.; Helgadottir, A.; Jonasdottir, A.; et al. A PRPH splice-donor variant associates with reduced sural nerve amplitude and risk of peripheral neuropathy. Nat. Commun. 2019, 10, 1777. [Google Scholar] [CrossRef] [Green Version]
- Boitard, C.; Villa, M.C.; Becourt, C.; Gia, H.P.; Huc, C.; Sempe, P.; Portier, M.M.; Bach, J.F. Peripherin: An islet antigen that is cross-reactive with nonobese diabetic mouse class II gene products. Proc. Natl. Acad. Sci. USA 1992, 89, 172–176. [Google Scholar] [CrossRef] [Green Version]
- Chamberlain, J.L.; Pittock, S.J.; Oprescu, A.M.; Dege, C.; Apiwattanakul, M.; Kryzer, T.J.; Lennon, V.A. Peripherin-IgG association with neurologic and endocrine autoimmunity. J. Autoimmun. 2010, 34, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Garabatos, N.; Alvarez, R.; Carrillo, J.; Carrascal, J.; Izquierdo, C.; Chapman, H.D.; Presa, M.; Mora, C.; Serreze, D.V.; Verdaguer, J.; et al. In vivo detection of peripherin-specific autoreactive B cells during type 1 diabetes pathogenesis. J. Immunol. 2014, 192, 3080–3090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puertas, M.C.; Carrillo, J.; Pastor, X.; Ampudia, R.M.; Planas, R.; Alba, A.; Bruno, R.; Pujol-Borrell, R.; Estanyol, J.M.; Vives-Pi, M.; et al. Peripherin is a relevant neuroendocrine autoantigen recognized by islet-infiltrating B lymphocytes. J. Immunol. 2007, 178, 6533–6539. [Google Scholar] [CrossRef]
- Doran, T.M.; Morimoto, J.; Simanski, S.; Koesema, E.J.; Clark, L.F.; Pels, K.; Stoops, S.L.; Pugliese, A.; Skyler, J.S.; Kodadek, T. Discovery of Phosphorylated Peripherin as a Major Humoral Autoantigen in Type 1 Diabetes Mellitus. Cell Chem. Biol. 2016, 23, 618–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehuen, A.; Diana, J.; Zaccone, P.; Cooke, A. Immune cell crosstalk in type 1 diabetes. Nat. Rev. Immunol. 2010, 10, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Racine, J.J.; Chapman, H.D.; Doty, R.; Cairns, B.M.; Hines, T.J.; Tadenev, A.L.D.; Anderson, L.C.; Green, T.; Dyer, M.E.; Wotton, J.M.; et al. T Cells from NOD-PerIg Mice Target Both Pancreatic and Neuronal Tissue. J. Immunol. 2020, 205, 2026–2038. [Google Scholar] [CrossRef]
- Liu, C.H.; Lan, C.T.; Chen, L.Y.; Liao, W.C.; Ko, M.H.; Tseng, T.J. Phosphorylation of extracellular signal-regulated kinase 1/2 in subepidermal nerve fibers mediates hyperalgesia following diabetic peripheral neuropathy. Neurotoxicology 2019, 71, 60–74. [Google Scholar] [CrossRef]
- Wang, S.M.; Liu, C.C.; Tseng, H.W.; Wang, J.R.; Huang, C.C.; Chen, Y.J.; Yang, Y.J.; Lin, S.J.; Yeh, T.F. Clinical spectrum of enterovirus 71 infection in children in southern Taiwan, with an emphasis on neurological complications. Clin. Infect. Dis. 1999, 29, 184–190. [Google Scholar] [CrossRef]
- Lim, Z.Q.; Ng, Q.Y.; Oo, Y.; Chu, J.J.H.; Ng, S.Y.; Sze, S.K.; Alonso, S. Enterovirus-A71 exploits peripherin and Rac1 to invade the central nervous system. EMBO Rep. 2021, 22, e51777. [Google Scholar] [CrossRef]
- Hwang, B.; Ise, H. Multimeric conformation of type III intermediate filaments but not the filamentous conformation exhibits high affinity to lipid bilayers. Genes Cells 2020, 25, 413–426. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, R.; Del Fiore, V.S.; Bucci, C. Role of the Intermediate Filament Protein Peripherin in Health and Disease. Int. J. Mol. Sci. 2022, 23, 15416. https://doi.org/10.3390/ijms232315416
Romano R, Del Fiore VS, Bucci C. Role of the Intermediate Filament Protein Peripherin in Health and Disease. International Journal of Molecular Sciences. 2022; 23(23):15416. https://doi.org/10.3390/ijms232315416
Chicago/Turabian StyleRomano, Roberta, Victoria Stefania Del Fiore, and Cecilia Bucci. 2022. "Role of the Intermediate Filament Protein Peripherin in Health and Disease" International Journal of Molecular Sciences 23, no. 23: 15416. https://doi.org/10.3390/ijms232315416
APA StyleRomano, R., Del Fiore, V. S., & Bucci, C. (2022). Role of the Intermediate Filament Protein Peripherin in Health and Disease. International Journal of Molecular Sciences, 23(23), 15416. https://doi.org/10.3390/ijms232315416






