Wild Wheat Rhizosphere-Associated Plant Growth-Promoting Bacteria Exudates: Effect on Root Development in Modern Wheat and Composition
Abstract
:1. Introduction
2. Results
2.1. Diazotrophic Bacteria: Origin and Characterization
2.2. PGPR Effect on a Wheat Elite Cultivar
2.3. Development of Culture Media Suitable for Metabolomics and Proteomics Analyses of Bacterial Exudates
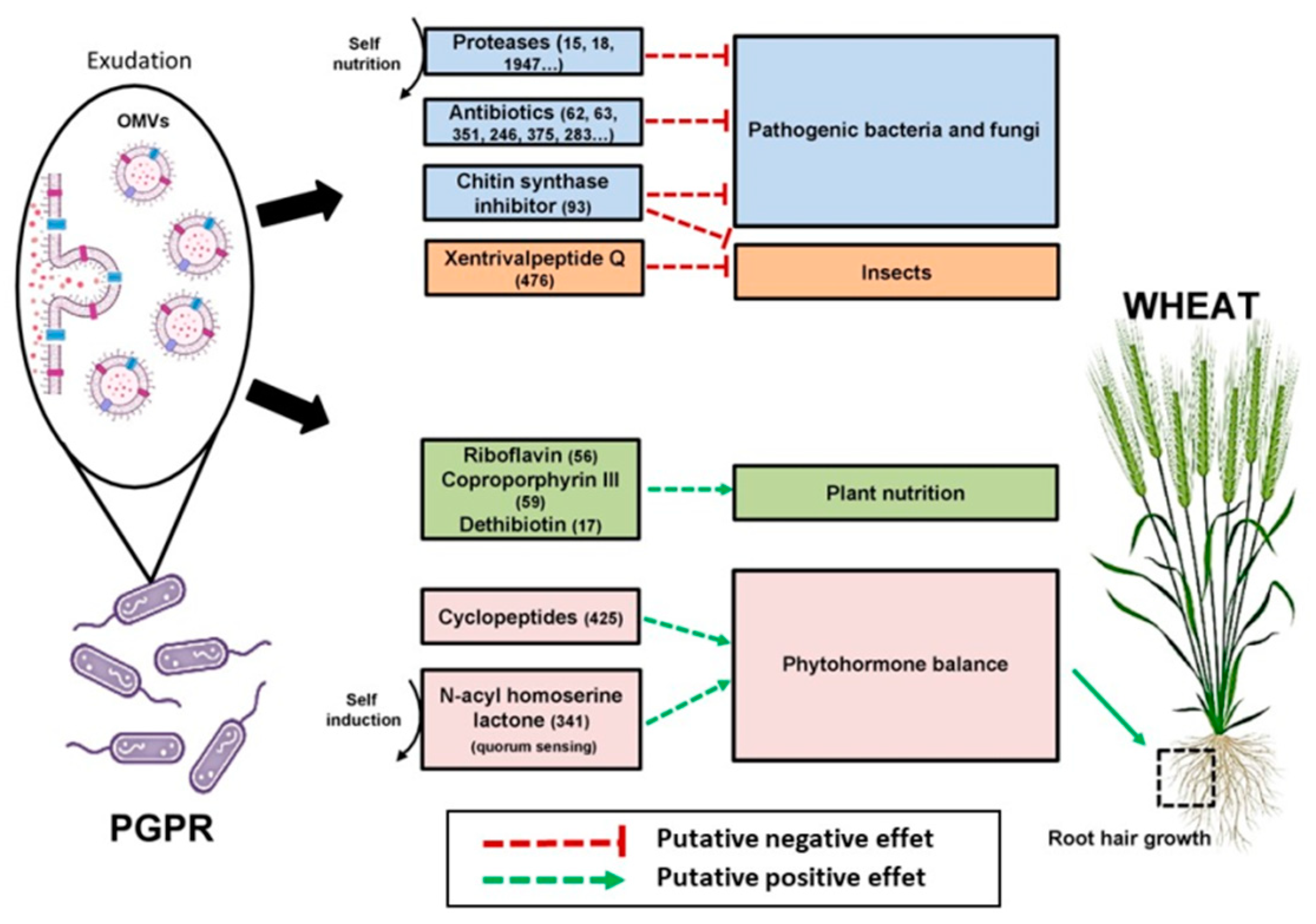
2.4. Bacterial Exudates Affect Root Development
2.5. Comparative Metabolomics Profiling of BPMP-PU-28 and BPMP-EL-40 Exudates
2.6. Comparative Proteomics Profiling of BPMP-PU-28 and BPMP-EL-40 Exudates
3. Discussion
3.1. Bacterial Strains Isolated from the Rhizosphere of a Wheat Ancestor Can Behave as Efficient PGPR in Modern Wheat Varieties
3.2. BPMP-PU-28 and BPMP-EL-40 Exudates Modify Root System Development
3.3. The High Complexity of the Exo-Metabolomes and Exo-Proteomes Offer Extensive Communication and Action Possibilities for Bacteria
3.4. Conclusions
4. Materials and Methods
4.1. Soil Sampling
4.2. Diazotrophic Bacteria Selection and Characterization
4.3. PGPR Effect on Growth and Development of an Elite Wheat Cultivar
4.4. Bacterial Culture Media for Metabolomic and Proteomic Profiling
4.5. Bacterial Growth Curve in Minimal Media for Metabolomic and Proteomic Analyses
4.6. Metabolomics and Proteomics Profiling of Bacterial Exudates
4.7. Effects of Bacterial Growth Supernatants on Wheat Development in Hydroponics Condition
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Álvarez-Rogel, J.; Barberá, G.; Maxwell, B.; Guerrero-Brotons, M.; Díaz-García, C.; Martínez-Sánchez, J.; Sallent, A.; Martínez-Ródenas, J.; González-Alcaraz, M.; Jiménez-Cárceles, F.; et al. The case of Mar Menor eutrophication: State of the art and description of tested Nature-Based Solutions. Ecol. Eng. 2020, 158, 106086. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Castro, R.; Campos-García, J.; López-Bucio, J. Pseudomonas putida and Pseudomonas fluorescens Influence Arabidopsis Root System Architecture Through an Auxin Response Mediated by Bioactive Cyclodipeptides. J. Plant Growth Regul. 2020, 39, 254–265. [Google Scholar] [CrossRef]
- Annapurna, K.; Kumar, A.; Kumar, L.V.; Govindasamy, V.; Bose, P.; Ramadoss, D. PGPR-Induced Systemic Resistance (ISR) in Plant Disease Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 405–425. [Google Scholar] [CrossRef]
- Rosier, A.; Medeiros, F.H.V.; Bais, H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 2018, 428, 35–55. [Google Scholar] [CrossRef] [Green Version]
- Calvo, P.; Zebelo, S.; McNear, D.; Kloepper, J.; Fadamiro, H. Plant growth-promoting rhizobacteria induce changes in Arabidopsis thaliana gene expression of nitrate and ammonium uptake genes. J. Plant Interactions 2019, 14, 224–231. [Google Scholar] [CrossRef] [Green Version]
- Grover, M.; Bodhankar, S.; Sharma, A.; Sharma, P.; Singh, J.; Nain, L. PGPR Mediated Alterations in Root Traits: Way Toward Sustainable Crop Production. Front. Sustain. Food Syst. 2021, 4, 618230. [Google Scholar] [CrossRef]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Wang, H.; Liu, R.; You, M.P.; Barbetti, M.J.; Chen, Y. Pathogen Biocontrol Using Plant Growth-Promoting Bacteria (PGPR): Role of Bacterial Diversity. Microorganisms 2021, 9, 1988. [Google Scholar] [CrossRef]
- Veresoglou, S.D.; Menexes, G. Impact of inoculation with Azospirillum spp. on growth properties and seed yield of wheat: A meta-analysis of studies in the ISI Web of Science from 1981 to 2008. Plant Soil 2010, 337, 469–480. [Google Scholar] [CrossRef]
- Etesami, H.; Alikhani, H.A. Co-inoculation with endophytic and rhizosphere bacteria allows reduced application rates of N-fertilizer for rice plant. Rhizosphere 2016, 2, 5–12. [Google Scholar] [CrossRef]
- Naseri, R.; Azadi, S.; Rahimi, M.J.; Maleki, A.; Mirzaei, A. Effects of inoculation with Azotobacter chroococcum and Pseudomonas putida on yield and some of the important agronomic traits in barley (Hordeum vulgare L.). Int. J. Agron. Plant Prod. 2013, 4, 1602–1610. [Google Scholar]
- Alves, G.C.; Videira, S.S.; Urquiaga, S.; Reis, V.M. Differential plant growth promotion and nitrogen fixation in two genotypes of maize by several Herbaspirillum inoculants. Plant Soil 2015, 387, 307–321. [Google Scholar] [CrossRef]
- Drogue, B.; Doré, H.; Borland, S.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res. Microbiol. 2012, 163, 500–510. [Google Scholar] [CrossRef]
- Chamam, A.; Sanguin, H.; Bellvert, F.; Meiffren, G.; Comte, G.; Wisniewski-Dyé, F.; Bertrand, C.; Prigent-Combaret, C. Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum–Oryza sativa association. Phytochemistry 2013, 87, 65–77. [Google Scholar] [CrossRef]
- Amaral, F.P.D.; Pankievicz, V.C.S.; Arisi, A.C.M.; de Souza, E.M.; Pedrosa, F.; Stacey, G. Differential growth responses of Brachypodium distachyon genotypes to inoculation with plant growth promoting rhizobacteria. Plant Mol. Biol. 2016, 90, 689–697. [Google Scholar] [CrossRef]
- Pankievicz, V.C.S.; Amaral, F.P.D.; Ané, J.-M.; Stacey, G. Diazotrophic Bacteria and Their Mechanisms to Interact and Benefit Cereals. Mol. Plant-Microbe Interactions 2021, 34, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.-M.; Farag, M.A.; Hu, C.-H.; Reddy, M.S.; Wei, H.-X.; Paré, P.W.; Kloepper, J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 4927–4932. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Luna, F.M.; López-Bucio, J.; Altamirano-Hernández, J.; Valencia-Cantero, E.; De La Cruz, H.R.; Macías-Rodríguez, L. Plant growth-promoting rhizobacteria modulate root-system architecture in Arabidopsis thaliana through volatile organic compound emission. Symbiosis 2010, 51, 75–83. [Google Scholar] [CrossRef]
- Ryu, C.-M.; Farag, M.A.; Pare, P.W.; Kloepper, J.W. Invisible Signals from the Underground: Bacterial Volatiles Elicit Plant Growth Promotion and Induce Systemic Resistance. Plant Pathol. J. 2005, 21, 7–12. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jaramillo, J.E.; Carrion, V.J.; De Hollander, M.; Raaijmakers, J.M. The wild side of plant microbiomes. Microbiome 2018, 6, 143. [Google Scholar] [CrossRef]
- Valente, J.; Gerin, F.; Le Gouis, J.; Moënne-Loccoz, Y.; Prigent–Combaret, C. Ancient wheat varieties have a higher ability to interact with plant growth-promoting rhizobacteria. Plant, Cell Environ. 2020, 43, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Spor, A.; Roucou, A.; Mounier, A.; Bru, D.; Breuil, M.-C.; Fort, F.; Vile, D.; Roumet, P.; Philippot, L.; Violle, C. Domestication-driven changes in plant traits associated with changes in the assembly of the rhizosphere microbiota in tetraploid wheat. Sci. Rep. 2020, 10, 12234. [Google Scholar] [CrossRef] [PubMed]
- Ezgi Özkurt, M.; Hassani, A.; Sesiz, U.; Künzel, S.; Dagan, T.; Özkan, H.; Stukenbrock, E.H. Higher stochasticity of microbiota composition in seedlings of domesticated wheat compared to wild wheat. bioRxiv 2019. [Google Scholar] [CrossRef]
- Martínez-Romero, E.; Aguirre-Noyola, J.L.; Taco-Taype, N.; Martínez-Romero, J.; Zuñiga-Dávila, D. Plant microbiota modified by plant domestication. Syst. Appl. Microbiol. 2020, 43, 126106. [Google Scholar] [CrossRef]
- Miller, S.B.; Heuberger, A.L.; Broeckling, C.D.; Jahn, C.E. Non-Targeted Metabolomics Reveals Sorghum Rhizosphere-Associated Exudates are Influenced by the Belowground Interaction of Substrate and Sorghum Genotype. Int. J. Mol. Sci. 2019, 20, 431. [Google Scholar] [CrossRef] [Green Version]
- Casas, M.E.; Matamoros, V. Analytical challenges and solutions for performing metabolomic analysis of root exudates. Trends Environ. Anal. Chem. 2021, 31, e00130. [Google Scholar] [CrossRef]
- Williams, A.; Langridge, H.; Straathof, A.L.; Muhamadali, H.; Hollywood, K.A.; Goodacre, R.; de Vries, F.T. Root functional traits explain root exudation rate and composition across a range of grassland species. J. Ecol. 2022, 110, 21–33. [Google Scholar] [CrossRef]
- Tyc, O.; Zweers, H.; de Boer, W.; Garbeva, P. Volatiles in Inter-Specific Bacterial Interactions. Front. Microbiol. 2015, 6, 1412. [Google Scholar] [CrossRef] [Green Version]
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 2015, 6, 151. [Google Scholar] [CrossRef] [Green Version]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Hanif, A.; Wu, L.; Colman, M.V.; Gao, X. Plant Growth Promotion by Volatile Organic Compounds Produced by Bacillus subtilis SYST2. Front. Microbiol. 2017, 8, 171. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, J.; Li, C.; Ma, Y. Antifungal and plant growth promotion activity of volatile organic compounds produced by Bacillus amyloliquefaciens. MicrobiologyOpen 2019, 8, e00813. [Google Scholar] [CrossRef] [Green Version]
- Yasmin, H.; Rashid, U.; Hassan, M.N.; Nosheen, A.; Naz, R.; Ilyas, N.; Sajjad, M.; Azmat, A.; Alyemeni, M.N. Volatile organic compounds produced by Pseudomonas pseudoalcaligenes alleviated drought stress by modulating defense system in maize ( Zea mays L.). Physiol. Plant. 2020, 172, 896–911. [Google Scholar] [CrossRef]
- Feitosa-Junior, O.R.; Stefanello, E.; Zaini, P.A.; Nascimento, R.; Pierry, P.M.; Dandekar, A.M.; Lindow, S.E.; da Silva, A.M. Proteomic and Metabolomic Analyses of Xylella fastidiosa OMV-Enriched Fractions Reveal Association with Virulence Factors and Signaling Molecules of the DSF Family. Phytopathology 2019, 109, 1344–1353. [Google Scholar] [CrossRef]
- Shavrukov, Y.; Genc, Y.; Hayes, J. The use of hydroponics in abiotic stress tolerance research. In Hydroponics—A Standard Methodology for Plant Biological Researches, 1st ed.; Toshiki, A., Ed.; InTech: Rijeka, Croatia, 2012; pp. 39–66. [Google Scholar] [CrossRef] [Green Version]
- Shane, M.W.; Dixon, K.W.; Lambers, H. The occurrence of dauciform roots amongst Western Australian reeds, rushes and sedges, and the impact of phosphorus supply on dauciform-root development in Schoenus unispiculatus (Cyperaceae). New Phytol. 2004, 165, 887–898. [Google Scholar] [CrossRef]
- Fraisier-Vannier, O.; Chervin, J.; Cabanac, G.; Puech-Pages, V.; Fournier, S.; Durand, V.; Amiel, A.; André, O.; Benamar, O.A.; Dumas, B.; et al. MS-CleanR: A Feature-Filtering Workflow for Untargeted LC–MS Based Metabolomics. Anal. Chem. 2020, 92, 9971–9981. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef]
- Afek, U.; Carmeli, S.; Aharoni, N. Columbianetin, a phytoalexin associated with celery resistance to pathogens during storage. Phytochemistry 1995, 39, 1347–1350. [Google Scholar] [CrossRef]
- Anttila, J.; Heinonen, P.; Nenonen, T.; Pino, A.; Iwaï, H.; Kauppi, E.; Soliymani, R.; Baumann, M.; Saksi, J.; Suni, N.; et al. Is coproporphyrin III a copper-acquisition compound in Paracoccus denitrificans? Biochim. Biophys. Acta 2011, 1807, 311–318. [Google Scholar] [CrossRef]
- Cleary, J.L.; Kolachina, S.; Wolfe, B.E.; Sanchez, L.M. Coproporphyrin III Produced by the Bacterium Glutamicibacter arilaitensis Binds Zinc and Is Upregulated by Fungi in Cheese Rinds. mSystems 2018, 3, e00036-18. [Google Scholar] [CrossRef] [Green Version]
- Schikora, A.; Schenk, S.T.; Hartmann, A. Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group. Plant Mol. Biol. 2016, 90, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.K.; Patriquin, D.G. Root hair deformation, bacterial attachment, and plant growth in wheat-Azospirillum associations. Appl. Environ. Microbiol. 1984, 48, 1208–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapulnik, Y.; Okon, Y.; Henis, Y. Changes in root morphology of wheat caused by Azospirillum inoculation. Can. J. Microbiol. 1985, 31, 881–887. [Google Scholar] [CrossRef]
- Dobbelaere, S.; Croonenborghs, A.; Thys, A.; Broek, A.V.; Vanderleyden, J. Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 1999, 212, 153–162. [Google Scholar] [CrossRef]
- Zhou, C.; Ma, Z.; Zhu, L.; Xiao, X.; Xie, Y.; Zhu, J.; Wang, J. Rhizobacterial Strain Bacillus megaterium BOFC15 Induces Cellular Polyamine Changes that Improve Plant Growth and Drought Resistance. Int. J. Mol. Sci. 2016, 17, 976. [Google Scholar] [CrossRef]
- Jochum, M.; McWilliams, K.; Borrego, E.J.; Kolomiets, M.V.; Niu, G.; Pierson, E.A.; Jo, Y.-K. Bioprospecting Plant Growth-Promoting Rhizobacteria That Mitigate Drought Stress in Grasses. Front. Microbiol. 2019, 10, 2106. [Google Scholar] [CrossRef]
- Mazzola, M.; Funnell, D.; Raaijmakers, J. Wheat Cultivar-Specific Selection of 2,4-Diacetylphloroglucinol-Producing Fluorescent Pseudomonas Species from Resident Soil Populations. Microb. Ecol. 2004, 48, 338–348. [Google Scholar] [CrossRef] [Green Version]
- Maketon, C.; Fortuna, A.-M.; Okubara, P.A. Cultivar-dependent transcript accumulation in wheat roots colonized by Pseudomonas fluorescens Q8r1-96 wild type and mutant strains. Biol. Control. 2012, 60, 216–224. [Google Scholar] [CrossRef]
- Sasaki, K.; Ikeda, S.; Eda, S.; Mitsui, H.; Hanzawa, E.; Kisara, C.; Kazama, Y.; Kushida, A.; Shinano, T.; Minamisawa, K.; et al. Impact of plant genotype and nitrogen level on rice growth response to inoculation with Azospirillum sp. strain B510 under paddy field conditions. Soil Sci. Plant Nutr. 2010, 56, 636–644. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [Green Version]
- Wintermans, P.C.A.; Bakker, P.A.H.M.; Pieterse, C.M.J. Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. 2016, 90, 623–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuluaga, M.Y.A.; de Oliveira, A.L.M.; Valentinuzzi, F.; Tiziani, R.; Pii, Y.; Mimmo, T.; Cesco, S. Can Inoculation With the Bacterial Biostimulant Enterobacter sp. Strain 15S Be an Approach for the Smarter P Fertilization of Maize and Cucumber Plants? Front. Plant Sci. 2021, 12, 719873. [Google Scholar] [CrossRef] [PubMed]
- Saubidet, M.I.; Fatta, N.; Barneix, A.J. The effect of inoculation with Azospirillum brasilense on growth and nitrogen utilization by wheat plants. Plant Soil 2002, 245, 215–222. [Google Scholar] [CrossRef]
- New, P.B.; Kennedy, I.R. Regional distribution and pH sensitivity of Azospirillum associated with wheat roots in Eastern Australia. Microb. Ecol. 1989, 17, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Cassán, F.; Diaz-Zorita, M. Azospirillum sp. in current agriculture: From the laboratory to the field. Soil Biol. Biochem. 2016, 103, 117–130. [Google Scholar] [CrossRef]
- Díaz-Zorita, M.; Fernández-Canigia, M.V. Field performance of a liquid formulation of Azospirillum brasilense on dryland wheat productivity. Eur. J. Soil Biol. 2009, 45, 3–11. [Google Scholar] [CrossRef]
- Hungria, M.; Campo, R.J.; Souza, E.M.; Pedrosa, F.O. Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 2010, 331, 413–425. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef]
- Cummings, S.P. The application of plant growth promoting rhizobacteria (PGPR) in low input and organic cultivation of graminaceous crops; potential and problems. Environ. Biotechnol. 2009, 5, 43–50. [Google Scholar]
- Sharafzadeh, S. Effects of pgpr on growth and nutrients uptake of tomato. Int. J. Adv. Eng. Technol. 2012, 2, 27–31. [Google Scholar]
- Verbon, E.H.; Liberman, L.M. Beneficial Microbes Affect Endogenous Mechanisms Controlling Root Development. Trends Plant Sci. 2016, 21, 218–229. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kim, M.-S.; Krishnamachari, V.; Payton, P.; Sun, Y.; Grimson, M.; Farag, M.A.; Ryu, C.-M.; Allen, R.; Melo, I.S.; et al. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 2007, 226, 839–851. [Google Scholar] [CrossRef]
- Contesto, C.; Milesi, S.; Mantelin, S.; Zancarini, A.; Desbrosses, G.; Varoquaux, F.; Bellini, C.; Kowalczyk, M.; Touraine, B. The auxin-signaling pathway is required for the lateral root response of Arabidopsis to the rhizobacterium Phyllobacterium brassicacearum. Planta 2010, 232, 1455–1470. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Mastranesti, P.; Dhonukshe, P.; Blilou, I.; Pieterse, C.M. Unraveling Root Developmental Programs Initiated by Beneficial Pseudomonas spp. Bacteria. Plant Physiol. 2013, 162, 304–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, Y.; Li, Y.; Wang, X.; Nan, W.; Hu, Y.; Zhang, H.; Zhao, C.; Wang, F.; Li, P.; et al. Endophytic microbes Bacillus sp. LZR216-regulated root development is dependent on polar auxin transport in Arabidopsis seedlings. Plant Cell Rep. 2015, 34, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Asari, S.; Tarkowska, D.; Rolcik, J.; Novak, O.; Palmero, D.V.; Bejai, S.; Meijer, J. Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta 2016, 245, 15–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poitout, A.; Martinière, A.; Kucharczyk, B.; Queruel, N.; Silva-Andia, J.; Mashkoor, S.; Gamet, L.; Varoquaux, F.; Paris, N.; Sentenac, H.; et al. Local signalling pathways regulate the Arabidopsis root developmental response to Mesorhizobium loti inoculation. J. Exp. Bot. 2017, 68, 1199–1211. [Google Scholar] [CrossRef] [Green Version]
- Dahmani, M.A.; Desrut, A.; Moumen, B.; Verdon, J.; Mermouri, L.; Kacem, M.; Coutos-Thévenot, P.; Kaid-Harche, M.; Bergès, T.; Vriet, C. Unearthing the plant growth-promoting traits of Bacillus megaterium RmBm31, an endophytic bacterium isolated from root nodules of Retama monosperma. Front. Plant Sci. 2020, 27, 124. [Google Scholar] [CrossRef]
- Lery, L.M.S.; Hemerly, A.S.; Nogueira, E.M.; von Krüger WM, A.; Bisch, P.M. Quantitative proteomic analysis of the interaction between the endophytic plant-growth-promoting bacterium gluconacetobacter diazotrophicus and sugarcane. APS 2011, 24, 562–576. [Google Scholar] [CrossRef] [Green Version]
- Kierul, K.; Voigt, B.; Albrecht, D.; Chen, X.-H.; Carvalhais, L.C.; Borriss, R. Influence of root exudates on the extracellular proteome of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Microbiology 2015, 161, 131–147. [Google Scholar] [CrossRef]
- Alberton, D.; Valdameri, G.; Moure, V.R.; Monteiro, R.A.; Pedrosa, F.D.O.; Müller-Santos, M.; De Souza, E.M. What Did We Learn From Plant Growth-Promoting Rhizobacteria (PGPR)-Grass Associations Studies Through Proteomic and Metabolomic Approaches? Front. Sustain. Food Syst. 2020, 4, 607343. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Piater, L.A.; Dubery, I.A.; Tugizimana, F.; Mhlongo, M.I. Rhizosphere Tripartite Interactions and PGPR-Mediated Metabolic Reprogramming towards ISR and Plant Priming: A Metabolomics Review. Biology 2022, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Rieusset, L.; Rey, M.; Wisniewski-Dyé, F.; Prigent-Combaret, C.; Comte, G. Wheat Metabolite Interferences on Fluorescent Pseudomonas Physiology Modify Wheat Metabolome through an Ecological Feedback. Metabolites 2022, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Babenko, L.M.; Kosakivska, I.V.; Romanenko, K.O. Molecular mechanisms of N-acyl homoserine lactone signals perception by plants. Cell Biol. Int. 2022, 46, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Moshynets, O.V.; Babenko, L.M.; Rogalsky, S.P.; Iungin, O.S.; Foster, J.; Kosakivska, I.V.; Potters, G.; Spiers, A.J. Priming winter wheat seeds with the bacterial quorum sensing signal N-hexanoyl-L-homoserine lactone (C6-HSL) shows potential to improve plant growth and seed yield. PLoS ONE 2019, 14, e0209460. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, A.; Klink, S.; Rothballer, M. Importance of N-Acyl-Homoserine Lactone-Based Quorum Sensing and Quorum Quenching in Pathogen Control and Plant Growth Promotion. Pathogens 2021, 10, 1561. [Google Scholar] [CrossRef]
- Wood, D.W.; Gong, F.; Daykin, M.M.; Williams, P.; Pierson, L.S. N-acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J. Bacteriol. 1997, 179, 7663–7670. [Google Scholar] [CrossRef] [Green Version]
- Mathesius, U.; Mulders, S.; Gao, M.; Teplitski, M.; Caetano-Anollés, G.; Rolfe, B.G.; Bauer, W.D. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. USA 2003, 100, 1444–1449. [Google Scholar] [CrossRef] [Green Version]
- Decker, H.; Walz, F.; Bormann, C.; Zahner, H.; Fiedler, H.P. Nikkomycins Wz and Wx, new chitin synthetase inhibitors from Streptomyces tendae. J. Antibiot. 1990, 1, 43–48. [Google Scholar] [CrossRef]
- Donabedian, H. Quorum Sensing and its Relevance to Infectious Diseases. J. Infect. 2003, 46, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Coquant, G.; Grill, J.-P.; Seksik, P. Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front. Immunol. 2020, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Constantin, O.E. Bacterial biofilms formation at air liquid interfaces. Innov. Rom. Food Biotechnol. 2009, 5, 18–22. [Google Scholar]
- Ortiz-Castro, R.; López-Bucio, J. Review: Phytostimulation and root architectural responses to quorum-sensing signals and related molecules from rhizobacteria. Plant Sci. 2019, 284, 135–142. [Google Scholar] [CrossRef]
- Zaytseva, Y.V.; Sidorov, A.V.; Marakaev, O.A.; Khmel, I.A. Plant-Microbial Interactions Involving Quorum Sensing Regulation. Microbiology 2019, 88, 523–533. [Google Scholar] [CrossRef]
- Song, G.C.; Choi, H.K.; Kim, Y.S.; Choi, J.S.; Ryu, C.-M. Seed defense biopriming with bacterial cyclodipeptides triggers immunity in cucumber and pepper. Sci. Rep. 2017, 7, 14209. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Castro, R.; Díaz-Pérez, C.; Martínez-Trujillo, M.; del Río, R.E.; Campos-García, J.; López-Bucio, J. Trans kingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 7253–7258. [Google Scholar] [CrossRef] [Green Version]
- Smets, D.; Loos, M.S.; Karamanou, S.; Economou, A. Protein Transport Across the Bacterial Plasma Membrane by the Sec Pathway. J. Protein Chem. 2019, 38, 262–273. [Google Scholar] [CrossRef]
- Qing, G.; Gong, N.; Chen, X.; Chen, J.; Zhang, H.; Wang, Y.; Wang, R.; Zhang, S.; Zhang, Z.; Zhao, X.; et al. Natural and engineered bacterial outer membrane vesicles. Biophys. Rep. 2019, 5, 184–198. [Google Scholar] [CrossRef] [Green Version]
- Dineshkumar, K.; Aparna, V.; Wu, L.; Wan, J.; Abdelaziz, M.H.; Su, Z.; Wang, S.; Xu, H. Bacterial bug-out bags: Outer membrane vesicles and their proteins and functions. J. Microbiol. 2020, 58, 531–542. [Google Scholar] [CrossRef]
- Furuyama, N.; Sircili, M.P. Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. BioMed Res. Int. 2021, 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mozaheb, N.; Mingeot-Leclercq, M.-P. Membrane Vesicle Production as a Bacterial Defense against Stress. Front. Microbiol. 2020, 11, 600221. [Google Scholar] [CrossRef] [PubMed]
- Avila-Calderón, E.D.; Ruiz-Palma, M.D.S.; Aguilera-Arreola, M.G.; Velázquez-Guadarrama, N.; Ruiz, E.A.; Gomez-Lunar, Z.; Witonsky, S.; Contreras-Rodríguez, A. Outer Membrane Vesicles of Gram-Negative Bacteria: An Outlook on Biogenesis. Front. Microbiol. 2021, 12, 557902. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Berleman, J.; Auer, M. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ. Microbiol. 2013, 15, 347–354. [Google Scholar] [CrossRef]
- Dell’Annunziata, F.; Folliero, V.; Giugliano, R.; De Filippis, A.; Santarcangelo, C.; Izzo, V.; Daglia, M.; Galdiero, M.; Arciola, C.; Franci, G. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 5985. [Google Scholar] [CrossRef]
- Estefan, G.; Sommer, R.; Ryan, J. Methods of Soil, Plant, and Water Analysis: A Manual for the West Asia and North Africa Region, 3rd ed.; International Center for Agricultural Research in the Dry Areas: Aleppo, Syria, 2013. [Google Scholar]
- Baldani, J.I.; Reis, V.M.; Videira, S.S.; Boddey, L.H.; Baldani, V.L.D. The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: A practical guide for microbiologists. Plant Soil 2014, 384, 413–431. [Google Scholar] [CrossRef]
- Louws, F.J.; Fulbright, D.W.; Stephens, C.T.; de Bruijn, F.J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 1994, 60, 2286–2295. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, L.; Ramos, P.L.; Manfio, G.P.; Barbosa, H.R.; Pavan, C.; Moreira-Filho, C.A. Molecular characterization of nitrogen-fixing bacteria isolated from brazilian agricultural plants at São Paulo state. Braz. J. Microbiol. 2008, 39, 414–422. [Google Scholar] [CrossRef] [Green Version]
- Pande, A.; Pandey, P.; Mehra, S.; Singh, M.; Kaushik, S. Phenotypic and genotypic characterization of phosphate solubilizing bacteria and their efficiency on the growth of maize. J. Genet. Eng. Biotechnol. 2017, 15, 379–391. [Google Scholar] [CrossRef]
- Rajawat, M.V.S.; Singh, S.; Tyagi, S.P.; Saxena, A.K. A Modified Plate Assay for Rapid Screening of Potassium-Solubilizing Bacteria. Pedosphere 2016, 26, 768–773. [Google Scholar] [CrossRef]
- Haque, M.; Mosharaf, K.; Khatun, M.; Haque, A.; Biswas, S.; Islam, S.; Islam, M.; Shozib, H.B.; Miah, M.U.; Molla, A.H.; et al. Biofilm Producing Rhizobacteria with Multiple Plant Growth-Promoting Traits Promote Growth of Tomato under Water-Deficit Stress. Front. Microbiol. 2020, 11, 542053. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorck, H. Production of Hydrocyanic Acid by Bacteria. Physiol. Plant. 1948, 1, 142–146. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yuxin, C.; Li, Z.; et al. SOAPnuke: A Map Reduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Cabrera-Bosquet, L.; Fournier, C.; Brichet, N.; Welcker, C.; Suard, B.; Tardieu, F. High-throughput estimation of incident light, light interception and radiation-use efficiency of thousands of plants in a phenotyping platform. New Phytol. 2016, 212, 269–281. [Google Scholar] [CrossRef] [Green Version]
- Brichet, N.; Fournier, C.; Turc, O.; Strauss, O.; Artzet, S.; Pradal, C.; Welcker, C.; Tardieu, F.; Cabrera-Bosquet, L. A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods 2017, 13, 96. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, S.; Vialaret, J.; Séveno, M.; Tiers, L.; Hirtz, C.; Lehmann, S.; Vialaret, J.; Séveno, M.; Tiers, L.; Gabelle, A.; et al. Comparison of hydrophobic, lipophilic and immunodepletion pre- fractionation methods for label-free LC-MS/MS identification of biomarkers in human cerebrospinal fluid. J. Proteomics Bioinform. 2021, S5. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification. Mol. Cell. Proteomics 2012, 11, M111.010587. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Zhou, G.; Ewald, J.; Le Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 178, 1735–1761. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhour, H.; Bray, F.; Dandache, I.; Marti, G.; Flament, S.; Perez, A.; Lis, M.; Cabrera-Bosquet, L.; Perez, T.; Fizames, C.; et al. Wild Wheat Rhizosphere-Associated Plant Growth-Promoting Bacteria Exudates: Effect on Root Development in Modern Wheat and Composition. Int. J. Mol. Sci. 2022, 23, 15248. https://doi.org/10.3390/ijms232315248
Zhour H, Bray F, Dandache I, Marti G, Flament S, Perez A, Lis M, Cabrera-Bosquet L, Perez T, Fizames C, et al. Wild Wheat Rhizosphere-Associated Plant Growth-Promoting Bacteria Exudates: Effect on Root Development in Modern Wheat and Composition. International Journal of Molecular Sciences. 2022; 23(23):15248. https://doi.org/10.3390/ijms232315248
Chicago/Turabian StyleZhour, Houssein, Fabrice Bray, Israa Dandache, Guillaume Marti, Stéphanie Flament, Amélie Perez, Maëlle Lis, Llorenç Cabrera-Bosquet, Thibaut Perez, Cécile Fizames, and et al. 2022. "Wild Wheat Rhizosphere-Associated Plant Growth-Promoting Bacteria Exudates: Effect on Root Development in Modern Wheat and Composition" International Journal of Molecular Sciences 23, no. 23: 15248. https://doi.org/10.3390/ijms232315248
APA StyleZhour, H., Bray, F., Dandache, I., Marti, G., Flament, S., Perez, A., Lis, M., Cabrera-Bosquet, L., Perez, T., Fizames, C., Baudoin, E., Madani, I., El Zein, L., Véry, A.-A., Rolando, C., Sentenac, H., Chokr, A., & Peltier, J.-B. (2022). Wild Wheat Rhizosphere-Associated Plant Growth-Promoting Bacteria Exudates: Effect on Root Development in Modern Wheat and Composition. International Journal of Molecular Sciences, 23(23), 15248. https://doi.org/10.3390/ijms232315248





