Rapid Detection of Brettanomyces bruxellensis in Wine by Polychromatic Flow Cytometry
Abstract
:1. Introduction
2. Results
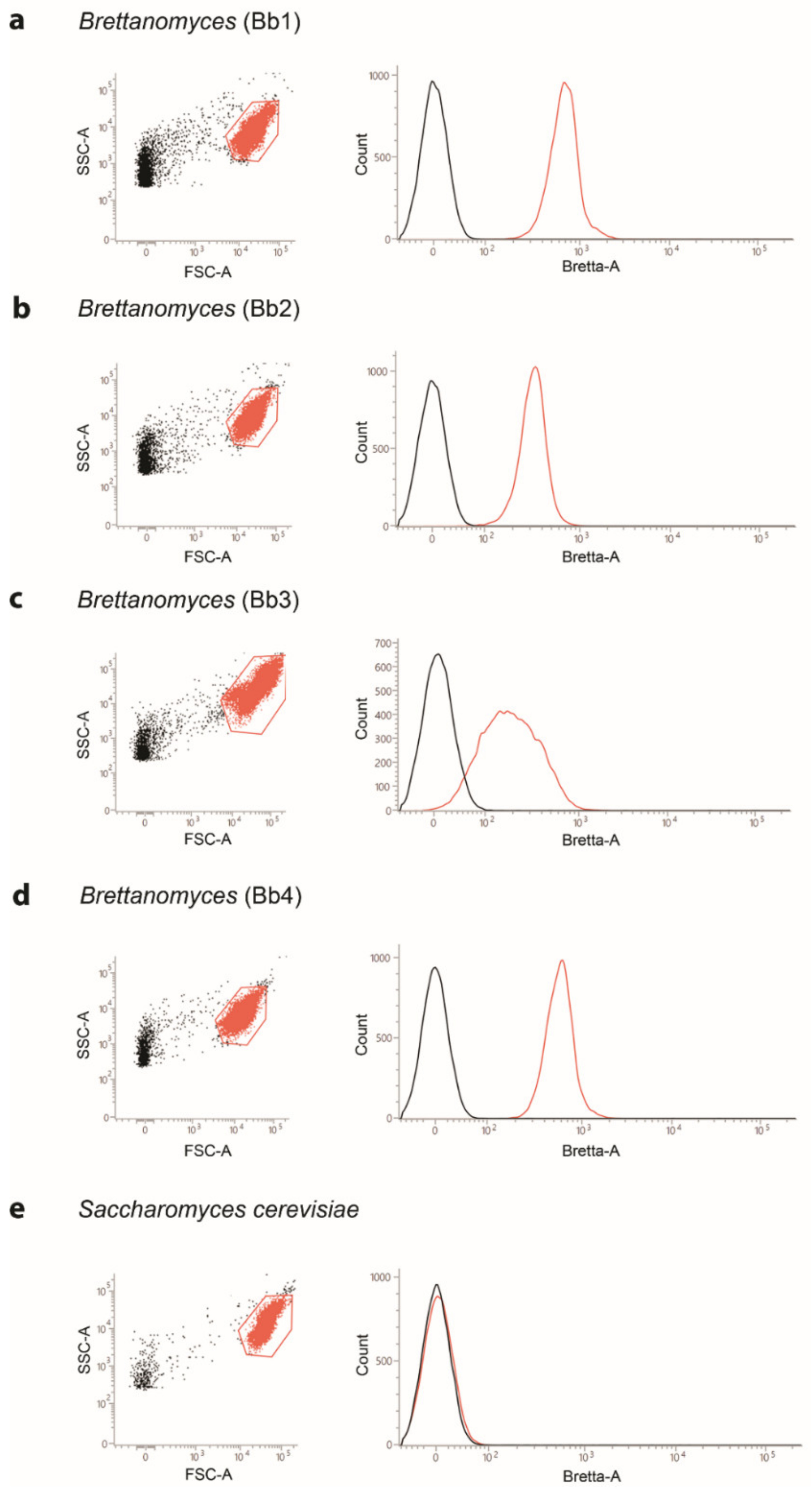
2.1. Specificity of the Flow Cytometry Measurements
2.2. Sensitivity of B. bruxellensis Flow Cytometry Measurements
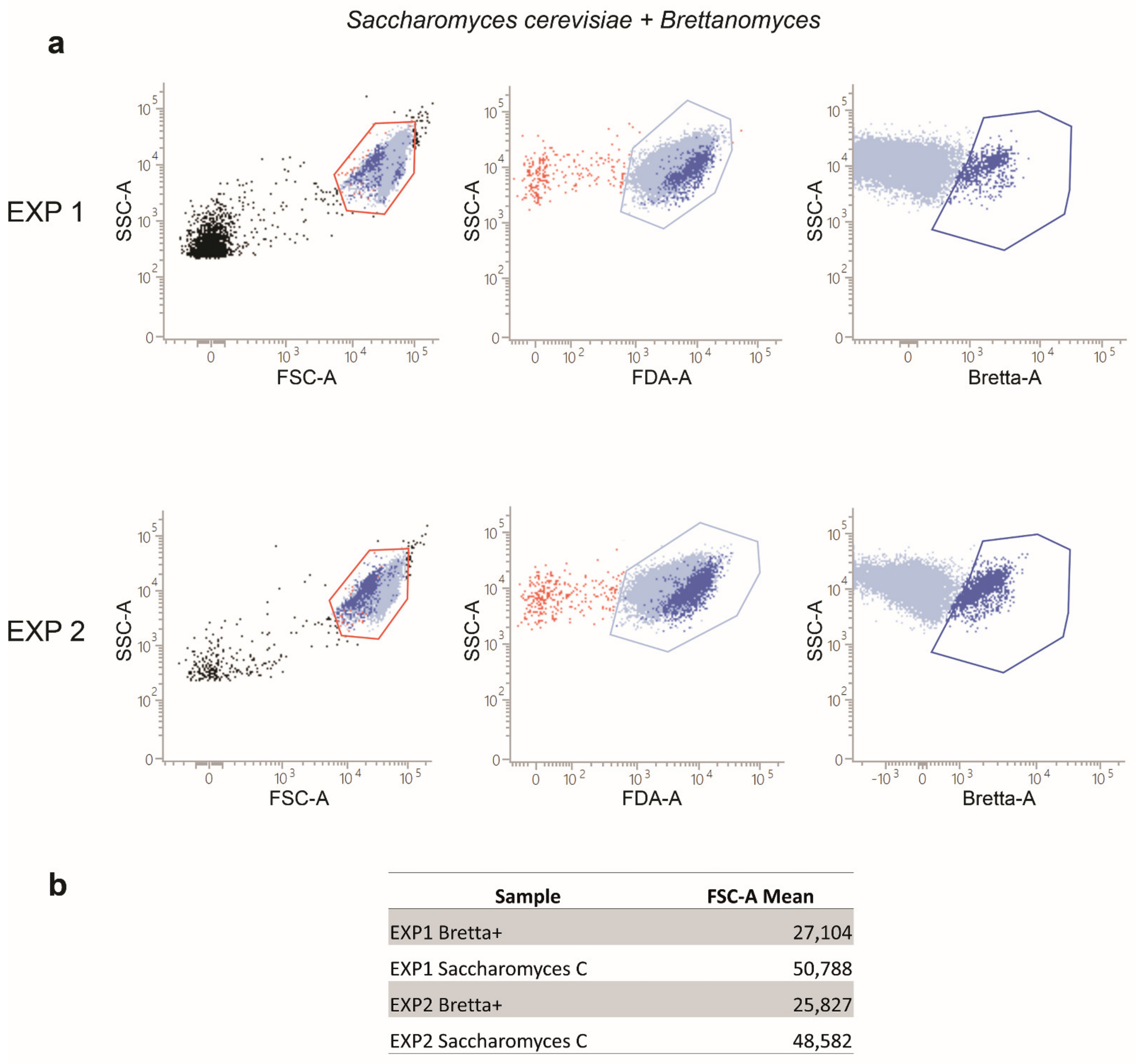
2.3. Flow Cytometry Analyses of S. cerevisiae and B. bruxellensis Mixed Cultures
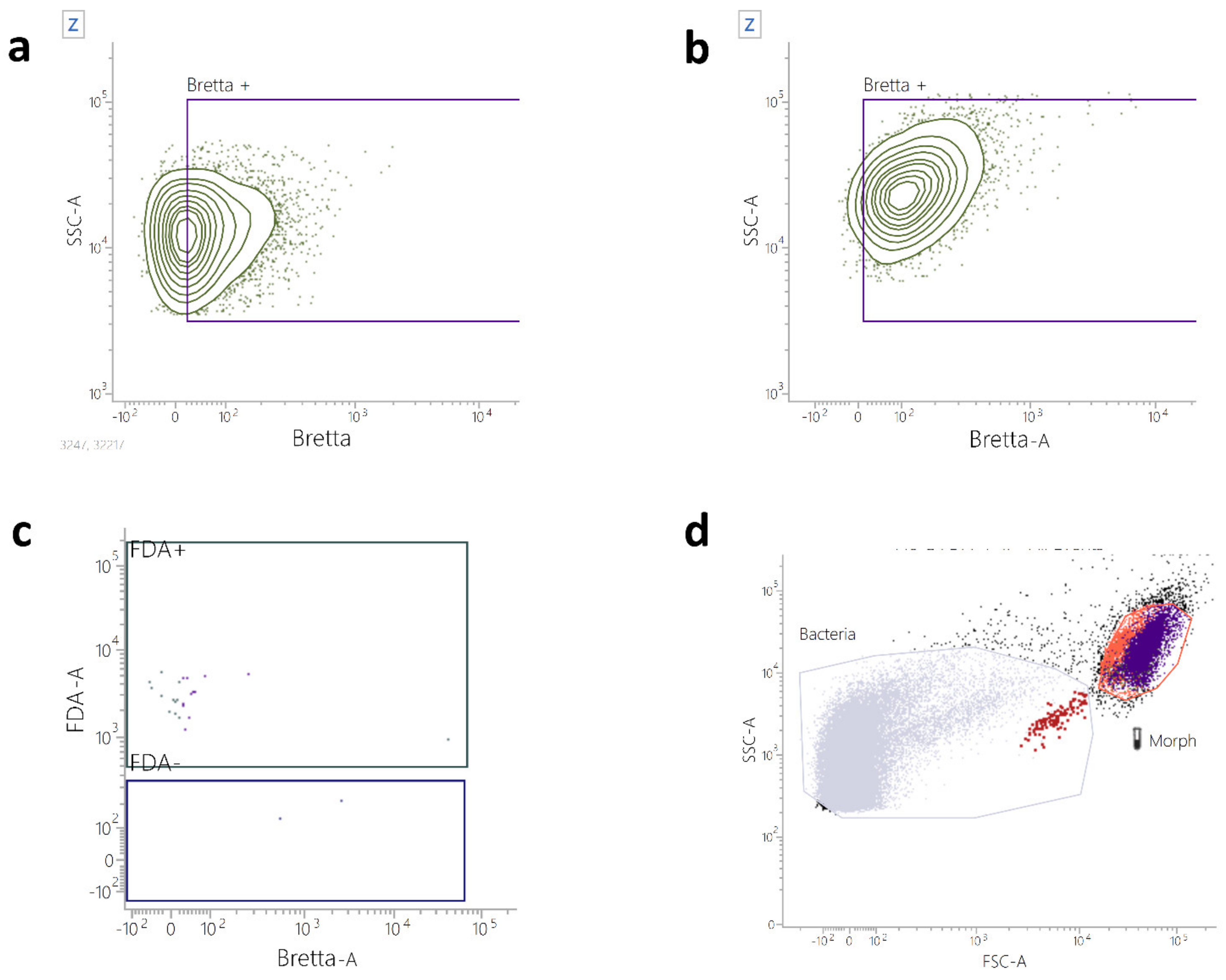
2.4. Flow Cytometry Identification and Count of B. bruxellensis in Wine Samples
3. Discussion
4. Materials and Methods
4.1. B. bruxellensis Cell Cultures
4.2. Wine Samples
4.3. Flow Cytometry Analyses
4.4. B. bruxellensis Counts
4.5. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier Science: London, UK; Burlington, MA, USA, 2011; Volume 1, ISBN 978-0-08-093127-2. [Google Scholar]
- Loureiro, V.; Malfeito-Ferreira, M. Spoilage Yeasts in the Wine Industry. Int. J. Food Microbiol. 2003, 86, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Agnolucci, M.; Tirelli, A.; Cocolin, L.; Toffanin, A. Brettanomyces bruxellensis Yeasts: Impact on Wine and Winemaking. World J. Microbiol. Biotechnol. 2017, 33, 180. [Google Scholar] [CrossRef] [PubMed]
- Henschke, P.; Curtin, C.; Grbin, P. Molecular Characterisation of the Wine Spoilage Yeast—Dekkera (Brettanomyces) bruxellensis. Microbiol. Aust. 2007, 28, 76. [Google Scholar] [CrossRef]
- Wedral, D.; Shewfelt, R.; Frank, J. The Challenge of Brettanomyces in Wine. LWT–Food Sci. Technol. 2010, 43, 1474–1479. [Google Scholar] [CrossRef]
- Oelofse, A.; Pretorius, I.S.; du Toit, M. Significance of Brettanomyces and Dekkera during Winemaking: A Synoptic Review. SAJEV 2008, 29. [Google Scholar] [CrossRef] [Green Version]
- Chatonnet, P.; Dubourdie, D.; Boidron, J.; Pons, M. The Origin of Ethylphenols in Wines. J. Sci. Food Agric. 1992, 60, 165–178. [Google Scholar] [CrossRef]
- Conterno, L.; Joseph, C.M.L.; Arvik, T.J.; Henick-Kling, T.; Bisson, L.F. Genetic and Physiological Characterization of Brettanomyces bruxellensis Strains Isolated from Wines. Am. J. Enol. Vitic. 2006, 57, 139. [Google Scholar] [CrossRef]
- Tubia, I.; Prasad, K.; Pérez-Lorenzo, E.; Abadín, C.; Zumárraga, M.; Oyanguren, I.; Barbero, F.; Paredes, J.; Arana, S. Beverage Spoilage Yeast Detection Methods and Control Technologies: A Review of Brettanomyces. Int. J. Food Microbiol. 2018, 283, 65–76. [Google Scholar] [CrossRef]
- Suárez, R.; Suárez-Lepe, J.A.; Morata, A.; Calderón, F. The Production of Ethylphenols in Wine by Yeasts of the Genera Brettanomyces and Dekkera: A Review. Food Chem. 2007, 102, 10–21. [Google Scholar] [CrossRef]
- Aguilar Uscanga, M.G.; Délia, M.-L.; Strehaiano, P. Brettanomyces bruxellensis: Effect of Oxygen on Growth and Acetic Acid Production. Appl. Microbiol. Biotechnol. 2003, 61, 157–162. [Google Scholar] [CrossRef]
- Serra Colomer, M.; Funch, B.; Forster, J. The Raise of Brettanomyces Yeast Species for Beer Production. Curr. Opin. Biotechnol. 2019, 56, 30–35. [Google Scholar] [CrossRef]
- Ji, J.; Henschen, C.W.; Nguyen, T.H.; Ma, L.; Waterhouse, A.L. Yeasts Induce Acetaldehyde Production in Wine Micro-Oxygenation Treatments. J. Agric. Food Chem. 2020, 68, 15216–15227. [Google Scholar] [CrossRef]
- Mansfield, A.K.; Zoecklein, B.W.; Whiton, R.S. Quantification of Glycosidase Activity in Selected Strains of Brettanomyces bruxellensis and Oenococcus oeni. Am. J. Enol. Vitic. 2002, 53, 303. [Google Scholar] [CrossRef]
- Pinto, L.; Baruzzi, F.; Cocolin, L.; Malfeito-Ferreira, M. Emerging Technologies to Control Brettanomyces spp. in Wine: Recent Advances and Future Trends. Trends Food Sci. Technol. 2020, 99, 88–100. [Google Scholar] [CrossRef]
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Zironi, R.; Comi, G. Molecular Detection and Identification of Brettanomyces/Dekkera bruxellensis and Brettanomyces/Dekkera anomalus in Spoiled Wines. Appl. Environ. Microbiol. 2004, 70, 1347–1355. [Google Scholar] [CrossRef] [Green Version]
- Tofalo, R.; Schirone, M.; Perpetuini, G.; Suzzi, G.; Corsetti, A. Development and Application of a Real-Time PCR-Based Assay to Enumerate Total Yeasts and Pichia anomala, Pichia guillermondii and Pichia kluyveri in Fermented Table Olives. Food Control 2012, 23, 356–362. [Google Scholar] [CrossRef]
- Tofalo, R.; Schirone, M.; Corsetti, A.; Suzzi, G. Detection of Brettanomyces spp. in Red Wines Using Real-Time PCR. J. Food Sci. 2012, 77, M545–M549. [Google Scholar] [CrossRef]
- Vendrame, M.; Manzano, M.; Comi, G.; Bertrand, J.; Iacumin, L. Use of Propidium Monoazide for the Enumeration of Viable Brettanomyces bruxellensis in Wine and Beer by Quantitative PCR. Food Microbiol 2014, 42, 196–204. [Google Scholar] [CrossRef]
- Van Wyk, S.; Silva, F. Enumeration of Brettanomyces in Wine Using Impedance. Appl. Microbiol. 2021, 1, 352–360. [Google Scholar] [CrossRef]
- Hong, D.; Lee, G.; Jung, N.C.; Jeon, M. Fast Automated Yeast Cell Counting Algorithm Using Bright-Field and Fluorescence Microscopic Images. Biol. Proced. Online 2013, 15, 13. [Google Scholar] [CrossRef]
- Capozzi, V.; Di Toro, M.R.; Grieco, F.; Michelotti, V.; Salma, M.; Lamontanara, A.; Russo, P.; Orrù, L.; Alexandre, H.; Spano, G. Viable But Not Culturable (VBNC) State of Brettanomyces bruxellensis in Wine: New Insights on Molecular Basis of VBNC Behaviour Using a Transcriptomic Approach. Food Microbiol. 2016, 59, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Cossarizza, A.; Chang, H.; Radbruch, A.; Acs, A.; Adam, D.; Adam-Klages, S.; Agace, W.W.; Aghaeepour, N.; Akdis, M.; Allez, M.; et al. Guidelines for the Use of Flow Cytometry and Cell Sorting in Immunological Studies (Second Edition). Eur. J. Immunol. 2019, 49, 1457–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares-Santos, V.; Pardo, I.; Ferrer, S. Cells-QPCR as a Direct Quantitative PCR Method to Avoid Microbial DNA Extractions in Grape Musts and Wines. Int. J. Food Microbiol. 2017, 261, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Longin, C.; Petitgonnet, C.; Guilloux-Benatier, M.; Rousseaux, S.; Alexandre, H. Application of Flow Cytometry to Wine Microorganisms. Food Microbiol. 2017, 62, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Davey, H.M. Life, Death, and In-Between: Meanings and Methods in Microbiology. Appl. Environ. Microbiol. 2011, 77, 5571–5576. [Google Scholar] [CrossRef] [Green Version]
- Branco, P.; Coutinho, R.; Malfeito-Ferreira, M.; Prista, C.; Albergaria, H. Wine Spoilage Control: Impact of Saccharomycin on Brettanomyces bruxellensis and Its Conjugated Effect with Sulfur Dioxide. Microorganisms 2021, 9, 2528. [Google Scholar] [CrossRef]
- Wei, R.; Chen, N.; Ding, Y.; Wang, L.; Liu, Y.; Gao, F.; Zhang, L.; Li, H.; Wang, H. Correlations between Microbiota with Physicochemical Properties and Volatile Compounds during the Spontaneous Fermentation of Cabernet Sauvignon (Vitis vinifera L.) Wine. LWT 2022, 163, 113529. [Google Scholar] [CrossRef]
- Granchi, L.; Bosco, M.; Messini, A.; Vincenzini, M. Rapid Detection and Quantification of Yeast Species during Spontaneous Wine Fermentation by PCR-RFLP Analysis of the RDNA ITS Region. J. Appl. Microbiol. 1999, 87, 949–956. [Google Scholar] [CrossRef]
- Rehan, M.; Younus, H. Effect of Organic Solvents on the Conformation and Interaction of Catalase and Anticatalase Antibodies. Int. J. Biol. Macromol. 2006, 38, 289–295. [Google Scholar] [CrossRef]
- Bonora, A.; Mares, D. A Simple Colorimetric Method for Detecting Cell Viability in Cultures of Eukaryotic Microorganisms. Curr. Microbiol. 1982, 7, 217–221. [Google Scholar] [CrossRef]
- Lanuti, P.; Simeone, P.; Rotta, G.; Almici, C.; Avvisati, G.; Azzaro, R.; Bologna, G.; Budillon, A.; Di Cerbo, M.; Di Gennaro, E.; et al. A Standardized Flow Cytometry Network Study for the Assessment of Circulating Endothelial Cell Physiological Ranges. Sci. Rep. 2018, 8, 5823. [Google Scholar] [CrossRef] [Green Version]
- Lanuti, P.; Rotta, G.; Almici, C.; Avvisati, G.; Budillon, A.; Doretto, P.; Malara, N.; Marini, M.; Neva, A.; Simeone, P.; et al. Endothelial Progenitor Cells, Defined by the Simultaneous Surface Expression of VEGFR2 and CD133, Are Not Detectable in Healthy Peripheral and Cord Blood. Cytom. Part A 2016, 89, 259–270. [Google Scholar] [CrossRef]
- Lanuti, P.; Ciccocioppo, F.; Bonanni, L.; Marchisio, M.; Lachmann, R.; Tabet, N.; Pierdomenico, L.; Santavenere, E.; Catinella, V.; Iacone, A.; et al. Amyloid-Specific T-Cells Differentiate Alzheimer’s Disease from Lewy Body Dementia. Neurobiol. Aging 2012, 33, 2599–2611. [Google Scholar] [CrossRef]
- Marchisio, M.; Simeone, P.; Bologna, G.; Ercolino, E.; Pierdomenico, L.; Pieragostino, D.; Ventrella, A.; Antonini, F.; Del Zotto, G.; Vergara, D.; et al. Flow Cytometry Analysis of Circulating Extracellular Vesicle Subtypes from Fresh Peripheral Blood Samples. Int. J. Mol. Sci. 2021, 22, 48. [Google Scholar] [CrossRef]
- Puca, V.; Ercolino, E.; Celia, C.; Bologna, G.; Di Marzio, L.; Mincione, G.; Marchisio, M.; Miscia, S.; Muraro, R.; Lanuti, P.; et al. Detection and Quantification of EDNA-Associated Bacterial Membrane Vesicles by Flow Cytometry. IJMS 2019, 20, 5307. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, N.; Goncalves, G.; Pereira-da-Silva, S.; Malfeito-Ferreira, M.; Loureiro, V. Development and Use of a New Medium to Detect Yeasts of the Genera Dekkera/Brettanomyces. J. Appl. Microbiol. 2001, 90, 588–599. [Google Scholar] [CrossRef] [Green Version]
- Cavazza, A.; Grando, M.; Zini, C. Rilevazione della Flora Microbica di Mosti e Vini. Vignevini 1992, 9, 17–20. [Google Scholar]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A Medium for the Cultivation of Lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]

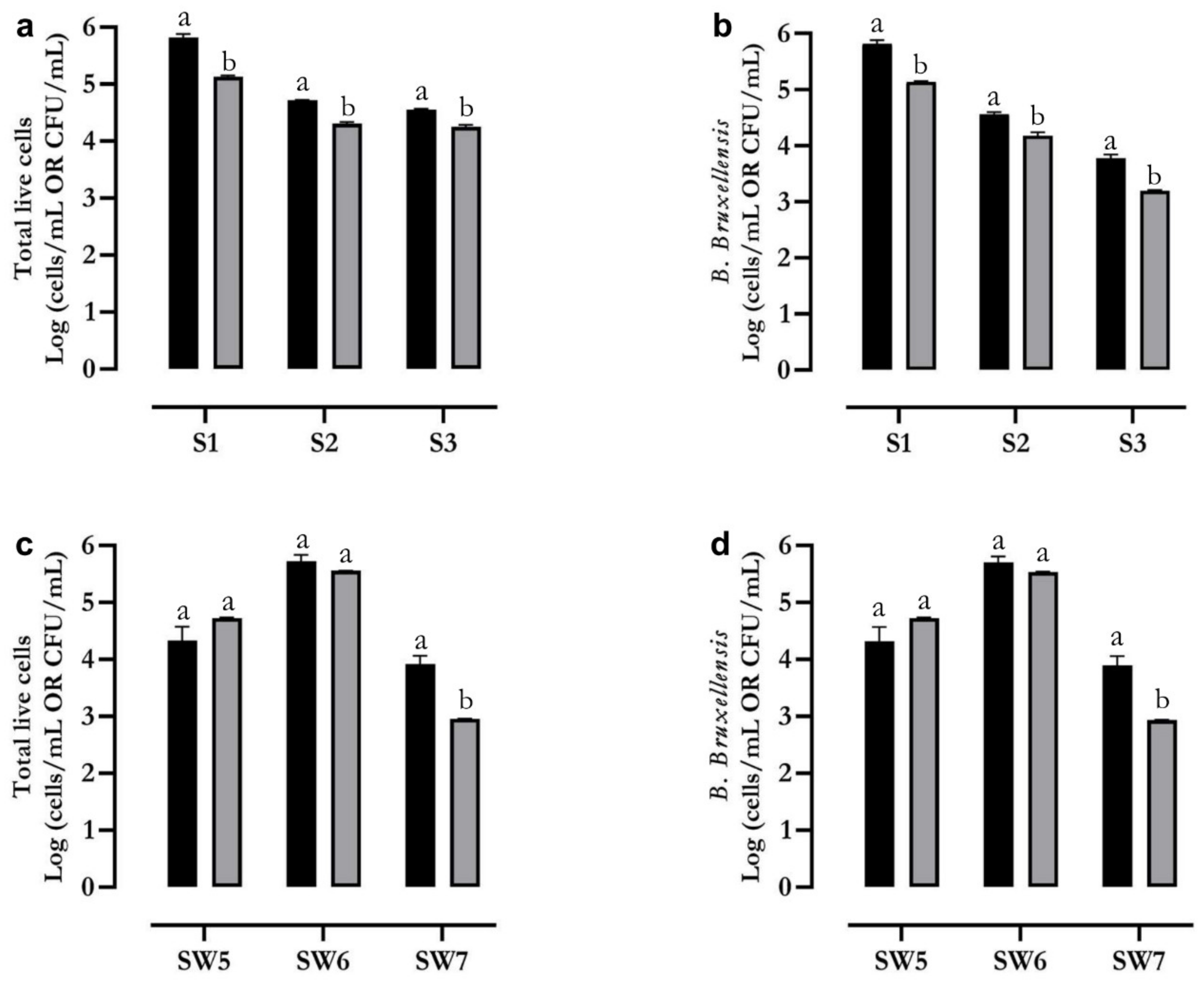
| Wine Sample | Inoculated Cells | Concentration (Cells/mL) |
|---|---|---|
| S1 | B. bruxellensis | 8 × 105 |
| S2 | B. bruxellensis | 8 × 104 |
| S. cerevisiae | 8 × 103 | |
| S3 | B. bruxellensis | 8 × 103 |
| S. cerevisiae | 8 × 103 | |
| Pichia spp. | 8 × 103 |
| Wine Sample | Inoculated Cells | Concentration (Cells/mL) |
|---|---|---|
| SW5 | B. bruxellensis | (6.0 ± 0.3) × 103 |
| SW6 | B. bruxellensis | (3.9 ± 0.1) × 104 |
| O. oeni | (3.3 ± 0.3) × 103 | |
| SW7 | B. bruxellensis | (9.4 ± 0.5) × 102 |
| Pichia spp. | (2.6 ± 0.7) × 102 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Bellis, D.; Di Stefano, A.; Simeone, P.; Catitti, G.; Vespa, S.; Patruno, A.; Marchisio, M.; Mari, E.; Granchi, L.; Viti, C.; et al. Rapid Detection of Brettanomyces bruxellensis in Wine by Polychromatic Flow Cytometry. Int. J. Mol. Sci. 2022, 23, 15091. https://doi.org/10.3390/ijms232315091
De Bellis D, Di Stefano A, Simeone P, Catitti G, Vespa S, Patruno A, Marchisio M, Mari E, Granchi L, Viti C, et al. Rapid Detection of Brettanomyces bruxellensis in Wine by Polychromatic Flow Cytometry. International Journal of Molecular Sciences. 2022; 23(23):15091. https://doi.org/10.3390/ijms232315091
Chicago/Turabian StyleDe Bellis, Domenico, Alessio Di Stefano, Pasquale Simeone, Giulia Catitti, Simone Vespa, Antonia Patruno, Marco Marchisio, Eleonora Mari, Lisa Granchi, Carlo Viti, and et al. 2022. "Rapid Detection of Brettanomyces bruxellensis in Wine by Polychromatic Flow Cytometry" International Journal of Molecular Sciences 23, no. 23: 15091. https://doi.org/10.3390/ijms232315091
APA StyleDe Bellis, D., Di Stefano, A., Simeone, P., Catitti, G., Vespa, S., Patruno, A., Marchisio, M., Mari, E., Granchi, L., Viti, C., Chiacchiaretta, P., Cichelli, A., Tofalo, R., & Lanuti, P. (2022). Rapid Detection of Brettanomyces bruxellensis in Wine by Polychromatic Flow Cytometry. International Journal of Molecular Sciences, 23(23), 15091. https://doi.org/10.3390/ijms232315091










