Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress
Abstract
1. Introduction
2. Results
2.1. Transcriptome and Proteome Analysis for Oil Palm Varieties under Low-Temperature Stress
2.2. Differentially Expressed Gene and Protein (DEG and DEP) Identification and Their Correlation in Three Oil Palm Varieties under Cold Stress
2.3. GO and Functional Classification and Correlation Analysis of DEGs and DEPs for Three Oil Palm Varieties in Response to Low-Temperature Stress
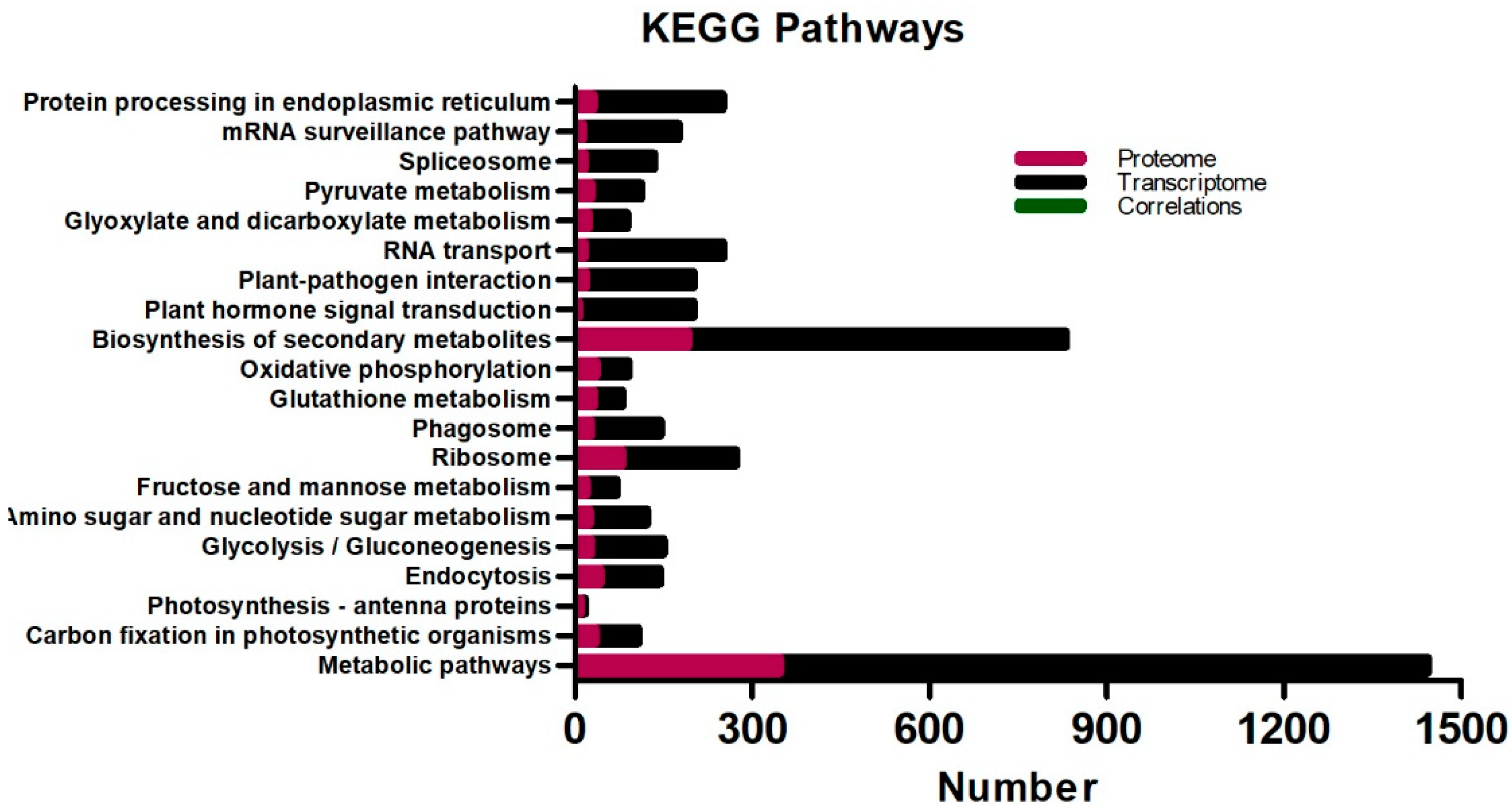
2.4. KEGG Pathway Enrichment and Correlation Analysis of Oil Palm DEGs and DEPs in Response to Low-Temperature Stress
2.5. Identification of Transcription Factors (TFs) in Three Oil Palm Varieties in Response to Low-Temperature Stress
2.6. Stress-Responsive DEPs in Three Oil Palm Varieties in Response to Low-Temperature Stress
2.7. DEPs Related to Respiration in Three Oil Palm Varieties in Response to Low-Temperature Stress
2.8. DEPs Related to Photosynthesis in Three Oil Palm Varieties under Low-Temperature Stress
3. Discussion
3.1. General Features and Correlation of the Transcriptome and Proteome of Three Oil Palm Varieties in Response to Low-Temperature Stress
3.2. Transcription Factors Regulation and Increased Abundance of Stress-Responsive Proteins among Three Oil Palm Varieties
3.3. Increased Abundance of Photosynthesis Proteins in BE Compared to Other Oil palm Varieties in Response to Low-Temperature Stress
3.4. Decreased Abundance of Respiratory Proteins in BE Variety under Low-Temperature Stress
4. Materials and Methods
4.1. Plant Materials and Low-Temperature Stress Treatments
4.2. RNA Extraction and RNA-seq Analysis in Oil Palm Varieties
4.3. Protein Extraction, Trypsin Digestion, and iTRAQ Labelling
4.4. Peptide Fractionation and LC-MS/MS Analysis
4.5. Identification and Quantification of Proteins
4.6. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mahlia, T.M.I.; Ismail, N.; Hossain, N.; Silitonga, A.S.; Shamsuddin, A.H. Palm oil and its wastes as bioenergy sources: A comprehensive review. Environ. Pollut. Res. 2019, 26, 14849–14866. [Google Scholar] [CrossRef] [PubMed]
- Bittencourt, C.B.; Carvalho da Silva, T.L.; Rodrigues Neto, J.C.; Vieira, L.R.; Leão, A.P.; de Aquino Ribeiro, J.A.; Abdelnur, P.V.; de Sousa, C.A.F.; Souza, M.T., Jr. Insights from a Multi-Omics Integration (MOI) Study in Oil Palm (Elaeis guineensis Jacq.) Response to Abiotic Stresses: Part One—Salinity. Plants 2022, 11, 1755. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.; Iqbal, A.; Qadri, R.; Shi, P.; Wang, Y.; Wu, Y.; Fan, H.; Wu, G. Correlation analysis of cold-related gene expression with physiological and biochemical indicators under cold stress in oil palm. PLoS ONE 2019, 14, e0225768. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Xiao, Y.; Xia, W.; Mason, A.S.; Yang, Y.; Ma, Z.; Peng, M. RNA-seq analysis of oil palm under cold stress reveals a different C-repeat binding factor (CBF) mediated gene expression pattern in Elaeis guineensis compared to other species. PLoS ONE 2014, 9, e114482. [Google Scholar] [CrossRef] [PubMed]
- Corley, R.H.V.; Tinker, P.B. Vegetative propagation and biotechnology. In The Oil Palm, 4th ed.; Blackwell Science: Oxford, UK, 2003; pp. 201–215. [Google Scholar]
- Yang, Y.; Saand, M.A.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Fan, H.; Wang, F. iTRAQ-based comparative proteomic analysis of two coconut varieties reveals aromatic coconut cold-sensitive in response to low temperature. J. Proteome 2020, 220, 103766. [Google Scholar] [CrossRef]
- Browse, J.; Xin, Z. Temperature sensing and cold acclimation. Curr. Opin. Plant Biol. 2001, 4, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ye, C.; Wang, M.; Chu, G. Identification of cold-stress responsive proteins in Anabasis aphylla seedlings via the iTRAQ proteomics technique. J. Plant Interact. 2017, 12, 505–519. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, N.; Deswal, R. The molecular biology of the low-temperature response in plants. Bioessays 2005, 27, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Lukatkin, A.S. Contribution of oxidative stress to the development of cold-induced damage to leaves of chilling-sensitive plants: 2. The activity of antioxidant enzymes during plant chilling. Russ. J. Plant Physiol. 2002, 49, 782–788. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, L.; Zhao, L.; Ren, Y.; Cui, D.; Chen, J.; Wang, Y.; Yu, P.; Chen, F. iTRAQ and virus-induced gene silencing revealed three proteins involved in cold response in bread wheat. Sci. Rep. 2017, 7, 7524. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stress. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of multi-omics technologies for crop improvement. Front. Plant Sci. 2021, 12, 1846. [Google Scholar] [CrossRef]
- Kok, S.Y.; Namasivayam, P.; Ee, G.C.L.; Ong-Abdullah, M. Comparative proteomic analysis of oil palm (Elaeis guineensis Jacq.) during early fruit development. J. Proteome 2021, 232, 104052. [Google Scholar] [CrossRef] [PubMed]
- Lau, B.Y.C.; Morton, D.J.; Deb-Choudhury, S.; Clerens, S.; Dyer, J.M.; Ramli, U.S. Differential expression analysis of oil palm fatty acid biosynthetic enzymes with gel-free quantitative proteomics. J. Oil Palm Res. 2017, 29, 23–34. [Google Scholar]
- Lau, B.Y.C.; Othman, A.; Ramli, U.S. Application of Proteomics Technologies in Oil Palm Research. Protein J. 2018, 37, 473–499. [Google Scholar] [CrossRef] [PubMed]
- Loei, H.; Lim, J.; Tan, M.; Lim, T.K.; Lin, Q.S.; Chew, F.T.; Kulaveerasingam, H.; Chung, M.C. Proteomic analysis of the oil palm fruit mesocarp reveals elevated oxidative phosphorylation activity is critical for increased storage oil production. J. Proteome Res. 2013, 12, 5096–5109. [Google Scholar] [CrossRef] [PubMed]
- Shearman, J.R.; Jantasuriyarat, C.; Sangsrakru, D.; Yoocha, T.; Vannavichit, A.; Tragoonrung, S.; Tangphatsornruang, S. Transcriptome analysis of normal and mantled developing oil palm flower and fruit. Genomics 2013, 101, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Othman, N.Q.; Sulaiman, S.; Lee, Y.P.; Tan, J.S. Transcriptomic data of mature oil palm basal trunk tissue infected with Ganoderma boninense. Data Brief 2019, 25, 104288. [Google Scholar] [CrossRef] [PubMed]
- Dussert, S.; Guerin, C.; Andersson, M.; Joët, T.; Tranbarger, T.J.; Pizot, M.; Sarah, G.; Omore, A.; Durand-Gasselin, T.; Morcillo, F. Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol. 2013, 162, 1337–1358. [Google Scholar] [CrossRef]
- Zhang, A.; Jin, L.; Yarra, R.; Cao, H.; Chen, P.; John Martin, J.J. Transcriptome analysis reveals key developmental and metabolic regulatory aspects of oil palm (Elaeis guineensis Jacq.) during zygotic embryo development. BMC Plant Biol. 2022, 22, 112. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 16, 2533–2544. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, Z.; Zhang, Y.; Chai, L.; Yi, H.; Deng, X. An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J. Exp. Bot. 2014, 65, 1651–1671. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Qin, Z.; Zhang, G.; Guo, Y.; Huang, J. Integration of the proteome and transcriptome reveals multiple levels of gene regulation in the rice dl2 mutant. Front. Plant Sci. 2015, 6, 351. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Htwe, Y.M.; Li, J.; Shi, P.; Zhang, D.; Zhao, Z.; Ihase, L.O. Integrative omics analysis on phytohormones involved in oil palm seed germination. BMC Plant Biol. 2019, 19, 363. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, P.; Aswathy, G.M.; Kumar, K.S.; Masilamani, P.; Kumar, V.; Ravi, V. Oil palm (Elaeis guineensis) genetic resources for abiotic stress tolerance: A review. Indian J. Agric. Sci. 2017, 87, 571–579. [Google Scholar]
- Barcelos, E.; Rios, S.D.A.; Cunha, R.N.; Lopes, R.; Motoike, S.Y.; Babiychuk, E.; Skirycz, A.; Kushnir, S. Oil palm natural diversity and the potential for yield improvement. Front. Plant Sci. 2015, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.N.; Babu, B.K.; Mathur, R.K.; Ramajayam, D. Genetic engineering of oil palm. In Genetic Engineering of Horticultural Crops; Rout, G.R., Peter, K.V., Eds.; Academic Press/Elsevier: Cambridge, MA, USA, 2018; pp. 169–191. [Google Scholar]
- Chapman, K.R.; Escobar, R.; Griffee, P. Cold tolerant or altitude adapted oil palm hybrid development initiatives in Asia/Pacific region. AU J.T. 2003, 6, 134–138. [Google Scholar]
- Silva, P.A.; Cosme, V.S.; Rodrigues, K.C.; Detmann, K.S.; Leão, F.M.; Cunha, R.L.; Festucci Buselli, R.A.; DaMatta, F.M.; Pinheiro, H.A. Drought tolerance in two oil palm hybrids as related to adjustments in carbon metabolism and vegetative growth. Act. Physiol. Planta 2017, 39, 58. [Google Scholar] [CrossRef]
- Bhat, K.A.; Mahajan, R.; Pakhtoon, M.M.; Urwat, U.; Bashir, Z.; Shah, A.A.; Agrawal, A.; Bhat, B.; Sofi, P.A.; Masi, A.; et al. Low Temperature Stress Tolerance: An Insight Into the Omics Approaches for Legume Crops. Front. Plant Sci. 2022, 13, 888710. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Fakher, B.; Ashraf, M.A.; Cheng, Y.; Wang, B.; Qin, Y. Plant low-temperature stress: Signaling and response. Agronomy 2022, 12, 702. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Petruccelli, R.; Bartolini, G.; Ganino, T.; Zelasco, S.; Lombardo, L.; Perri, E.; Durante, M.; Bernardi, R. Cold Stress, Freezing Adaptation, Varietal Susceptibility of Olea europaea L. A Review. Plants 2022, 11, 1367. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBF s in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Shen, X.; Li, H.; Liu, X.; Niu, C.; et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.X.; Li, X.Y.; Chen, J.W. Comparative transcriptome profiling of freezing stress responses in loquat (Eriobotrya japonica) fruitlets. J. Plant Res. 2017, 130, 893–907. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Kumar, R. The expanding roles of APETALA2/Ethylene Responsive Factors and their potential applications in crop improvement. Brief. Funct. Genome 2019, 18, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zheng, G.; Dong, X.; Li, H.; Liu, S.; Wang, Y.; Liu, Z. Integration of transcriptome and proteome analysis reveals the mechanism of freezing tolerance in winter rapeseed. Plant Growth Regul. 2022, 96, 103–118. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zhao, L.; Wang, Y.; Shen, J.; Zhang, Y.; Jia, S.; Li, Y.; Ding, Z. Integrated RNA-Seq and sRNA-Seq analysis identifies chilling and freezing responsive key molecular players and pathways in tea plant (Camellia sinensis). PLoS ONE 2015, 10, e0125031. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- Huang, G.Q.; Li, W.; Zhou, W.; Zhang, J.M.; Li, D.D.; Gong, S.Y.; Li, X.B. Seven cotton genes encoding putative NAC domain proteins are preferentially expressed in roots and in responses to abiotic stress during root development. Plant Growth Regul. 2013, 71, 101–112. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Lee, B.H.; Henderson, D.A.; Zhu, J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, H.; Ren, J.; Dong, J.; Zhao, X.; Wang, X.; Wang, J.; Zhong, C.; Zhao, S.; Liu, X.; et al. Comparative transcriptome-based mining and expression profiling of transcription factors related to cold tolerance in peanut. Int. J. Mol. Sci. 2020, 21, 1921. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Nayak, D.K.; Behera, L.; Mohapatra, T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019, 19, 352. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, X.; Cheng, F.; Cao, H.; Liang, H.; Lu, J.; Kong, Q.; Bie, Z. iTRAQ-based quantitative proteomics analysis of cold stress-induced mechanisms in grafted watermelon seedlings. J. Proteome 2019, 192, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.T.; Ma, L.; Duan, W.; Wang, B.C.; Li, J.H.; Xu, H.G.; Yan, X.Q.; Yan, B.F.; Li, S.H.; Wang, L.J. Differential proteomic analysis of grapevine leaves by iTRAQ reveals responses to heat stress and subsequent recovery. BMC Plant Biol. 2014, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Rurek, M.; Czołpińska, M.; Pawłowski, T.A.; Staszak, A.M.; Nowak, W.; Krzesiński, W.; Spiżewski, T. Mitochondrial biogenesis in diverse cauliflower cultivars under mild and severe drought. Impaired coordination of selected transcript and proteomic responses, and regulation of various multifunctional proteins. Int. J. Mol. Sci. 2018, 19, 1130. [Google Scholar] [CrossRef]
- Wu, X.; Yan, J.; Wu, Y.; Zhang, H.; Mo, S.; Xu, X.; Zhou, F.; Ding, H. Proteomic analysis by iTRAQ-PRM provides integrated insight into mechanisms of resistance in pepper to Bemisia tabaci (Gennadius). BMC Plant Biol. 2019, 19, 270. [Google Scholar] [CrossRef]
- Long, R.; Gao, Y.; Sun, H.; Zhang, T.; Li, X.; Li, M.; Sun, Y.; Kang, J.; Wang, Z.; Ding, W.; et al. Quantitative proteomic analysis using iTRAQ to identify salt-responsive proteins during the germination stage of two Medicago species. Sci. Rep. 2018, 8, 9553. [Google Scholar] [CrossRef]
- Huh, S.M.; Noh, E.K.; Kim, H.G.; Jeon, B.W.; Bae, K.; Hu, H.C.; Kwak, J.M.; Park, O.K. Arabidopsis annexins AnnAt1 and AnnAt4 interact with each other and regulate drought and salt stress responses. Plant Cell Physiol. 2010, 51, 1499–1514. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Cheng, T.; Chen, J.; Ef, A.; Wang, P.; Wang, G.; Hu, X.; Shi, J. Quantitative proteomics analysis reveals that S-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta 2015, 242, 1361–1390. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef]
- Wang, X.; Shan, X.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Han, J.; Xue, C.; Yuan, Y. iTRAQ-based quantitative proteomic analysis reveals new metabolic pathways responding to chilling stress in maize seedlings. J. Proteom. 2016, 146, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Dixon, D.P.; Walbot, V. Plant glutathione S-transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000, 5, 193–198. [Google Scholar] [CrossRef]
- Song, W.; Tang, F.; Cai, W.; Zhang, Q.; Zhou, F.; Ning, M.; Tian, H.; Shan, C. iTRAQ-based quantitative proteomics analysis of cantaloupe (Cucumis melo var. saccharinus) after cold storage. BMC Genom. 2020, 21, 390. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J. Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 2011, 15, 1129–1159. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Huang, X.; Zhang, L.; Zhao, N.; Yang, D.; Zhang, K. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2009, 85, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Rajan, V.B.V.; D’Silva, P. Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genom. 2009, 9, 433–446. [Google Scholar] [CrossRef]
- Lin, Q.; Xie, Y.; Guan, W.; Duan, Y.; Wang, Z.; Sun, C. Combined transcriptomic and proteomic analysis of cold stress induced sugar accumulation and heat shock proteins expression during postharvest potato tuber storage. Food Chem. 2019, 297, 124991. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Cheung, M.Y.; Li, M.W.; Fu, Y.; Sun, Z.; Sun, S.M.; Lam, H.M. Rice hypersensitive induced reaction protein 1 (OsHIR1) associates with plasma membrane and triggers hypersensitive cell death. BMC Plant Biol. 2010, 10, 290. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yao, L.; Li, B.; Meng, Y.; Ma, X.; Lai, Y.; Si, E.; Ren, P.; Yang, K.; Shang, X.; et al. Comparative proteomic analysis of cultured suspension cells of the halophyte Halogeton glomeratus by iTRAQ provides insights into response mechanisms to salt stress. Front. Plant Sci. 2016, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.S.; Tang, L.; Li, Y.Y.; Wang, W.C.; Yang, Z.; Li, X.G.; Zeng, C. Differential proteomic analysis reveals the mechanism of Musa paradisiaca responding to salt stress. Mol. Biol. Rep. 2019, 46, 1057–1068. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Ruelland, E.; Vaultier, M.N.; Zachowski, A.; Hurry, V. Cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar]
- Li, Q.; Chang, R.; Sun, Y.; Li, B. iTRAQ-based quantitative proteomic analysis of Spirulina platensis in response to low temperature stress. PLoS ONE 2016, 11, e0166876. [Google Scholar] [CrossRef] [PubMed]
- Gharechahi, J.; Alizadeh, H.; Naghavi, M.R.; Sharifi, G. A proteomic analysis to identify cold acclimation associated proteins in wild wheat (Triticum urartu L.). Mol. Biol. Rep. 2014, 41, 3897–3905. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Han, G.; Shang, C.; Li, J.; Zhang, H.; Liu, F.; Wang, J.; Liu, H.; Zhang, Y. Proteomic analyses reveal differences in cold acclimation mechanisms in freezing-tolerant and freezing-sensitive cultivars of alfalfa. Front. Plant Sci. 2015, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Gołębiowska-Pikania, G.; Kopeć, P.; Surówka, E.; Krzewska, M.; Dubas, E.; Nowicka, A.; Rapacz, M.; Wójcik-Jagła, M.; Malaga, S.; Żur, I. Changes in protein abundance and activity involved in freezing tolerance acquisition in winter barley (Hordeum vulgare L.). J. Proteom. 2017, 169, 58–72. [Google Scholar] [CrossRef]
- Xu, Y.; Zeng, X.; Wu, J.; Zhang, F.; Li, C.; Jiang, J.; Wang, Y.; Sun, W. iTRAQ-based quantitative proteome revealed metabolic changes in winter turnip rape (Brassica rapa L.) under cold stress. Int. J. Mol. Sci. 2018, 19, 3346. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Qin, Y.Y.; Luo, M.R.; Li, Z.; Ma, D.N.; Wang, W.H.; Zheng, H.L. Proteome analysis reveals a systematic response of cold-acclimated seedlings of an exotic mangrove plant Sonneratia apetala to chilling stress. J. Proteom. 2021, 248, 104349. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. iTRAQ-based comparative proteomic analysis of the roots of TWO winter turnip rapes (Brassica rapa L.) with different freezing-tolerance. Int. J. Mol. Sci. 2018, 19, 4077. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, J.; Zack, D.J.; Qian, J. Computational analysis of tissue-specific combinatorial gene regulation: Predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006, 34, 4925–4936. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNAseq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saand, M.A.; Li, J.; Wu, Y.; Zhou, L.; Cao, H.; Yang, Y. Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress. Int. J. Mol. Sci. 2022, 23, 14926. https://doi.org/10.3390/ijms232314926
Saand MA, Li J, Wu Y, Zhou L, Cao H, Yang Y. Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress. International Journal of Molecular Sciences. 2022; 23(23):14926. https://doi.org/10.3390/ijms232314926
Chicago/Turabian StyleSaand, Mumtaz Ali, Jing Li, Yi Wu, Lixia Zhou, Hongxing Cao, and Yaodong Yang. 2022. "Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress" International Journal of Molecular Sciences 23, no. 23: 14926. https://doi.org/10.3390/ijms232314926
APA StyleSaand, M. A., Li, J., Wu, Y., Zhou, L., Cao, H., & Yang, Y. (2022). Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress. International Journal of Molecular Sciences, 23(23), 14926. https://doi.org/10.3390/ijms232314926






