An Intestinal Symbiotic Bacterial Strain of Oscheius chongmingensis Modulates Host Viability at Both Global and Post-Transcriptional Levels
Abstract
:1. Introduction
2. Results and Discussion
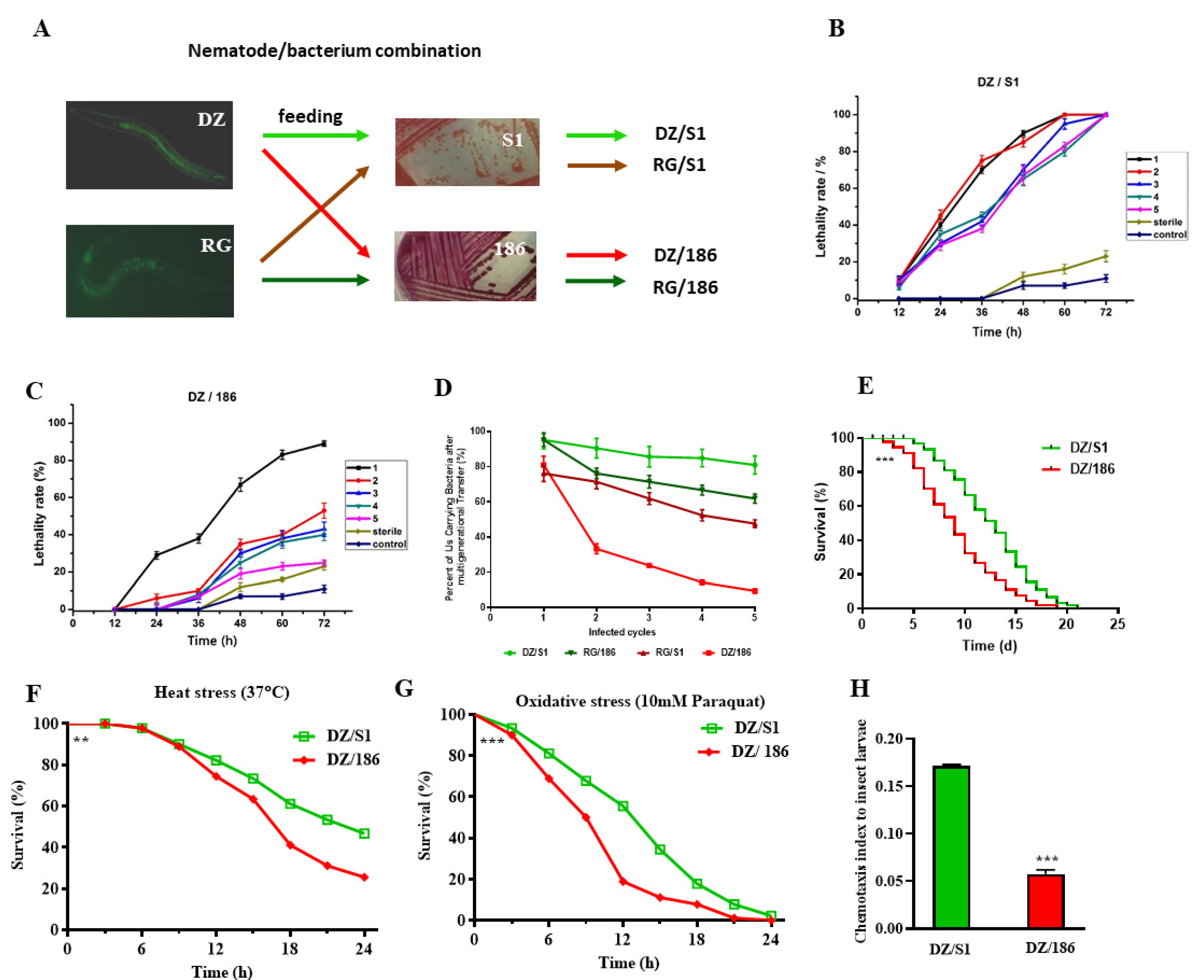
2.1. The Pathogenicity of Oscheius chongmingensis Is Correlated with Symbiotic Bacteria Carriage Rate and Declines Sharply with the Non-Native Strain Serratia Nematodiphila DR186 after Multigenerational Transfer
2.2. The Non-Native Bacterial Strain 186 Reduces the Fecundity of O. chongmingensis, Shortens Its Longevity, Impairs Its Stress Tolerance, and Decreases Its Olfactory Chemotaxis
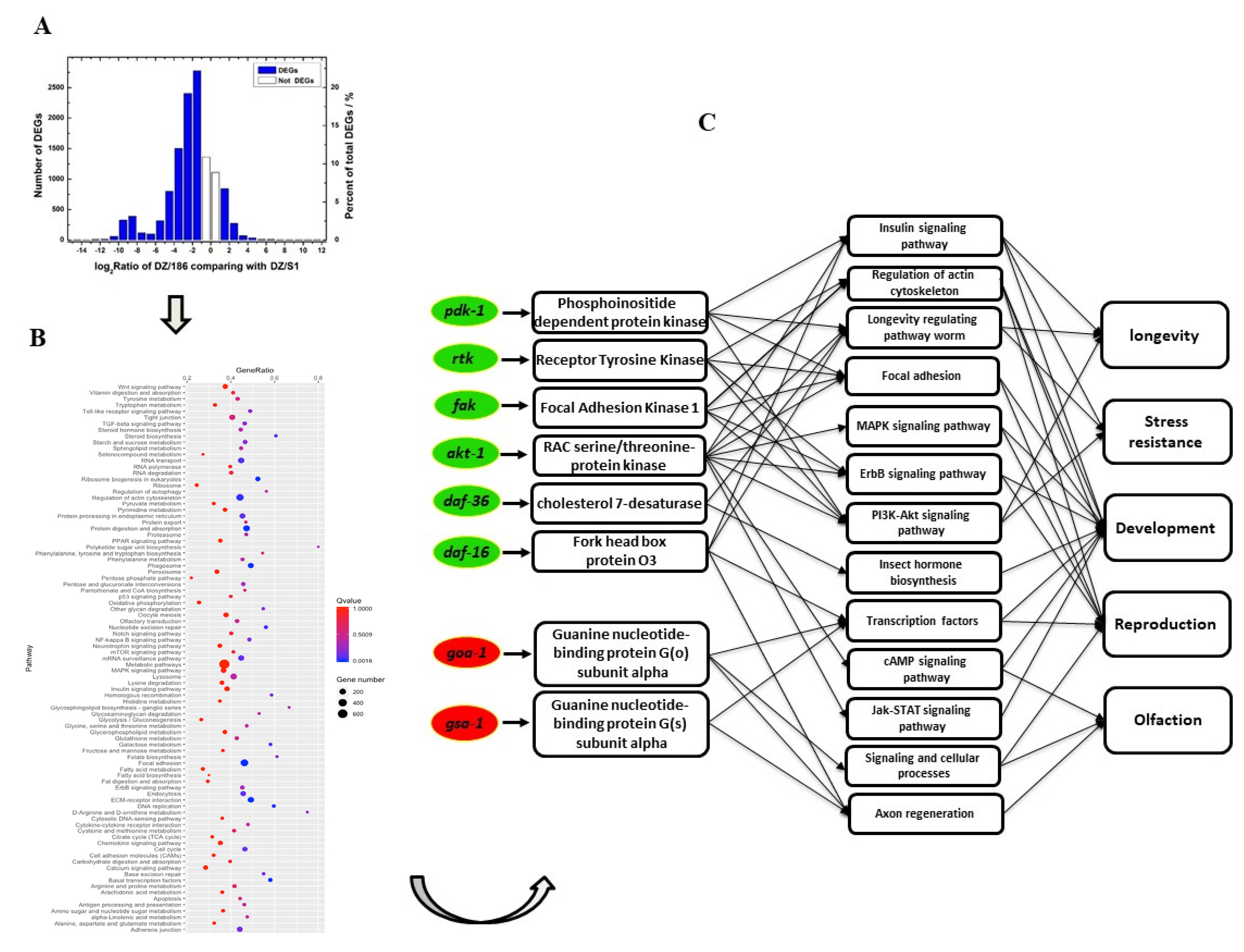
2.3. Expression Levels of the Majority of Genes Involved in Biological Processes and Related Pathways in O. chongmingensis Associated with Strain 186 Are Downregulated Compared with Those in the Native Combination
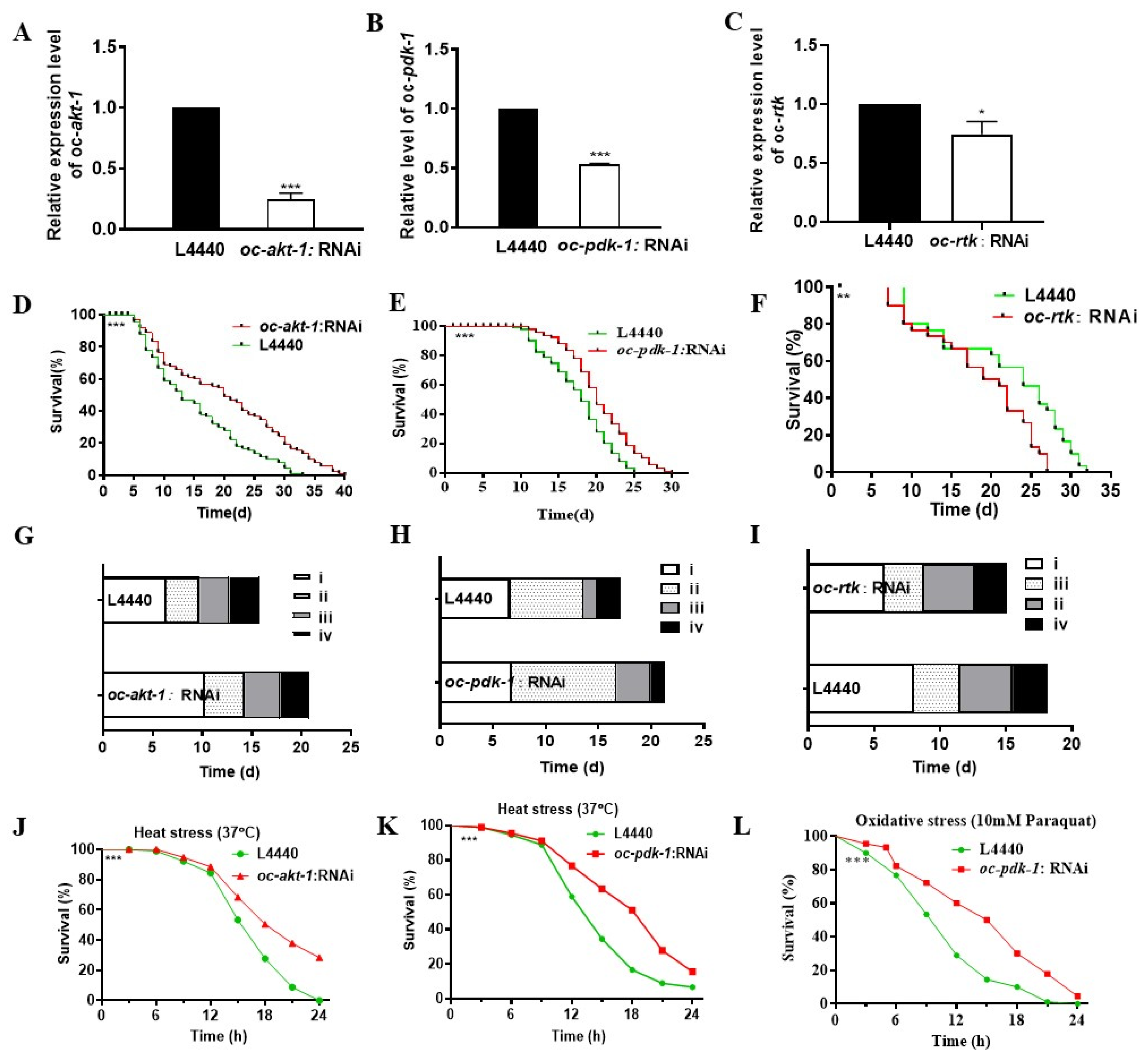
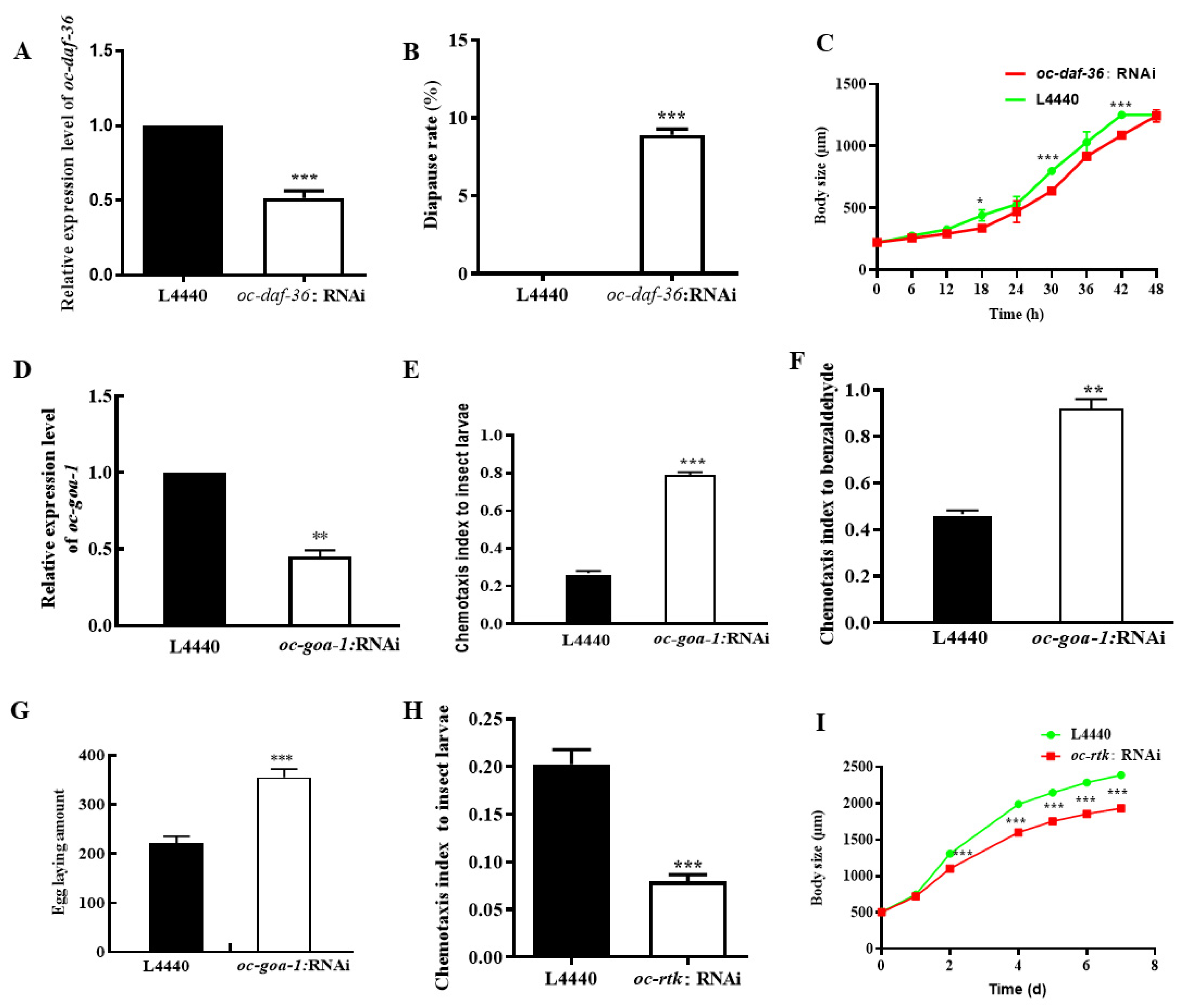
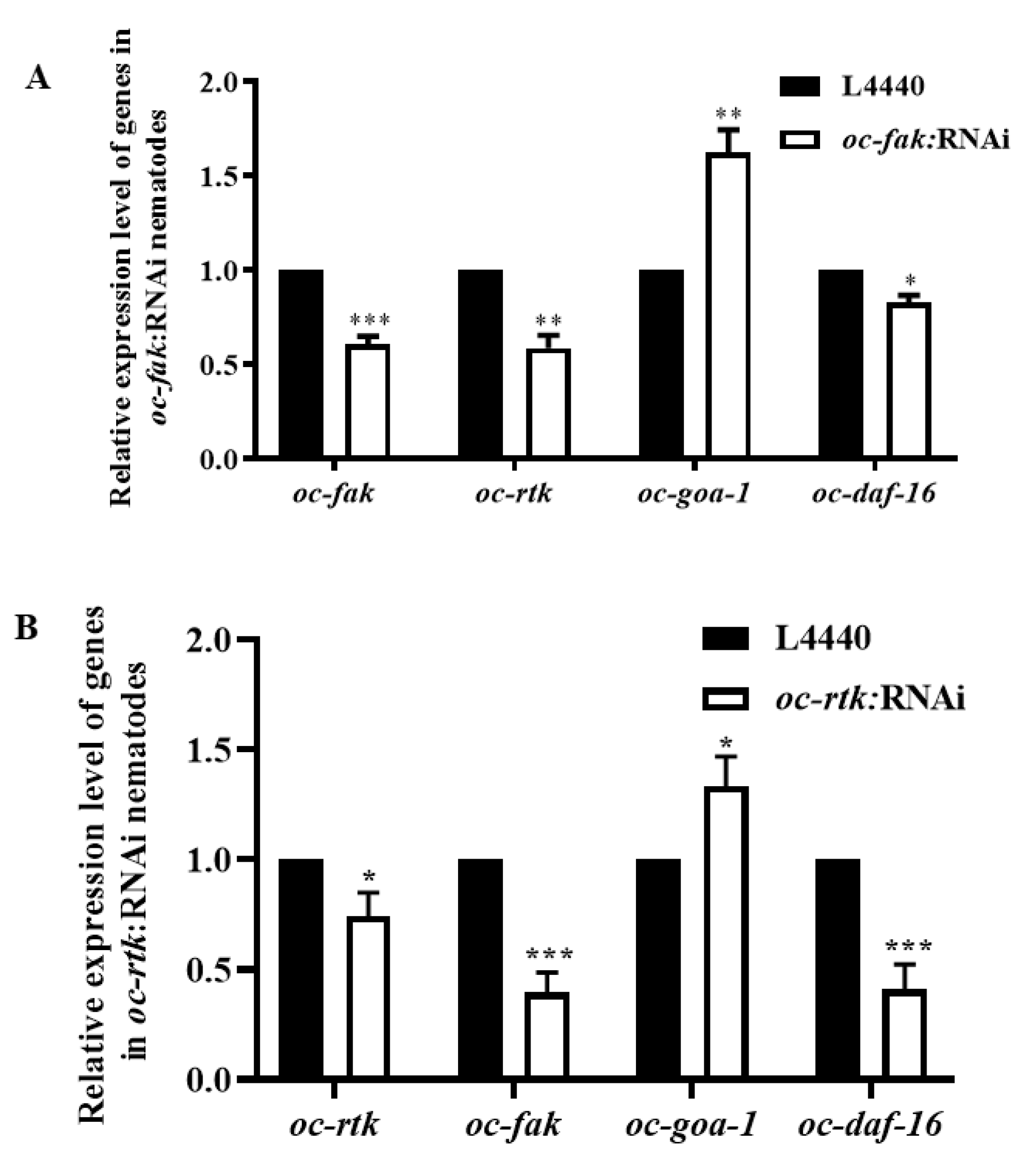
2.4. Downregulated Oc-akt-1, Oc-fak, Oc-rtk, and Oc-pdk-1 as well as Three Transcription Factors, Oc-daf-16, Oc-goa-1, and Oc-gsa-1, Were Involved in the Regulatory Mechanism of Decreased Viability of O. chongmingensis
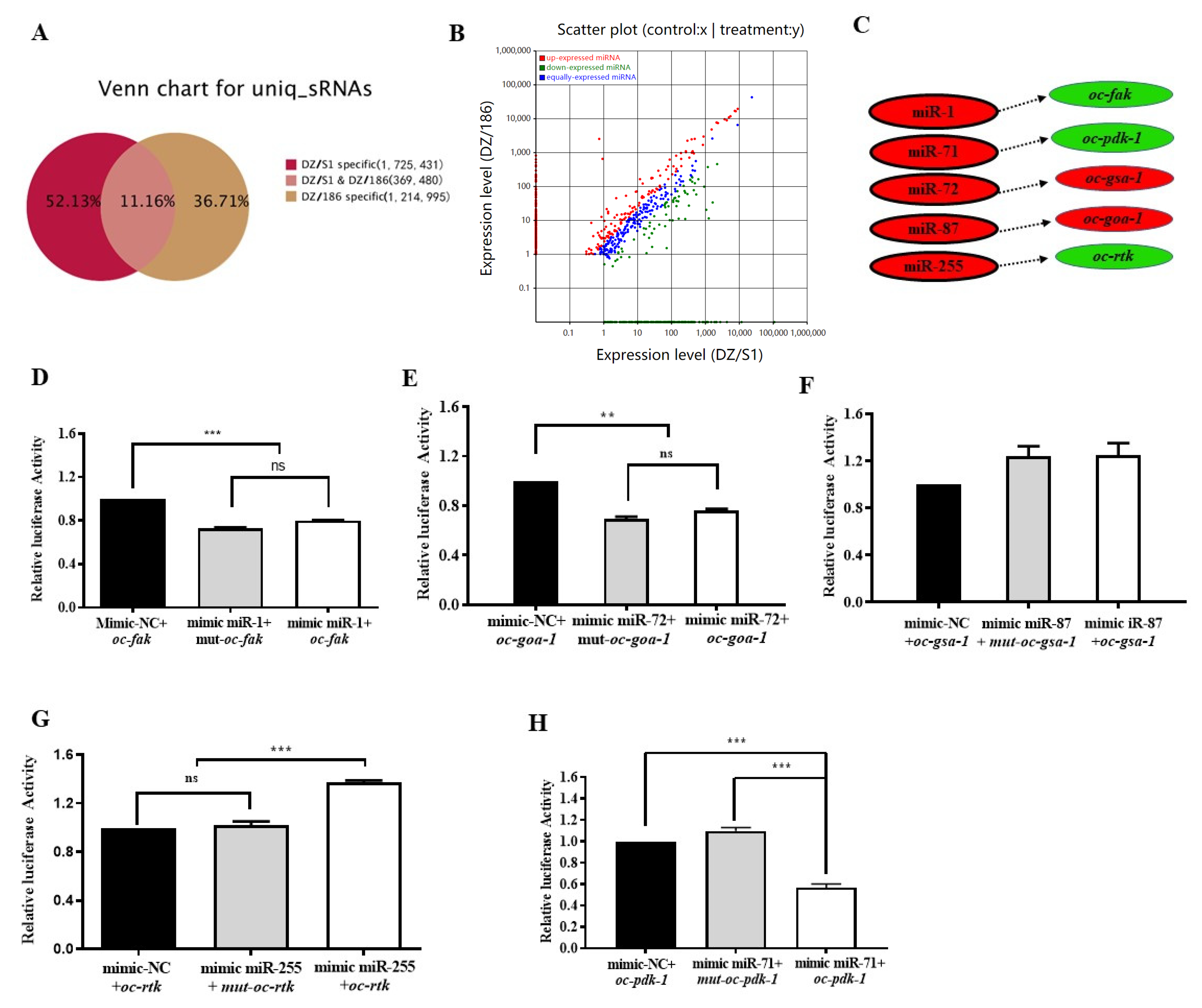
2.5. High-Throughput Sequencing Indicates More Than 88.84% MicroRNAs in the Two Combinations Were Significantly Differentially Expressed
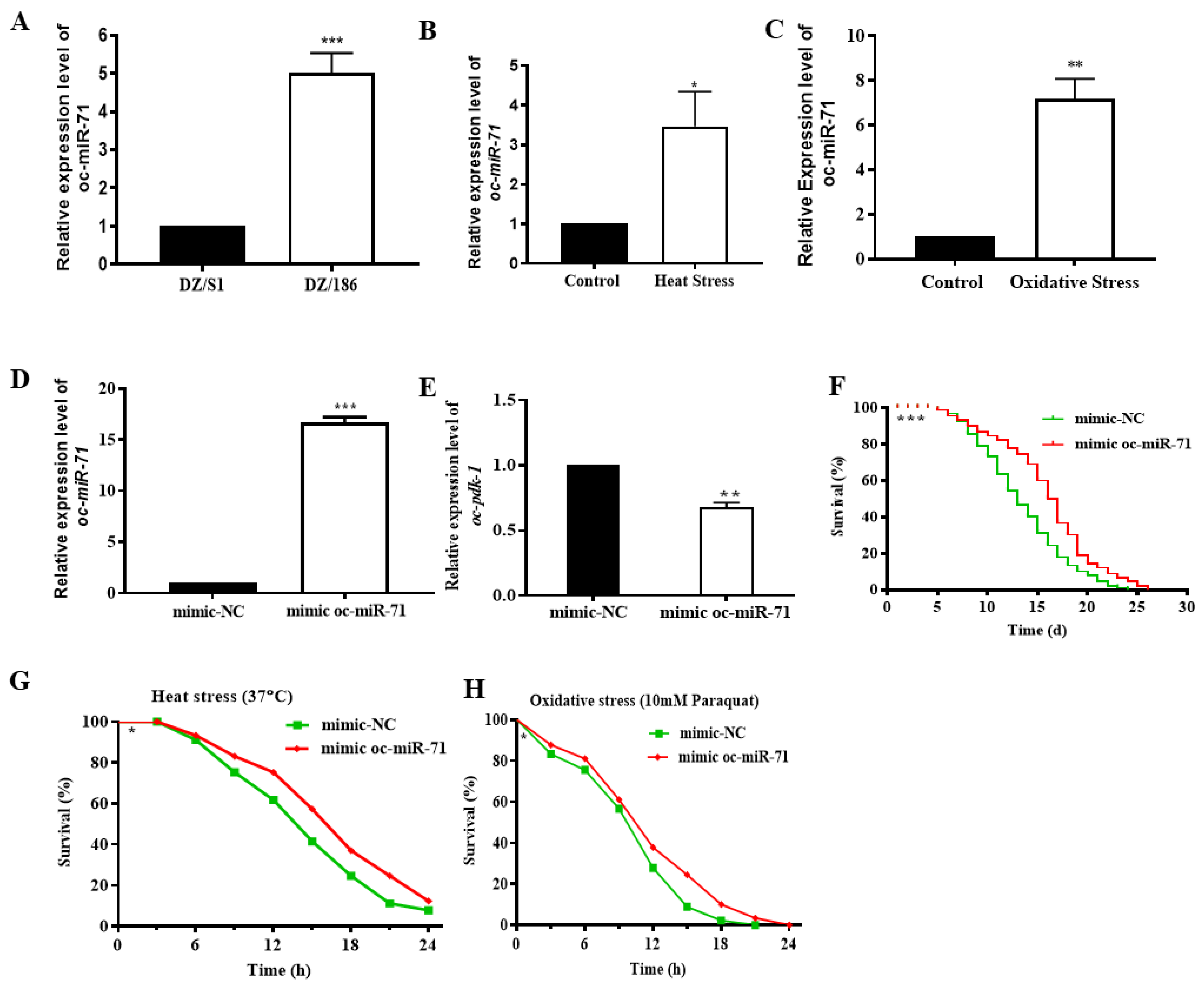
2.6. Oc-miR-71 Mediates the Longevity and Stress Tolerance of O. chongmingensis by Targeting Oc-pdk-1
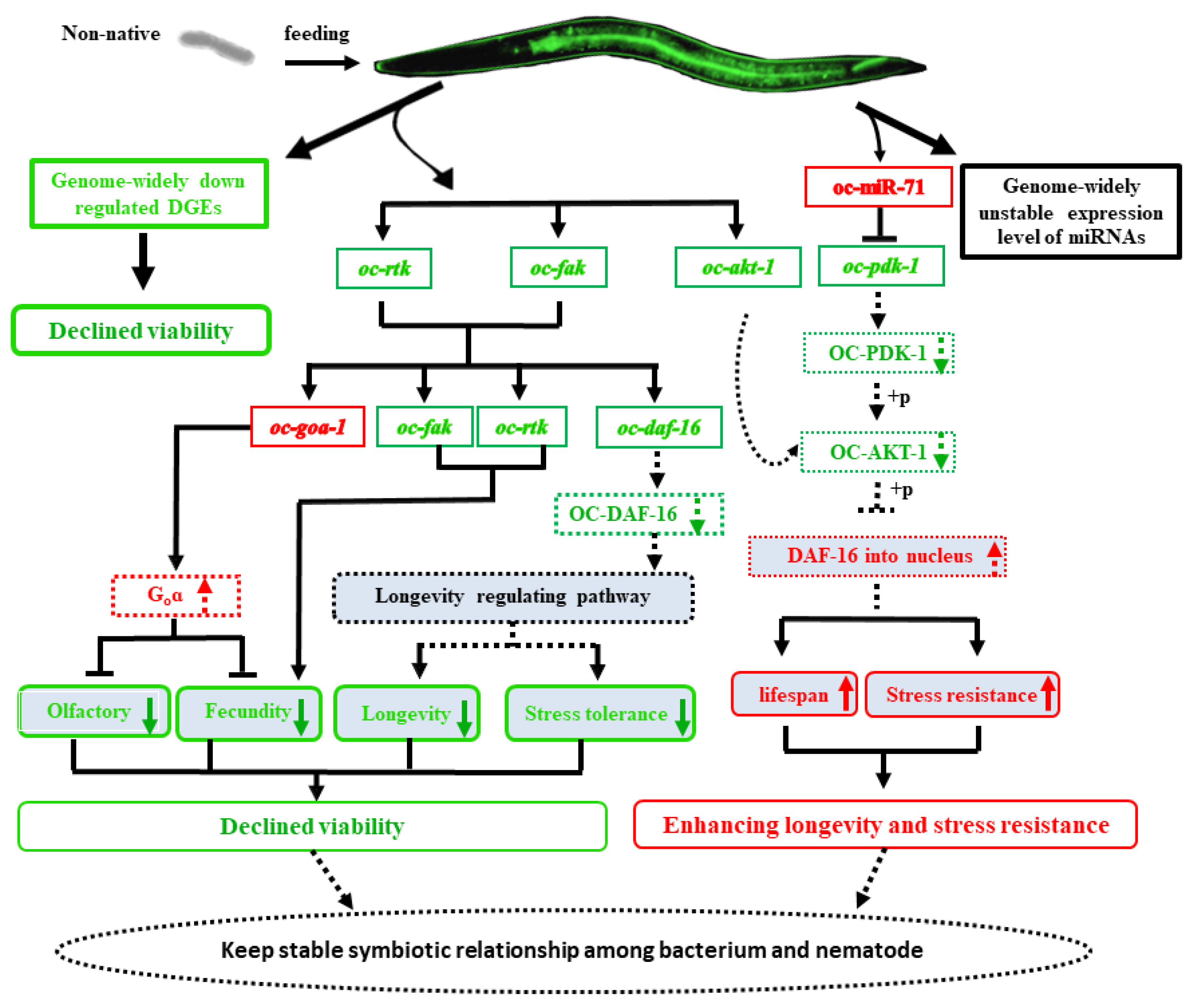
2.7. The Non-Native Intestinal Bacterial Strain 186 Mediates O. Chongmingensis’ Viability and Symbiotic Regulatory Response at Both Global and Post-Transcriptional Levels
3. Materials and Methods
3.1. Nematodes and Symbiotic Bacterial Strains
3.2. Axenic IJs and Combinations of Monoxenic Nematodes with Native and Non-Native Bacterial Strains
3.3. Pathogenicity of IJs from Different Infection Cycles of the Four Monoxenic Combinations against Waxworms
3.4. Bacterial Carriage Rate of the Four Nematode–Bacterium Combinations
3.5. Influence of S. Nematodiphila Strains on the Development and Reproduction of Two Species of Oscheius
3.6. Longevity and Stress Tolerance Analysis
3.7. Determination of Olfactory Chemotaxis by Nematodes
3.8. Screening for Differentially Expressed Genes and Functional Classification
3.9. RT-qPCR Validation of DEG Expression Levels
3.10. SRNA Library Construction and Differential Expression Analysis of MiRNAs in DZ/S1 and DZ/186
3.11. Bioinformatic Prediction and Annotation of Potential Targets of Differentially Expressed MiRNAs
3.12. Luciferase Reporter Assay Validation of In Vitro MiRNA Targets
3.13. Transfection of Mimic MiRNA into Nematode and Function Validation In Vivo
3.14. RNAi by Feeding
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blacher, E.; Bashiardes, S.; Shapiro, H.; Rothschild, D.; Mor, U.; Dori-Bachash, M.; Kleimeyer, C.; Moresi, C.; Harnik, Y.; Zur, M.; et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 2019, 572, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Woelk, C.H.; Snyder, A. Modulating gut microbiota to treat cancer. Science 2021, 371, 573–574. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Sivaramakrishnan, P.; Lin, C.-C.J.; Neve, I.A.; He, J.; Tay, L.W.R.; Sowa, J.N.; Sizovs, A.; Du, G.; Wang, J.; et al. Microbial Genetic Composition Tunes Host Longevity. Cell 2017, 169, 1249–1262.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, T.A.; Quintaneiro, L.M.; Norvaisas, P.; Lui, P.P.; Wilson, M.P.; Leung, K.-Y.; Herrera-Dominguez, L.; Sudiwala, S.; Pessia, A.; Clayton, P.T.; et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell 2017, 169, 442–456.e18. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Sun, G.; Feng, T.; Zhang, J.; Huang, X.; Wang, T.; Xie, Z.; Chu, X.; Yang, J.; Wang, H.; et al. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression. Cell Res. 2019, 29, 787–803. [Google Scholar] [CrossRef] [Green Version]
- Le Roy, T.; Lécuyer, E.; Chassaing, B.; Rhimi, M.; Lhomme, M.; Boudebbouze, S.; Ichou, F.; Haro Barceló, J.; Huby, T.; Guerin, M.; et al. The Intestinal Microbiota Regulates Host Cholesterol Homeostasis. BMC Biol. 2019, 17, 94. [Google Scholar] [CrossRef] [Green Version]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef] [Green Version]
- Akhurst, R.J. Morphological and Functional Dimorphism in Xenorhabdus spp., Bacteria Symbiotically Associated with the Insect Pathogenic Nematodes Neoaplectana and Heterorhabditis. Microbiology 1980, 121, 303–309. [Google Scholar] [CrossRef] [Green Version]
- Thomas, G.M.; Poinar, G.O., Jr. Xenorhabdus gen. nov., a genus of entomopathogenic, nematophilic bacteria of the family Enterobacteriaceae. Int. J. Syst. Evol. Microbiol. 1979, 29, 352–360. [Google Scholar] [CrossRef] [Green Version]
- Boemare, N.E.; Akhurst, R.J.; Mourant, R.G. DNA Relatedness between Xenorhabdus spp. (Enterobacteriaceae), Symbiotic Bacteria of Entomopathogenic Nematodes, and a Proposal to Transfer Xenorhabdus luminescens to a New Genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 1993, 43, 249–255. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Xu, M.; Sun, J.; Yang, S.; An, X.; Gao, G.; Lin, M.; Lai, R.; He, Z.; et al. Heterorhabditidoides chongmingensis gen. nov., sp. nov. (Rhabditida: Rhabditidae), a novel member of the entomopathogenic nematodes. J. Invertebr. Pathol. 2008, 98, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.X.; Yang, S.Y.; Xu, M.X.; Sun, J.; Liu, H.; Liu, J.R.; Liu, H.; Kan, F.; Sun, J.; Lai, R.; et al. Serratia nematodiphila sp. nov., associated symbiotically with the entomopathogenic nematode Heterorhabditidoides chongmingensis (Rhabditida: Rhabditidae). Int. J. Syst. Evol. Microbiol. 2009, 59, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Dillman, A.R.; Chaston, J.M.; Adams, B.J.; Ciche, T.A.; Goodrich-Blair, H.; Stock, S.P.; Sternberg, P.W. An Entomopathogenic Nematode by Any Other Name. PLoS Pathog. 2012, 8, e1002527. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.Y. Symbiotic specialization, stability and pathogenicity relationships among rhabditid entomopathogenic nematodes Heterorhabditidoides and their symbiotic bacteria. J. Nematol. 2014, 46, 259. [Google Scholar]
- Chandra, H.; Khandelwal, P.; Khattri, A.; Banerjee, N. Type 1 fimbriae of insecticidal bacterium Xenorhabdus nematophila is necessary for growth and colonization of its symbiotic host nematode Steinernema carpocapsiae. Environ. Microbiol. 2008, 10, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Chapuis, E.; Emelianoff, V.; Paulmier, V.; Le Brun, N.; Pagès, S.; Sicard, M.; Ferdy, J.-B. Manifold aspects of specificity in a nematode-bacterium mutualism. J. Evol. Biol. 2009, 22, 2104–2117. [Google Scholar] [CrossRef] [PubMed]
- Somvanshi, V.S.; Sloup, R.E.; Crawford, J.M.; Martin, A.R.; Heidt, A.J.; Kim, K.-S.; Clardy, J.; Ciche, T.A. A Single Promoter Inversion Switches Photorhabdus between Pathogenic and Mutualistic States. Science 2012, 337, 88–93. [Google Scholar] [CrossRef] [Green Version]
- Bedding, R.A.; Molyneux, A.S.; Akhurst, R.J. Heterorhabditis spp., Neoaplectana spp., and Steinernema kraussei: Interspecific and intraspecific differences in infectivity for insects. Exp. Parasitol. 1983, 55, 249–257. [Google Scholar] [CrossRef]
- Akhurst, R. Neoaplectana species: Specificity of association with bacteria of the genus Xenorhabdus. Exp. Parasitol. 1983, 55, 258–263. [Google Scholar] [CrossRef]
- Han, R.; Ehlers, R.-U. Pathogenicity, Development, and Reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under Axenic In Vivo Conditions. J. Invertebr. Pathol. 2000, 75, 55–58. [Google Scholar] [CrossRef]
- Cao, L.; Qiu, X.; Liu, X.; Liu, X.; Han, R. Nutrient potential of various Xenorhabdus and Photorhabdus bacteria for a free-living nematode Panagrellus redivivus. Nematology 2008, 10, 79–85. [Google Scholar] [CrossRef]
- Zhang, K.; Baiocchi, T.; Lu, D.; Chang, D.Z.; Dillman, A.R. Differentiating between scavengers and entomopathogenic nematodes: Which is Oscheius chongmingensis? J. Invertebr. Pathol. 2019, 167, 107245. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.J.; Fodor, A.; Koppenhöfer, H.S.; Stackebrandt, E.; Stock, S.P.; Klein, M.G. Biodiversity and systematics of nematode–bacterium entomopathogens. Biol. Control 2006, 37, 32–49. [Google Scholar] [CrossRef]
- Sicard, M.; Le Brun, N.; Pages, S.; Godelle, B.; Moulia, C. Effect of native Xenorhabdus on the fitness of their Steinernema hosts: Contrasting types of interaction. Parasitol. Res. 2003, 91, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Mitani, D.K.; Kaya, H.K.; Goodrich-Blair, H. Comparative study of the entomopathogenic nematode, Steinernema carpocapsae, reared on mutant and wild-type Xenorhabdus nematophila. Biol. Control 2004, 29, 382–391. [Google Scholar] [CrossRef]
- Gerritsen, L.J.M.; Smits, P.H. The influence of Photorhabdus luminescens strains and form vatiants on the reproduction and bacterial retention of Heterorhabditis megidis. Fundam. Appl. Nematol. 1997, 20, 317–322. [Google Scholar]
- Dillman, A.R.; Macchietto, M.; Porter, C.F.; Rogers, A.; Williams, B.; Antoshechkin, I.; Lee, M.-M.; Goodwin, Z.; Lu, X.; Lewis, E.E.; et al. Comparative genomics of Steinernema reveals deeply conserved gene regulatory networks. Genome Biol. 2015, 16, 200. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Adams, B.J.; Ciche, T.A.; Clifton, S.; Gaugler, R.; Kim, K.-S.; Spieth, J.; Sternberg, P.W.; Wilson, R.K.; Grewal, P.S. A Lover and a Fighter: The Genome Sequence of an Entomopathogenic Nematode Heterorhabditis bacteriophora. PLoS ONE 2013, 8, e69618. [Google Scholar] [CrossRef] [Green Version]
- Lefoulon, E.; McMullen, J.G.; Stock, S.P. Transcriptomic Analysis of Steinernema Nematodes Highlights Metabolic Costs Associated to Xenorhabdus Endosymbiont Association and Rearing Conditions. Front. Physiol. 2022, 13, 821845. [Google Scholar] [CrossRef]
- Neubacher, N.; Tobias, N.J.; Huber, M.; Cai, X.; Glatter, T.; Pidot, S.J.; Stinear, T.P.; Lütticke, A.L.; Papenfort, K.; Bode, H.B. Symbiosis, virulence and natural-product biosynthesis in entomopathogenic bacteria are regulated by a small RNA. Nat. Microbiol. 2020, 5, 1481–1489. [Google Scholar] [CrossRef]
- Roder, A.C.; Stock, S.P. Influence of Xenorhabdus (Gamma-Proteobacteria: Enterobacteriaceae) symbionts on gonad postembryonic development in Steinernema (Nematoda: Steinernematidae) nematodes. J. Invertebr. Pathol. 2018, 153, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C. The plasticity of aging: Insights from long–lived mutants. Cell 2005, 120, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Wolkow, C.A.; Kimura, K.D.; Lee, M.-S.; Ruvkun, G. Regulation of C. elegans Life-Span by Insulinlike Signaling in the Nervous System. Science 2000, 290, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, T.; Iino, Y. Neural circuit-dependent odor adaptation in C. elegans is regulated by the Ras-MAPK pathway. Genes Cells 2005, 10, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Jones, F.S.; Jones, P.L. The tenascin family of ECM glycoproteins: Structure, function, and regulation during embryonic development and tissue remodeling. Dev. Dyn. 2000, 218, 235–259. [Google Scholar] [CrossRef]
- Zhan, C.; Xiong, Y.; Zhang, T.; Zhang, K. Functional identification of daf-16 in Heterorhabditidoides chongmingensis (Rhabditida: Rhabditidae). Acta Entomol. Sin. 2018, 61, 932–940. [Google Scholar]
- Guo, D.; Bao, H.; Dou, Z.; Zhang, K. Functional identification of fak gene in Heterorhabditidoides chongmingensis. J. Nanjing Agric. Univ. 2021, 44, 287–295. [Google Scholar] [CrossRef]
- Ségalat, L.; Elkes, D.A.; Kaplan, J.M. Modulation of Serotonin-Controlled Behaviors by Go in Caenorhabditis elegans. Science 1995, 267, 1648–1651. [Google Scholar] [CrossRef]
- Miller, K.G.; Emerson, M.D.; Rand, J.B. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron 1999, 24, 323–333. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Huang, J.; Hu, T.; Huang, X.; Zhang, K. Functional identification of gsa-1 gene in Heterorhabditidoides chongmingensis. J. Nanjing Agric. Univ. 2020, 43, 89–97. [Google Scholar] [CrossRef]
- Colombo, K.; Grill, S.W.; Kimple, R.J.; Willard, F.S.; Siderovski, D.P.; Gönczy, P. Translation of Polarity Cues into Asymmetric Spindle Positioning in Caenorhabditis elegans Embryos. Science 2003, 300, 1957–1961. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuhashi, T.; Sakamoto, K. FoxO/Daf-16 restored thrashing movement reduced by heat stress in Caenorhabditis elegans. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2014, 170, 26–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulias, K.; Horvitz, H.R. The C. elegans MicroRNA mir-71 Acts in Neurons to Promote Germline-Mediated Longevity through Regulation of DAF-16/FOXO. Cell Metab. 2012, 15, 439–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Hisamoto, N.; Nix, P.; Kanao, S.; Mizuno, T.; Bastiani, M.; Matsumoto, K. The growth factor SVH-1 regulates axon regeneration in C. elegans via the JNK MAPK cascade. Nat. Neurosci. 2012, 15, 551–557. [Google Scholar] [CrossRef]
- Pastuhov, S.I.; Fujiki, K.; Nix, P.; Kanao, S.; Bastiani, M.; Matsumoto, K.; Hisamoto, N. Endocannabinoid-Goα signalling inhibits axon regeneration in Caenorhabditis elegans by antagonizing Gqα-PKC-JNK signalling. Nat. Commun. 2012, 3, 1136. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Hsin, H.; Libina, N.; Kenyon, C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 2001, 28, 139–145. [Google Scholar] [CrossRef]
- Libina, N.; Berman, J.R.; Kenyon, C. Tissue–specific activities of C. elegans DAF–16 in the regulation of lifespan. Cell 2003, 115, 489–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaletsky, R.; Lakhina, V.; Arey, R.; Williams, A.; Landis, J.; Ashraf, J.; Murphy, C.T. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 2015, 529, 92–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, A.; Grabbe, C.; Lee, J.; Lee, J.; Palmer, R.H.; Wu, C.-F. Mutation of Drosophila focal adhesion kinase induces bang-sensitive behavior and disrupts glial function, axonal conduction and synaptic transmission. Eur. J. Neurosci. 2008, 27, 2860–2870. [Google Scholar] [CrossRef]
- Wolfson, M.; Budovsky, A.; Tacutu, R.; Fraifeld, V. The signaling hubs at the crossroad of longevity and age-related disease networks. Int. J. Biochem. Cell Biol. 2009, 41, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Johnson, T.E. Life extension and stress resistance in Caenorhabditis elegans. modulated by the tkr-1 gene. Curr. Biol. 1998, 8, 1091–1094, S1–S4. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Johnson, T.E. The OLD-1 positive regulator of longevity and stress resistance is under DAF-16 regulation in Caenorhabditis elegans. Curr. Biol. 2001, 11, 1517–1523. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.Y.; Liu, X.H.; Tan, J.; Wang, Y.; Qiao, L.; Yedid, G.; Dai, C.S.; Qiu, R.L.; Yan, X.W.; Tan, H.W.; et al. Heterorhabditidoides rugaoensis n. sp. (Rhabditida: Rhabditidae), a Novel Highly Pathogenic Entomopathogenic Nematode Member of Rhabditidae. J. Nematol. 2012, 44, 348–360. [Google Scholar]
- Ehlers, R.-U.; Han, R. Cultivation of Axenic Heterorhabditis spp. Dauer Juveniles and Their Response to Non-Specific Photorhabdus luminescens Food Signals. Nematologica 1998, 44, 425–435. [Google Scholar] [CrossRef]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Bonifassi, E.; Saux, M.F.-L.; Boemare, N.; Lanois, A.; Laumond, C.; Smart, G. Gnotobiological Study of Infective Juveniles and Symbionts of Steinernema scapterisci: A Model to Clarify the Concept of the Natural Occurrence of Monoxenic Associations in Entomopathogenic Nematodes. J. Invertebr. Pathol. 1999, 74, 164–172. [Google Scholar] [CrossRef]
- Li, J.; Ebata, A.; Dong, Y.; Rizki, G.; Iwata, T.; Lee, S.S. Caenorhabditis elegans HCF-1 Functions in Longevity Maintenance as a DAF-16 Regulator. PLoS Biol. 2008, 6, e233. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Xiong, C.; Kornfeld, K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2004, 101, 8084–8089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, R.; Wilson, L.; Sturrock, M.; Warnock, N.D.; Carrizo, D.; Cox, D.; Maule, A.G.; Dalzell, J.J. A neuropeptide modulates sensory perception in the entomopathogenic nematode Steinernema carpocapsae. PLoS Pathog. 2017, 13, e1006185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Bao, Y.-Y.; Li, B.-L.; Cheng, Y.-B.; Peng, Z.-Y.; Liu, H.; Xu, H.-J.; Zhu, Z.-R.; Lou, Y.-G.; Cheng, J.-A.; et al. Transcriptome Analysis of the Brown Planthopper Nilaparvata lugens. PLoS ONE 2010, 5, e14233. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.-M. The Significance of Digital Gene Expression Profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Adhikari, B.N.; Lin, C.-Y.; Bai, X.; Ciche, T.A.; Grewal, P.S.; Dillman, A.R.; Chaston, J.M.; Shapiro-Ilan, D.I.; Bilgrami, A.L.; Gaugler, R.; et al. Transcriptional profiling of trait deterioration in the insect pathogenic nematode Heterorhabditis bacteriophora. BMC Genom. 2009, 10, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34 (Suppl. S2), W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.; Srivastava, M.; Williams, N.J.; Papa, I.; Lee, S.K.; Dinh, X.T.; Hutloff, A.; Jordan, M.A.; Zhao, J.L.; Casellas, R.; et al. MicroRNA-146a regulates ICOS–ICOSL signalling to limit accumulation of T follicular helper cells and germinal centres. Nat. Commun. 2015, 6, 6436. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Liu, J.; Xie, F.; Gao, X.; Ye, J.-H.; Sun, L.-Y.; Wei, R.; Ai, J. miR-124/ATF-6, Novel lifespan extension pathway of astragalus polysaccharide in Caenorhabditis elegans. J. Cell. Biochem. 2014, 116, 242–251. [Google Scholar] [CrossRef]
- Timmons, L.; Fire, A. Specific interference by ingested dsRNA. Nature 1998, 395, 854. [Google Scholar] [CrossRef]
| Combination | Developmental Rate (%) | Number of Second Generation IJs per Insect Larva | IJs Size | |
|---|---|---|---|---|
| Length (μm) | Max Width (μm) | |||
| DZ/S1 | 85.43 | 6782 | 425.7 ± 25.43 | 22.6 ± 3.1 |
| DZ/186 | 26.72 | 386 | 398.52 ± 35.25 | 20.3 ± 4.6 |
| RG/S1 | 75.19 | 5873 | 569.4 ± 36.72 | 27.38 ± 3.61 |
| RG/186 | 83.52 | 4950 | 582.7 ± 38.32 | 28.62 ± 2.65 |
| Groups | No. of Worms | Mean Lifespan (Days) | Max Lifespan (Days) | Median Lifespan | Log-Rank Test |
|---|---|---|---|---|---|
| DZ/S1 | 90 | 13.10 | 24 | 14 | p = 0.0005 |
| DZ/186 | 90 | 11.28 | 22 | 10 | p = 0.0005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, C.; Chen, L.; Guo, D.; Sun, J.; Duan, Y.; Zhang, P.; Li, P.; Ma, L.; Xu, M.; Wang, Y.; et al. An Intestinal Symbiotic Bacterial Strain of Oscheius chongmingensis Modulates Host Viability at Both Global and Post-Transcriptional Levels. Int. J. Mol. Sci. 2022, 23, 14692. https://doi.org/10.3390/ijms232314692
Zhan C, Chen L, Guo D, Sun J, Duan Y, Zhang P, Li P, Ma L, Xu M, Wang Y, et al. An Intestinal Symbiotic Bacterial Strain of Oscheius chongmingensis Modulates Host Viability at Both Global and Post-Transcriptional Levels. International Journal of Molecular Sciences. 2022; 23(23):14692. https://doi.org/10.3390/ijms232314692
Chicago/Turabian StyleZhan, Chengxiu, Long Chen, Dandan Guo, Jing Sun, Yunbin Duan, Panjie Zhang, Pengpeng Li, Lijun Ma, Man Xu, Ying Wang, and et al. 2022. "An Intestinal Symbiotic Bacterial Strain of Oscheius chongmingensis Modulates Host Viability at Both Global and Post-Transcriptional Levels" International Journal of Molecular Sciences 23, no. 23: 14692. https://doi.org/10.3390/ijms232314692





