Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of New Drugs against Human Topoisomerase I Receptor
Abstract
:1. Introduction
2. Results
2.1. In Silico Molecular Docking
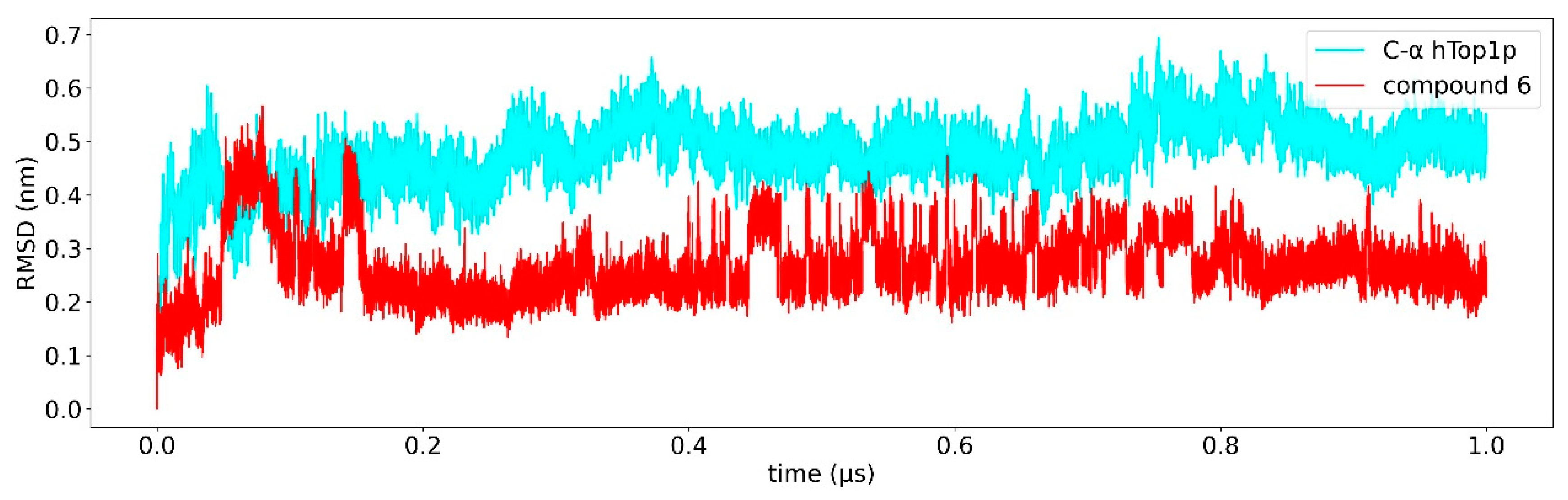
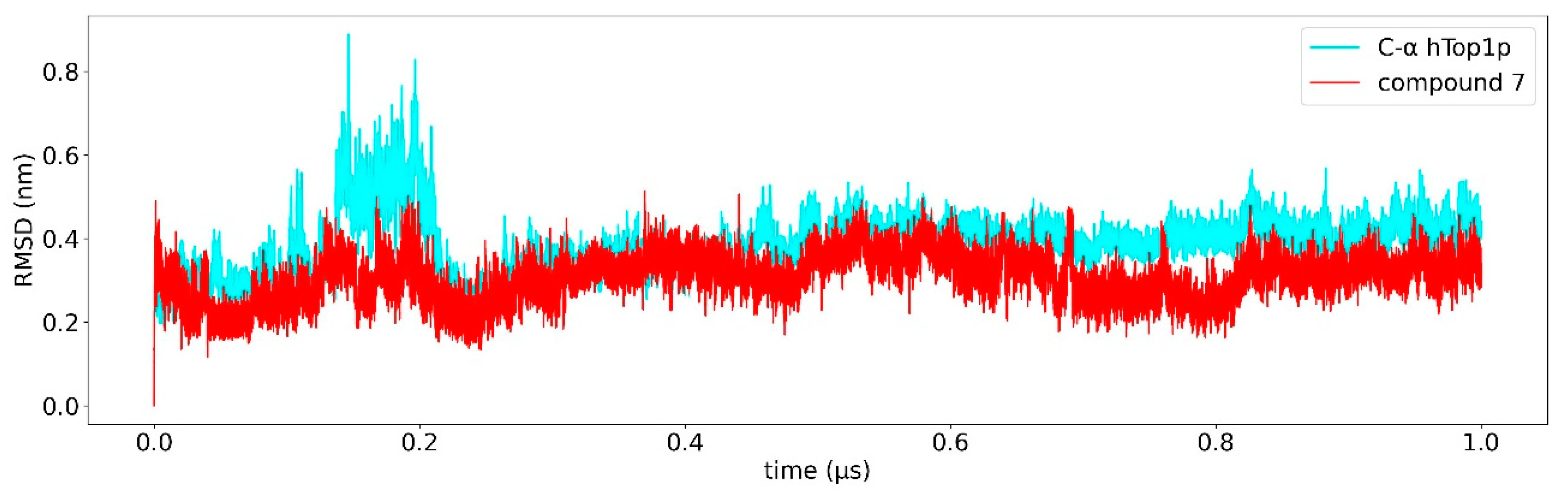
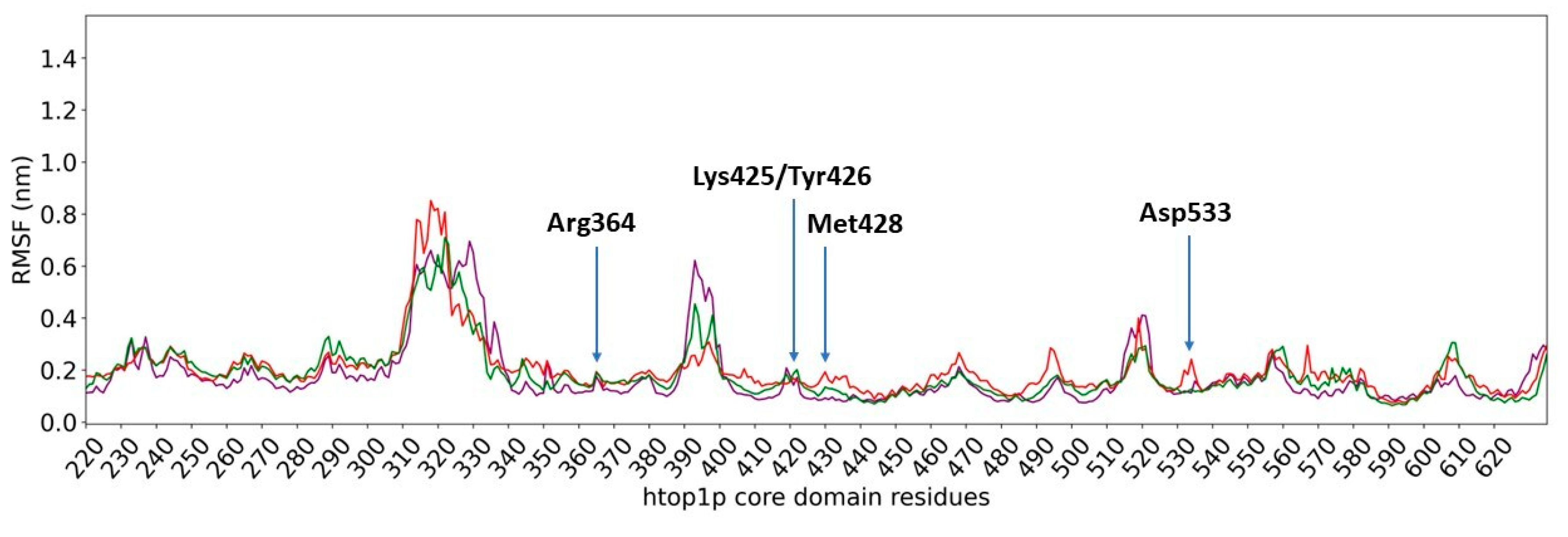
2.2. Classical Molecular Dynamics Simulation
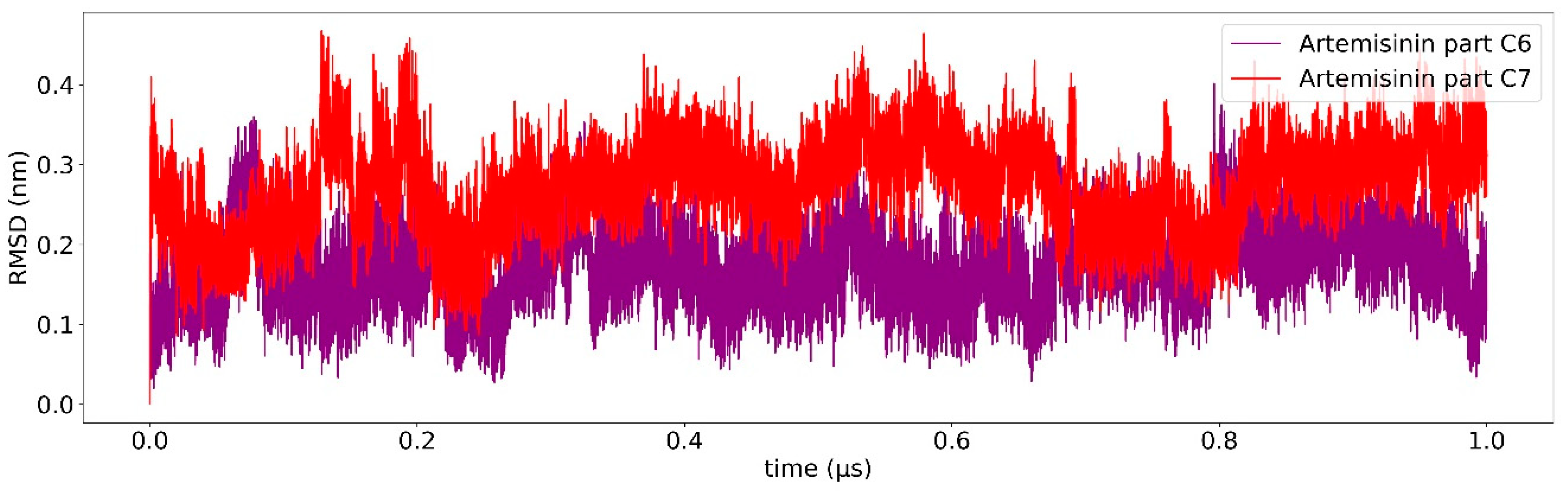
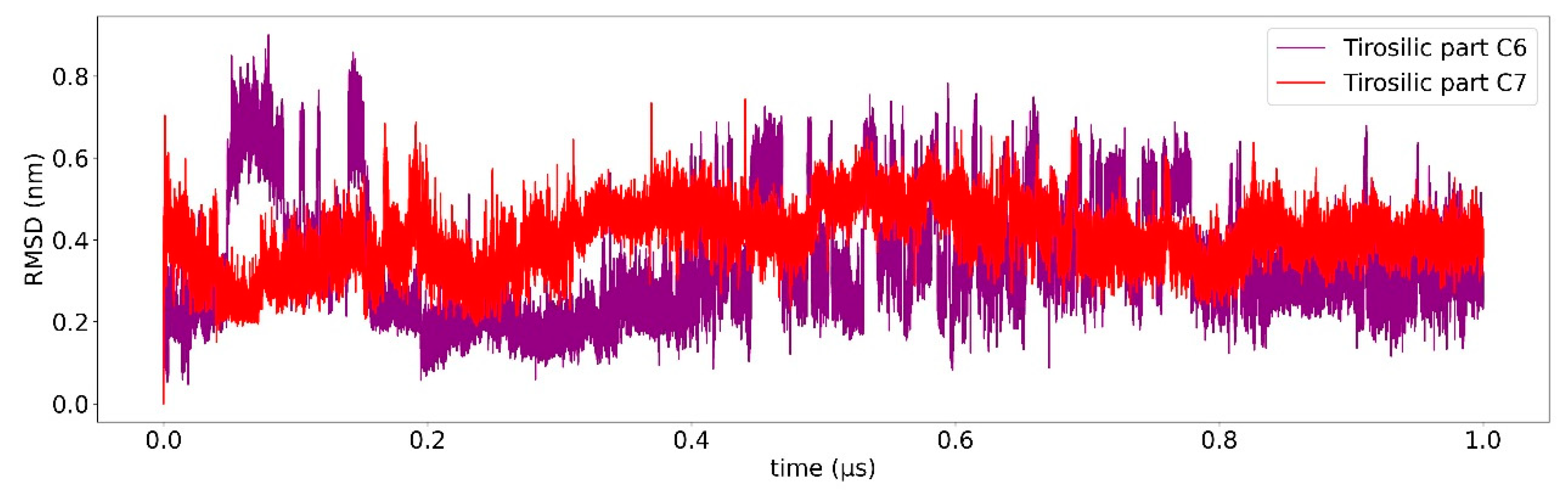
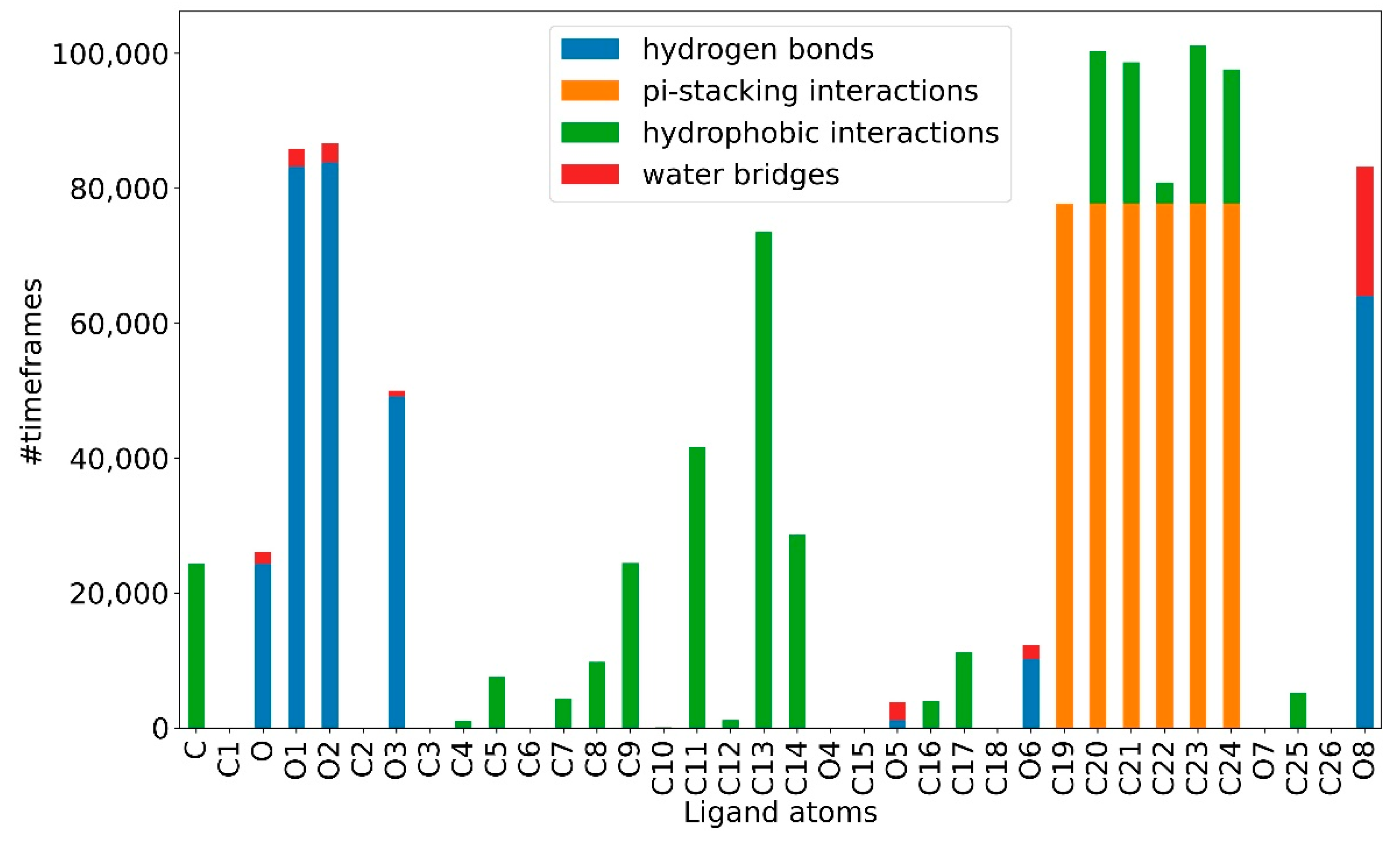
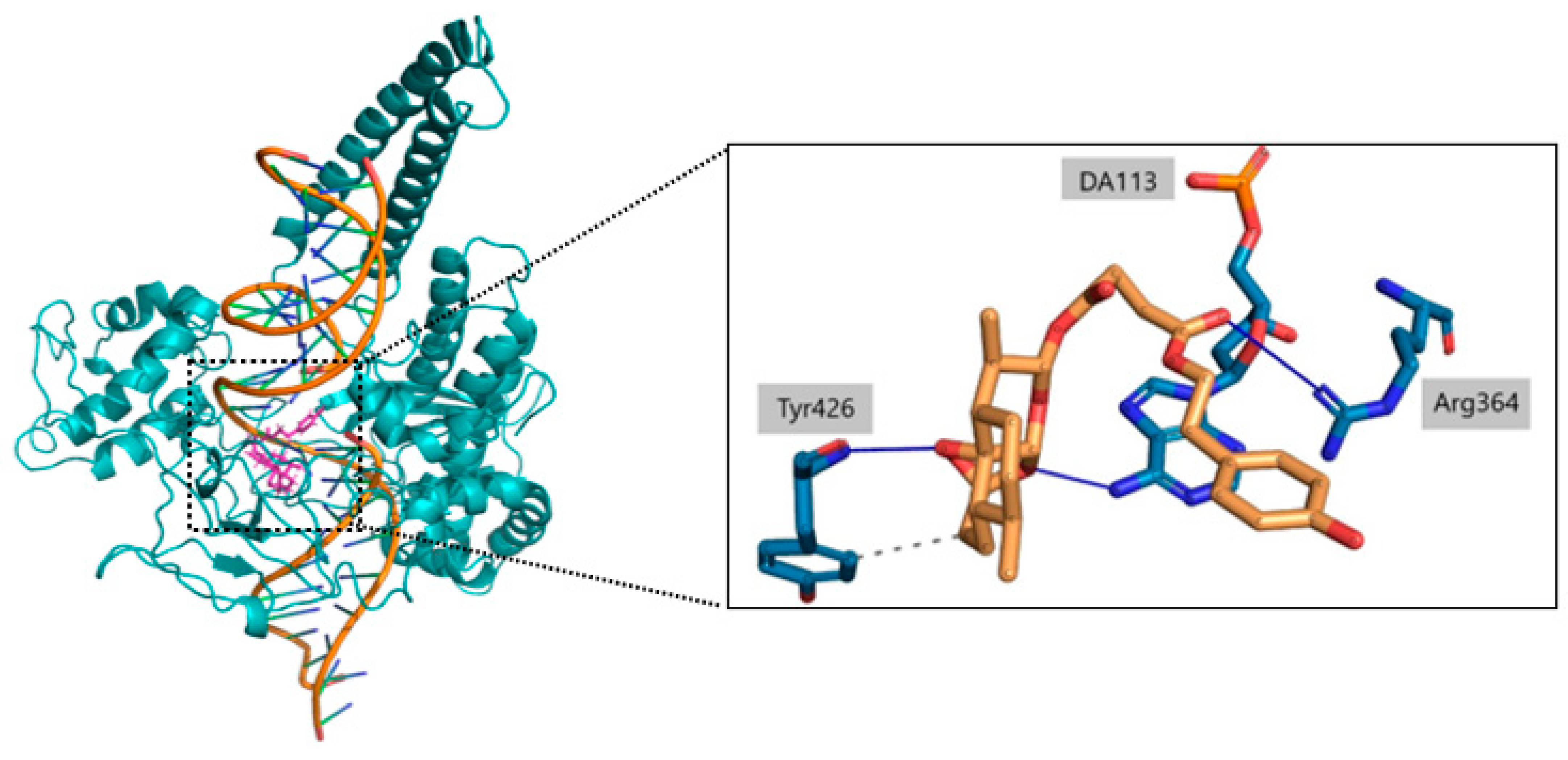
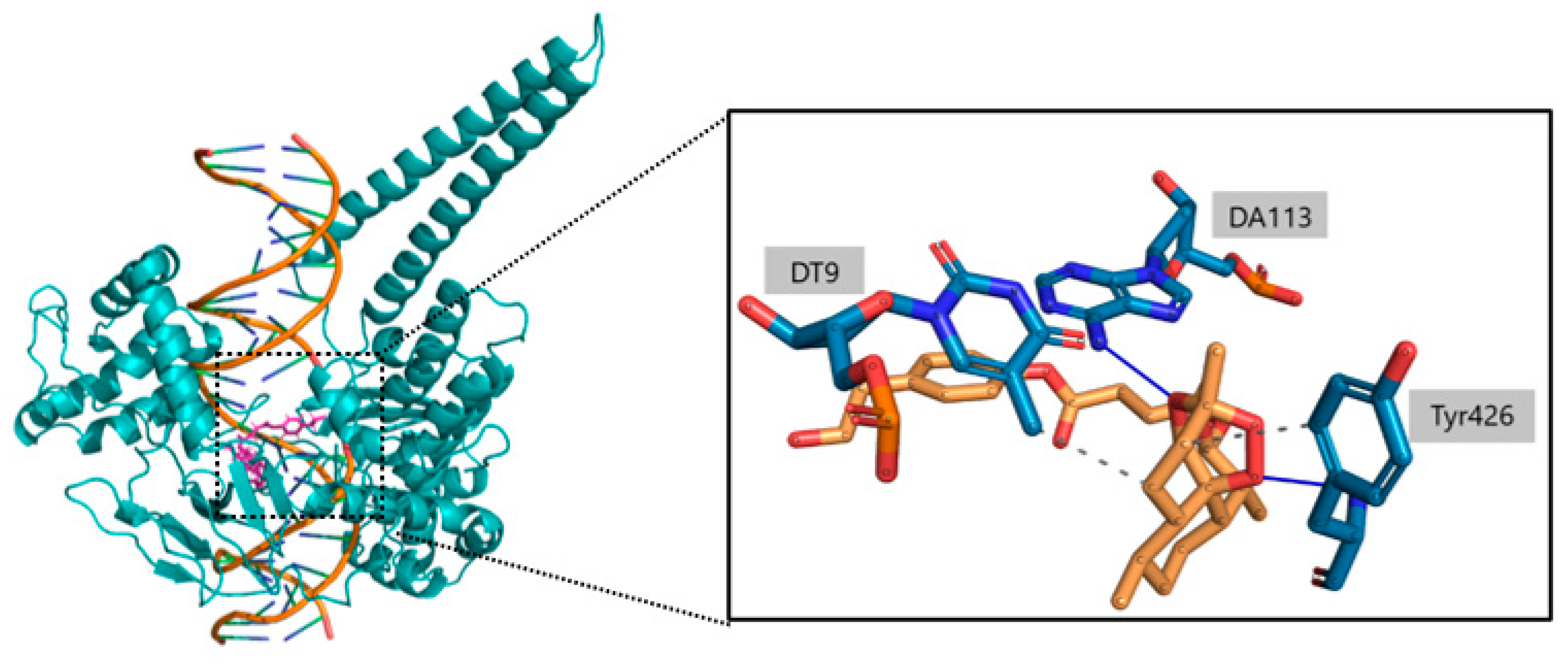
2.3. Ligand Receptor Binding Interactions as a Function of the Time
3. Discussion
4. Materials and Methods
4.1. Data Preparation
4.2. In Silico Molecular Docking
4.3. Classical Molecular Dynamics Simulation
4.4. Analysis of Non-Covalent Interactions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McClendon, A.K.; Rodriguez, A.C.; Osheroff, N. Human Topoisomerase Iiα Rapidly Relaxes Positively Supercoiled Dna: Implications For Enzyme Action Ahead of Replication Forks. J. Biol. Chem. 2005, 280, 39337–39345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chrencik, J.E.; Staker, B.L.; Burgin, A.B.; Pourquier, P.; Pommier, Y.; Stewart, L.; Redinbo, M.R. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004, 339, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Chillemi, G.; Redinbo, M.; Bruselles, A.; Desideri, A. Role of the linker domain and the 203–214 N-terminal residues in the human topoisomerase I DNA complex dynamics. Biophys. J. 2004, 87, 4087–4097. [Google Scholar] [CrossRef]
- Palle, K.; Pattarello, L.; Van Der Merwe, M.; Losasso, C.; Benedetti, P.; Bjornsti, M.A. Disulfide cross-links reveal conserved features of DNA topoisomerase I architecture and a role for the N terminus in clamp closure. J. Biol. Chem. 2008, 283, 27767–27775. [Google Scholar] [CrossRef] [Green Version]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, L.; Ireton, G.C.; Champoux, J.J. The Domain Organization of Human Topoisomerase I. J. Biol. Chem. 1996, 271, 7602–7608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eng, W.K.; Faucette, L.; Johnson, R.K.; Sternglanz, R.O.L.F. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol. Pharmacol. 1988, 34, 755–760. [Google Scholar] [PubMed]
- Li, M.; Liu, Y. Topoisomerase I in human disease pathogenesis and treatments. Genom. Proteom. Bioinform. 2016, 14, 166–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Her, C. Inhibition of topoisomerase (DNA) I (TOP1): DNA damage repair and anticancer therapy. Biomolecules 2015, 5, 1652–1670. [Google Scholar] [CrossRef] [Green Version]
- Haldar, K.; Bhattacharjee, S.; Safeukui, I. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 2018, 16, 156–170. [Google Scholar] [CrossRef]
- Takatani-Nakase, T.; Matsui, C.; Hosotani, M.; Omura, M.; Takahashi, K.; Nakase, I. Hypoxia enhances motility and EMT through the Na+/H+ exchanger NHE-1 in MDA-MB-231 breast cancer cells. Exp. Cell Res. 2022, 412, 13006. [Google Scholar] [CrossRef] [PubMed]
- Botta, L.; Filippi, S.; Zippilli, C.; Cesarini, S.; Bizzarri, B.M.; Cirigliano, A.; Rinaldi, T.; Paiardini, A.; Fiorucci, D.; Saladino, R.; et al. Artemisinin derivatives with antimelanoma activity show inhibitory effect against Human DNA topoisomerase 1. ACS Med. Chem. Lett. 2020, 11, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Botta, L.; Filippi, S.; Bizzarri, B.M.; Zippilli, C.; Meschini, R.; Pogni, R.; Baratto, M.C.; Villanova, L.; Saladino, R. Synthesis and evaluation of artemisinin-based hybrid and dimer derivatives as antimelanoma agents. ACS Omega 2019, 5, 243–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castrignanò, T.; Chillemi, G.; Desideri, A. Structure and hydration of BamHI DNA recognition site: A molecular dynamics investigation. Biophys. J. 2000, 79, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Chillemi, G.; Castrignanò, T.; Desideri, A. Structure and hydration of the DNA-human topoisomerase I covalent complex. Biophys. J. 2001, 81, 490–500. [Google Scholar] [CrossRef] [Green Version]
- Castrignanò, T.; Chillemi, G.; Varani, G.; Desideri, A. Molecular dynamics simulation of the RNA complex of a double-stranded RNA-binding domain reveals dynamic features of the intermolecular interface and its hydration. Biophys. J. 2002, 83, 3542–3552. [Google Scholar] [CrossRef] [Green Version]
- Bò, L.; Miotto, M.; Di Rienzo, L.; Milanetti, E.; Ruocco, G. Exploring the association between sialic acid and SARS-CoV-2 spike protein through a molecular dynamics-based approach. Front. Med. Technol. 2021, 2, 614652. [Google Scholar] [CrossRef]
- Komatsu, T.S.; Okimoto, N.; Koyama, Y.M.; Hirano, Y.; Morimoto, G.; Ohno, Y.; Taiji, M. Drug binding dynamics of the dimeric SARS-CoV-2 main protease, determined by molecular dynamics simulation. Sci. Rep. 2020, 10, 16986. [Google Scholar] [CrossRef]
- Keretsu, S.; Bhujbal, S.P.; Cho, S.J. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci. Rep. 2020, 10, 17716. [Google Scholar] [CrossRef]
- Salmaso, V.; Moro, S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Luo, Y.; Weng, Q.; He, Q.; Lu, W.; Yang, B. CPT21, a novel compound with anti-proliferative effect against gastric cancer cell SGC7901. Investig. New Drugs 2008, 26, 517–524. [Google Scholar] [CrossRef]
- Li, Q.Y.; Zu, Y.G.; Shi, R.Z.; Yao, L.P. Review camptothecin: Current perspectives. Curr. Med. Chem. 2006, 13, 2021–2039. [Google Scholar] [CrossRef] [PubMed]
- Gabellone, S.; Piccinino, D.; Filippi, S.; Castrignanò, T.; Zippilli, C.; Del Buono, D.; Saladino, R. Lignin Nanoparticles Deliver Novel Thymine Biomimetic Photo-Adducts with Antimelanoma Activity. Int. J. Mol. Sci. 2022, 23, 915. [Google Scholar] [CrossRef]
- Staker, B.L.; Feese, M.D.; Cushman, M.; Pommier, Y.; Zembower, D.; Stewart, L.; Burgin, A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005, 48, 2336–2345. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Kohlhagen, G.; Kohn, K.W.; Leteurtre, F.; Wani, M.C.; Wall, M.E. Interaction of an alkylating camptothecin derivative with a DNA base at topoisomerase I-DNA cleavage sites. Proc. Natl. Acad. Sci. USA 1995, 92, 8861–8865. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huey, R.; Morris, G.M.; Forli, S. Using AutoDock 4 and AutoDock Vina with AutoDockTools: A tutorial. Scripps Res. Inst. Mol. Graph. Lab. 2012, 10550, 92037. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loschwitz, J.; Jäckering, A.; Keutmann, M.; Olagunju, M.; Olubiyi, O.O.; Strodel, B. Dataset of AMBER force field parameters of drugs, natural products and steroids for simulations using GROMACS. Data Brief 2021, 35, 106948. [Google Scholar] [CrossRef] [PubMed]
- Sousa, D.; Alan, W.; Vranken, W.F. Acpype-Antechamber python parser interface. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [Green Version]
- Mark, P.; Lennart, N. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Castrignanò, T.; Gioiosa, S.; Flati, T.; Cestari, M.; Picardi, E.; Chiara, M.; Fratelli, M.; Amente, S.; Cirilli, M.; Tangaro, M.A.; et al. ELIXIR-IT HPC@ CINECA: High performance computing resources for the bioinformatics community. BMC Bioinform. 2020, 21, 352. [Google Scholar] [CrossRef]



| Compound Name | 2D Structure | Smiles Stringes | Binding Affinity (kcal/mol) | Reference |
|---|---|---|---|---|
| c0 |  | C[C@]1(OO2)O[C@](O[C@](OC(CCC(OCC3=CC[C@H](C(C)=C)CC3)=O)=O)([H])[C@@H]4C)([H])[C@]2([C@@]4([H])CC[C@H]5C)[C@@]5([H])CC1 | −8.2 | [12] |
| c1 | 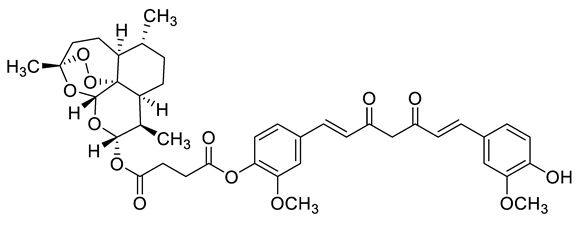 | C[C@]1(OO2)O[C@](O[C@](OC(CCC(OC3=C(OC)C=C(/C=C/C(CC(/C=C/C4=CC(OC)=C(O)C=C4)=O)=O)C=C3)=O)=O)([H])[C@@H]5C)([H])[C@]2([C@@]5([H])CC[C@H]6C)[C@@]6([H])CC1 | −7.7 | [12] |
| c2 | 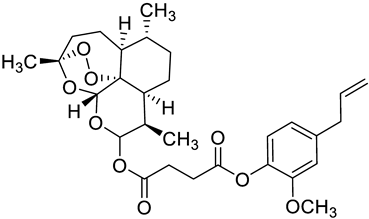 | C[C@]1(OO2)O[C@](OC(OC(CCC(OC(C(OC)=C3)=CC=C3CC=C)=O)=O)[C@@H]4C)([H])[C@]2([C@@]4([H])CC[C@H]5C)[C@@]5([H])CC1 | −7.7 | [12] |
| c3 | 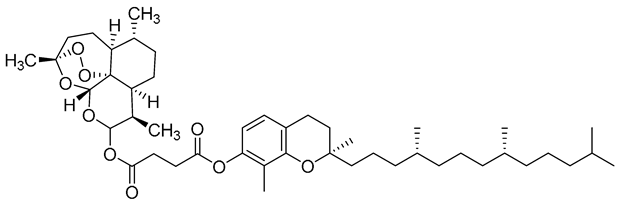 | C[C@]1(OO2)O[C@](OC(OC(CCC(OC3=C©C4=C(CC[C@](CCC[C@H]©CCC[C@H]©CCCC©C)©O4)C=C3)=O)=O)[C@@H]5C)([H])[C@]2([C@@]5([H])CC[C@H]6C)[C@@]6([H])CC1 | −7.0 | [12] |
| c4 |  | C[C@]1(OO2)O[C@](O[C@](OCCC3=CC=C(O)C=C3)([H])[C@@H]4C)([H])[C@]2([C@@]4([H])CC[C@H]5C)[C@@]5([H])CC1 | −7.8 | [12] |
| c5 | 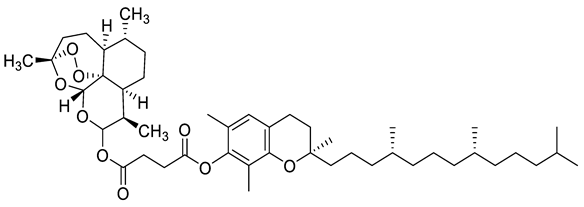 | CC1=C(OC(CCC(OC([C@@H]2C)O[C@]3([H])O[C@](C)(OO4)CC[C@]5([H])[C@@]34[C@@]2([H])CC[C@H]5C)=O)=O)C(C)=CC6=C1O[C@@](CCC[C@H](C)CCC[C@H](C)CCCC(C)C)(C)CC6 | −6.1 | [12] |
| c6 | 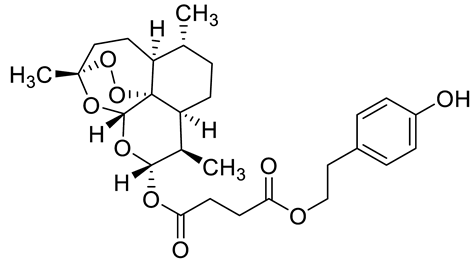 | C[C@]1(OO2)O[C@](O[C@](OC(CCC(OCCC3=CC=C(O)C=C3)=O)=O)([H])[C@@H]4C)([H])[C@]2([C@@]4([H])CC[C@H]5C)[C@@]5([H])CC1 | −9.1 | [12] |
| c7 | 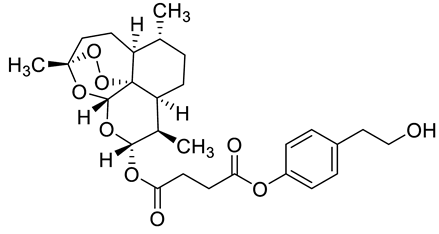 | C[C@]1(OO2)O[C@](O[C@](OC(CCC(OC3=CC=C(CCO)C=C3)=O)=O)([H])[C@@H]4C)([H])[C@]2([C@@]4([H])CC[C@H]5C)[C@@]5([H])CC1 | −8.7 | [12] |
| CPT |  | CCC1(C2=C(COC1=O)C(=O)N3CC4=CC5=CC=CC=C5N=C4C3=C2)O | −8.5 | [7] |
| CPT21 | 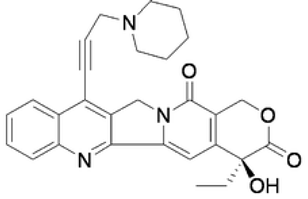 | CC[C@@]1(C2=C(COC1=O)C(=O)N3CC4=C(C3=C2)N=C5C(=C4)C=CC=C5C#CCC6CCCNC6)O | −7.6 | [22] |
| Topotecan | 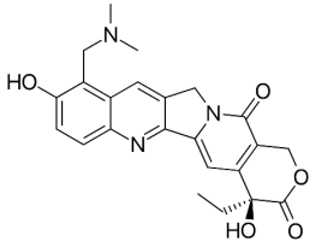 | CC[C@@]1(C2=C(COC1=O)C(=O)N3CC4=CC5=C(C=CC(=C5CN(C)C)O)N=C4C3=C2)O | −8.7 | [23] |
| SN-38 | 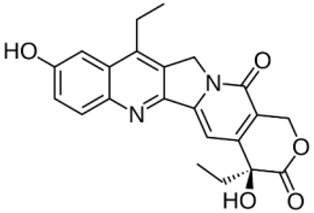 | CCC1=C2CN3C(=CC4=C(C3=O)COC(=O)[C@@]4(CC)O)C2=NC5=C1C=C(C=C5)O | −8.1 | [23] |
| Hydrogen Bonds | pi-Stacking Interactions | Hydrophobic Interactions | Water Bridges | |
|---|---|---|---|---|
| Compound c6 | 100% | 45.1% | 98.4% | 30.6% |
| Compound c7 | 99.8% | 77.8% | 99.5% | 27.6% |
| Glu356 | Arg364 | Lys425 | Tyr426 | DT9 | DC111 | DC112 | DA113 | DA114 | |
|---|---|---|---|---|---|---|---|---|---|
| O | - | - | 10% | - | - | - | - | - | - |
| O1 | - | - | - | 98% | - | - | - | - | - |
| O2 | - | - | - | - | - | - | - | 93% | 35% |
| O5 | 16% | - | - | - | - | - | 10% | - | - |
| O6 | - | 73% | - | - | - | - | - | - | - |
| O8 | - | - | - | - | - | 14% | - | - | - |
| C13 | - | - | - | - | 56% | - | - | - | - |
| Asn352 | Arg364 | Tyr426 | Lys532 | Asp533 | DT9 | DT10 | DA113 | DA114 | |
|---|---|---|---|---|---|---|---|---|---|
| O | - | - | 18% | - | - | - | - | - | - |
| O1 | 15% | - | 76% | - | - | - | - | - | - |
| O2 | - | - | - | - | - | - | - | 54% | 51% |
| O3 | - | - | - | - | - | - | - | 45% | - |
| O6 | - | 11% | - | - | - | - | - | - | - |
| O8 | - | 22% | - | 14% | 19% | - | 29% | - | - |
| C13 | - | - | - | - | - | 72% | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madeddu, F.; Di Martino, J.; Pieroni, M.; Del Buono, D.; Bottoni, P.; Botta, L.; Castrignanò, T.; Saladino, R. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of New Drugs against Human Topoisomerase I Receptor. Int. J. Mol. Sci. 2022, 23, 14652. https://doi.org/10.3390/ijms232314652
Madeddu F, Di Martino J, Pieroni M, Del Buono D, Bottoni P, Botta L, Castrignanò T, Saladino R. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of New Drugs against Human Topoisomerase I Receptor. International Journal of Molecular Sciences. 2022; 23(23):14652. https://doi.org/10.3390/ijms232314652
Chicago/Turabian StyleMadeddu, Francesco, Jessica Di Martino, Michele Pieroni, Davide Del Buono, Paolo Bottoni, Lorenzo Botta, Tiziana Castrignanò, and Raffaele Saladino. 2022. "Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of New Drugs against Human Topoisomerase I Receptor" International Journal of Molecular Sciences 23, no. 23: 14652. https://doi.org/10.3390/ijms232314652
APA StyleMadeddu, F., Di Martino, J., Pieroni, M., Del Buono, D., Bottoni, P., Botta, L., Castrignanò, T., & Saladino, R. (2022). Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of New Drugs against Human Topoisomerase I Receptor. International Journal of Molecular Sciences, 23(23), 14652. https://doi.org/10.3390/ijms232314652









