Inhibition of the MAPK/c-Jun-EGR1 Pathway Decreases Photoreceptor Cell Death in the rd1 Mouse Model for Inherited Retinal Degeneration
Abstract
:1. Introduction
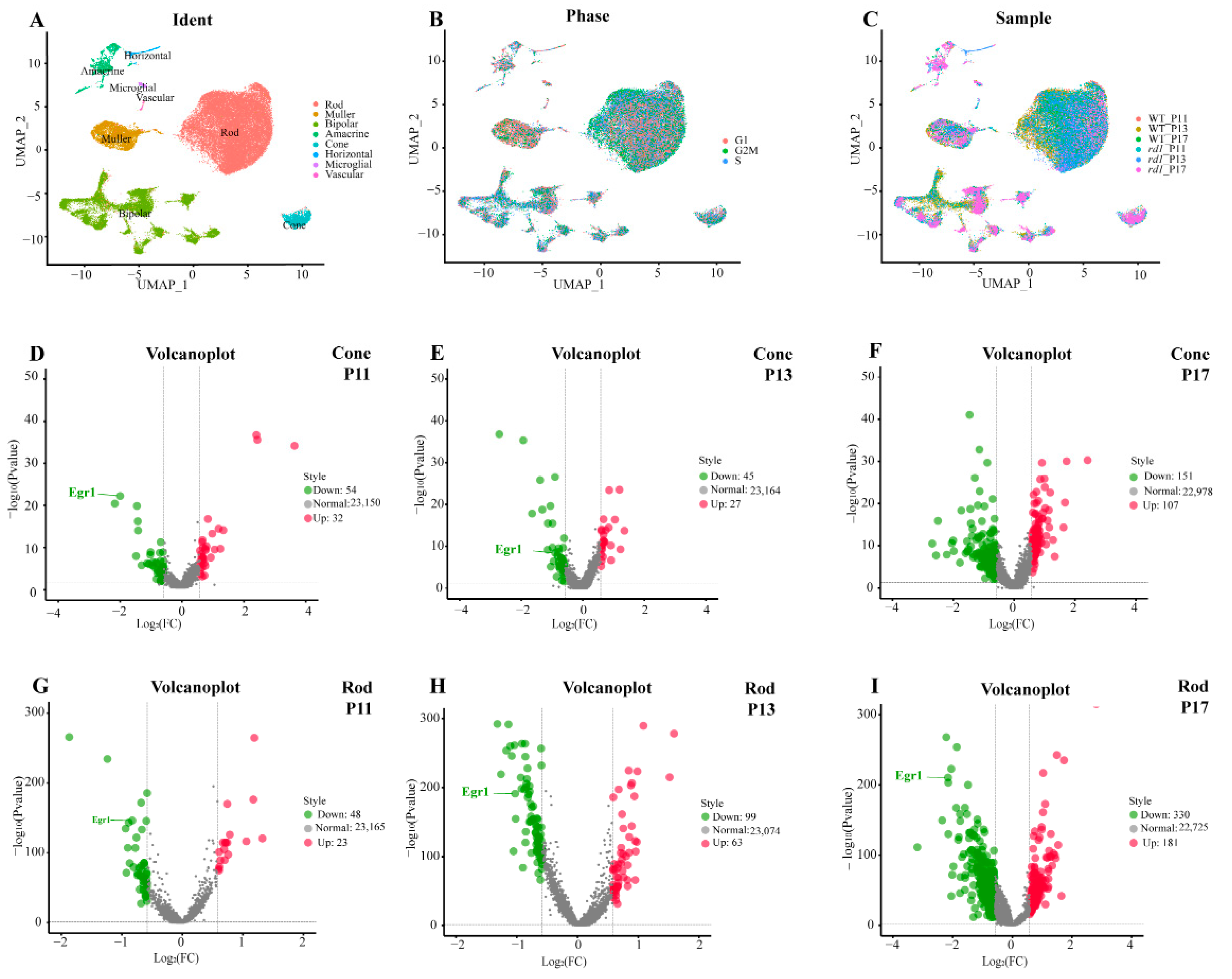
2. Results
2.1. EGR1 Binds to the PARP1 Promoter Region
2.2. Suppression of EGR1 Expression Inhibited Photoreceptor Cell Death
2.3. Inhibition of MAPK/c-Jun Pathway Activation Suppressed Photoreceptor Cell Death
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neal, T.B.; Luther, E.E. Retinitis Pigmentosa; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Campochiaro, P.A.; Mir, T.A. The mechanism of cone cell death in Retinitis Pigmentosa. Prog. Retin. Eye Res. 2018, 62, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Bowes, C.; Li, T.; Frankel, W.N.; Danciger, M.; Coffin, J.M.; Applebury, M.L.; Farber, D.B. Localization of a retroviral element within the rd gene coding for the beta subunit of cGMP phosphodiesterase. Proc. Natl. Acad. Sci. USA 1993, 90, 2955–2959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cote, R.H.; Gupta, R.; Irwin, M.J.; Wang, X. Photoreceptor Phosphodiesterase (PDE6): Structure, Regulatory Mechanisms, and Implications for Treatment of Retinal Diseases. Adv. Exp. Med. Biol. 2022, 1371, 33–59. [Google Scholar] [CrossRef] [PubMed]
- Pentia, D.C.; Hosier, S.; Collupy, R.A.; Valeriani, B.A.; Cote, R.H. Purification of PDE6 isozymes from mammalian retina. Methods Mol. Biol. 2005, 307, 125–140. [Google Scholar] [CrossRef]
- Gopalakrishna, K.N.; Boyd, K.; Artemyev, N.O. Mechanisms of mutant PDE6 proteins underlying retinal diseases. Cell. Signal. 2017, 37, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Dvir, L.; Srour, G.; Abu-Ras, R.; Miller, B.; Shalev, S.A.; Ben-Yosef, T. Autosomal-recessive early-onset retinitis pigmentosa caused by a mutation in PDE6G, the gene encoding the gamma subunit of rod cGMP phosphodiesterase. Am. J. Hum. Genet. 2010, 87, 258–264. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Huang, Q.; Du, X.; Xu, S.; Li, M.; Ma, S. Early activation of Egr-1 promotes neuroinflammation and dopaminergic neurodegeneration in an experimental model of Parkinson’s disease. Exp. Neurol. 2018, 302, 145–154. [Google Scholar] [CrossRef]
- Pagel, J.I.; Deindl, E. Early growth response 1—A transcription factor in the crossfire of signal transduction cascades. Indian J. Biochem. Biophys. 2011, 48, 226–235. [Google Scholar]
- Lee, S.M.; Park, M.S.; Park, S.Y.; Choi, Y.D.; Chung, J.O.; Kim, D.H.; Jung, Y.D.; Kim, H.S. Primary bile acid activates Egr1 expression through the MAPK signaling pathway in gastric cancer. Mol. Med. Rep. 2022, 25, 129. [Google Scholar] [CrossRef]
- Yen, J.H.; Lin, C.Y.; Chuang, C.H.; Chin, H.K.; Wu, M.J.; Chen, P.Y. Nobiletin Promotes Megakaryocytic Differentiation through the MAPK/ERK-Dependent EGR1 Expression and Exerts Anti-Leukemic Effects in Human Chronic Myeloid Leukemia (CML) K562 Cells. Cells 2020, 9, 877. [Google Scholar] [CrossRef] [Green Version]
- Barutcu, S.A.; Girnius, N.; Vernia, S.; Davis, R.J. Role of the MAPK/cJun NH2-terminal kinase signaling pathway in starvation-induced autophagy. Autophagy 2018, 14, 1586–1595. [Google Scholar] [CrossRef]
- Yi, E.H.; Xu, F.; Li, P. (3R)-5,6,7-trihydroxy-3-isopropyl-3-methylisochroman-1-one alleviates lipoteichoic acid-induced photoreceptor cell damage. Cutan. Ocul. Toxicol. 2018, 37, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Huang, S.W.; Zheng, Y.J.; Dong, Y.R. Angiotensin II type 1 receptor blockade suppresses H2O2-induced retinal degeneration in photoreceptor cells. Cutan. Ocul. Toxicol. 2015, 34, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Paquet-Durand, F.; Hauck, S.M.; van Veen, T.; Ueffing, M.; Ekström, P. PKG activity causes photoreceptor cell death in two retinitis pigmentosa models. J. Neurochem. 2009, 108, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Power, M.; Das, S.; Schütze, K.; Marigo, V.; Ekström, P.; Paquet-Durand, F. Cellular mechanisms of hereditary photoreceptor degeneration—Focus on cGMP. Prog. Retin. Eye Res. 2020, 74, 100772. [Google Scholar] [CrossRef]
- Tolone, A.; Belhadj, S.; Rentsch, A.; Schwede, F.; Paquet-Durand, F. The cGMP Pathway and Inherited Photoreceptor Degeneration: Targets, Compounds, and Biomarkers. Genes 2019, 10, 453. [Google Scholar] [CrossRef] [Green Version]
- Bushnell, H.L.; Feiler, C.E.; Ketosugbo, K.F.; Hellerman, M.B.; Nazzaro, V.L.; Johnson, R.I. JNK is antagonized to ensure the correct number of interommatidial cells pattern the Drosophila retina. Dev. Biol. 2018, 433, 94–107. [Google Scholar] [CrossRef]
- Liang, X.; Brooks, M.J.; Swaroop, A. Developmental genome-wide occupancy analysis of bZIP transcription factor NRL uncovers the role of c-Jun in early differentiation of rod photoreceptors in the mammalian retina. Hum. Mol. Genet. 2022, 31, 3914–3933. [Google Scholar] [CrossRef]
- Jiao, K.; Sahaboglu, A.; Zrenner, E.; Ueffing, M.; Ekstrom, P.A.; Paquet-Durand, F. Efficacy of PARP inhibition in Pde6a mutant mouse models for retinitis pigmentosa depends on the quality and composition of individual human mutations. Cell Death Discov. 2016, 2, 16040. [Google Scholar] [CrossRef]
- Yeo, H.; Lee, Y.; Ahn, S.; Jung, E.; Lim, Y.; Shin, S. Chrysin Inhibits TNFα-Induced TSLP Expression through Downregulation of EGR1 Expression in Keratinocytes. Int. J. Mol. Sci. 2021, 22, 4350. [Google Scholar] [CrossRef]
- Hwang, D.; Kim, S.; Won, D.; Kim, C.; Shin, Y.; Park, J.; Chun, Y.; Lim, K.; Yun, J. Egr1 Gene Expression as a Potential Biomarker for In Vitro Prediction of Ocular Toxicity. Pharmaceutics 2021, 13, 1584. [Google Scholar] [CrossRef]
- Portera-Cailliau, C.; Sung, C.H.; Nathans, J.; Adler, R. Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 1994, 91, 974–978. [Google Scholar] [CrossRef] [Green Version]
- Dias, M.F.; Joo, K.; Kemp, J.A.; Fialho, S.L.; da Silva Cunha, A., Jr.; Woo, S.J.; Kwon, Y.J. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog. Retin. Eye Res. 2018, 63, 107–131. [Google Scholar] [CrossRef]
- Vighi, E.; Trifunović, D.; Veiga-Crespo, P.; Rentsch, A.; Hoffmann, D.; Sahaboglu, A.; Strasser, T.; Kulkarni, M.; Bertolotti, E.; van den Heuvel, A.; et al. Combination of cGMP analogue and drug delivery system provides functional protection in hereditary retinal degeneration. Proc. Natl. Acad. Sci. USA 2018, 115, E2997–E3006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Popp, V.; Power, M.; Groeneveld, K.; Yan, J.; Melle, C.; Rogerson, L.; Achury, M.; Schwede, F.; Strasser, T.; et al. Redefining the role of Ca2+-permeable channels in photoreceptor degeneration using diltiazem. Cell Death Discov. 2022, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Tolone, A.; Hilhorst, R.; Groten, J.; Tomar, T.; Paquet-Durand, F. Kinase activity profiling identifies putative downstream targets of cGMP/PKG signaling in inherited retinal neurodegeneration. Cell Death Discov. 2022, 8, 93. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Marigo, V.; Ekström, P. RD Genes Associated with High Photoreceptor cGMP-Levels (Mini-Review). Adv. Exp. Med. Biol. 2019, 1185, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Arango-Gonzalez, B.; Trifunović, D.; Sahaboglu, A.; Kranz, K.; Michalakis, S.; Farinelli, P.; Koch, S.; Koch, F.; Cottet, S.; Janssen-Bienhold, U.; et al. Identification of a common non-apoptotic cell death mechanism in hereditary retinal degeneration. PLoS ONE 2014, 9, e112142. [Google Scholar] [CrossRef]
- Sothilingam, V.; Garcia Garrido, M.; Jiao, K.; Buena-Atienza, E.; Sahaboglu, A.; Trifunović, D.; Balendran, S.; Koepfli, T.; Mühlfriedel, R.; Schön, C.; et al. Retinitis pigmentosa: Impact of different Pde6a point mutations on the disease phenotype. Hum. Mol. Genet. 2015, 24, 5486–5499. [Google Scholar] [CrossRef] [Green Version]
- Lolley, R.N.; Farber, D.B.; Rayborn, M.E.; Hollyfield, J.G. Cyclic GMP accumulation causes degeneration of photoreceptor cells: Simulation of an inherited disease. Science 1977, 196, 664–666. [Google Scholar] [CrossRef]
- Huang, J.; Siragy, H.M. Sodium depletion enhances renal expression of (pro)renin receptor via cyclic GMP-protein kinase G signaling pathway. Hypertension 2012, 59, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Spigolon, G.; Cavaccini, A.; Trusel, M.; Tonini, R.; Fisone, G. cJun N-terminal kinase (JNK) mediates cortico-striatal signaling in a model of Parkinson’s disease. Neurobiol. Dis. 2018, 110, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.P.; Kamath, S.P.; Choolani, M.; Lu, J.; Dheen, S.T. microRNA-200b modulates microglia-mediated neuroinflammation via the cJun/MAPK pathway. J. Neurochem. 2014, 130, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Kim, E.K.; Bae, J.Y.; Moon, S.; Kim, J. Human telomerase reverse transcriptase (hTERT) promotes cancer invasion by modulating cathepsin D via early growth response (EGR)-1. Cancer Lett. 2016, 370, 222–231. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Lee, J.M.; Lim, Y.; Lee, Y.H. Transcriptional regulation of the growth-regulated oncogene alpha gene by early growth response protein-1 in response to tumor necrosis factor alpha stimulation. Biochim. Et Biophys. Acta BBA Gene Regul. Mech. 2013, 1829, 1066–1074. [Google Scholar] [CrossRef]
- Kaur, J.; Mencl, S.; Sahaboglu, A.; Farinelli, P.; van Veen, T.; Zrenner, E.; Ekström, P.; Paquet-Durand, F.; Arango-Gonzalez, B. Calpain and PARP activation during photoreceptor cell death in P23H and S334ter rhodopsin mutant rats. PLoS ONE 2011, 6, e22181. [Google Scholar] [CrossRef] [Green Version]
- Sahaboglu, A.; Sharif, A.; Feng, L.; Secer, E.; Zrenner, E.; Paquet-Durand, F. Temporal progression of PARP activity in the Prph2 mutant rd2 mouse: Neuroprotective effects of the PARP inhibitor PJ34. PLoS ONE 2017, 12, e0181374. [Google Scholar] [CrossRef]
- Wucherpfennig, S.; Haq, W.; Popp, V.; Kesh, S.; Das, S.; Melle, C.; Rentsch, A.; Schwede, F.; Paquet-Durand, F.; Nache, V. cGMP Analogues with Opposing Actions on CNG Channels Selectively Modulate Rod or Cone Photoreceptor Function. Pharmaceutics 2022, 14, 2102. [Google Scholar] [CrossRef]
- Yan, J.; Chen, Y.; Zhu, Y.; Paquet-Durand, F. Programmed Non-Apoptotic Cell Death in Hereditary Retinal Degeneration: Crosstalk between cGMP-Dependent Pathways and PARthanatos? Int. J. Mol. Sci. 2021, 22, 10567. [Google Scholar] [CrossRef]
- Tanimoto, N.; Sothilingam, V.; Seeliger, M.W. Functional phenotyping of mouse models with ERG. Methods Mol. Biol. 2013, 935, 69–78. [Google Scholar] [CrossRef]
- Himawan, E.; Ekström, P.; Buzgo, M.; Gaillard, P.; Stefánsson, E.; Marigo, V.; Loftsson, T.; Paquet-Durand, F. Drug delivery to retinal photoreceptors. Drug Discov. Today 2019, 24, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Qin, Y.; Huang, Y.; Gao, P.; Li, J.; Yu, S.; Jia, D.; Chen, X.; Lv, Y.; Tu, J.; et al. Rod genesis driven by mafba in an nrl knockout zebrafish model with altered photoreceptor composition and progressive retinal degeneration. PLoS Genet. 2022, 18, e1009841. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.E29. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Y.; Xu, W.; Li, Y.; Wei, C.; Hu, Y.; Hu, Z.; Paquet-Durand, F.; Jiao, K. Inhibition of the MAPK/c-Jun-EGR1 Pathway Decreases Photoreceptor Cell Death in the rd1 Mouse Model for Inherited Retinal Degeneration. Int. J. Mol. Sci. 2022, 23, 14600. https://doi.org/10.3390/ijms232314600
Dong Y, Xu W, Li Y, Wei C, Hu Y, Hu Z, Paquet-Durand F, Jiao K. Inhibition of the MAPK/c-Jun-EGR1 Pathway Decreases Photoreceptor Cell Death in the rd1 Mouse Model for Inherited Retinal Degeneration. International Journal of Molecular Sciences. 2022; 23(23):14600. https://doi.org/10.3390/ijms232314600
Chicago/Turabian StyleDong, Yujie, Wenrong Xu, Yan Li, Chunling Wei, Yunzhang Hu, Zhulin Hu, François Paquet-Durand, and Kangwei Jiao. 2022. "Inhibition of the MAPK/c-Jun-EGR1 Pathway Decreases Photoreceptor Cell Death in the rd1 Mouse Model for Inherited Retinal Degeneration" International Journal of Molecular Sciences 23, no. 23: 14600. https://doi.org/10.3390/ijms232314600
APA StyleDong, Y., Xu, W., Li, Y., Wei, C., Hu, Y., Hu, Z., Paquet-Durand, F., & Jiao, K. (2022). Inhibition of the MAPK/c-Jun-EGR1 Pathway Decreases Photoreceptor Cell Death in the rd1 Mouse Model for Inherited Retinal Degeneration. International Journal of Molecular Sciences, 23(23), 14600. https://doi.org/10.3390/ijms232314600




