G Protein-Coupled Receptor 37L1 Modulates Epigenetic Changes in Human Renal Proximal Tubule Cells
Abstract
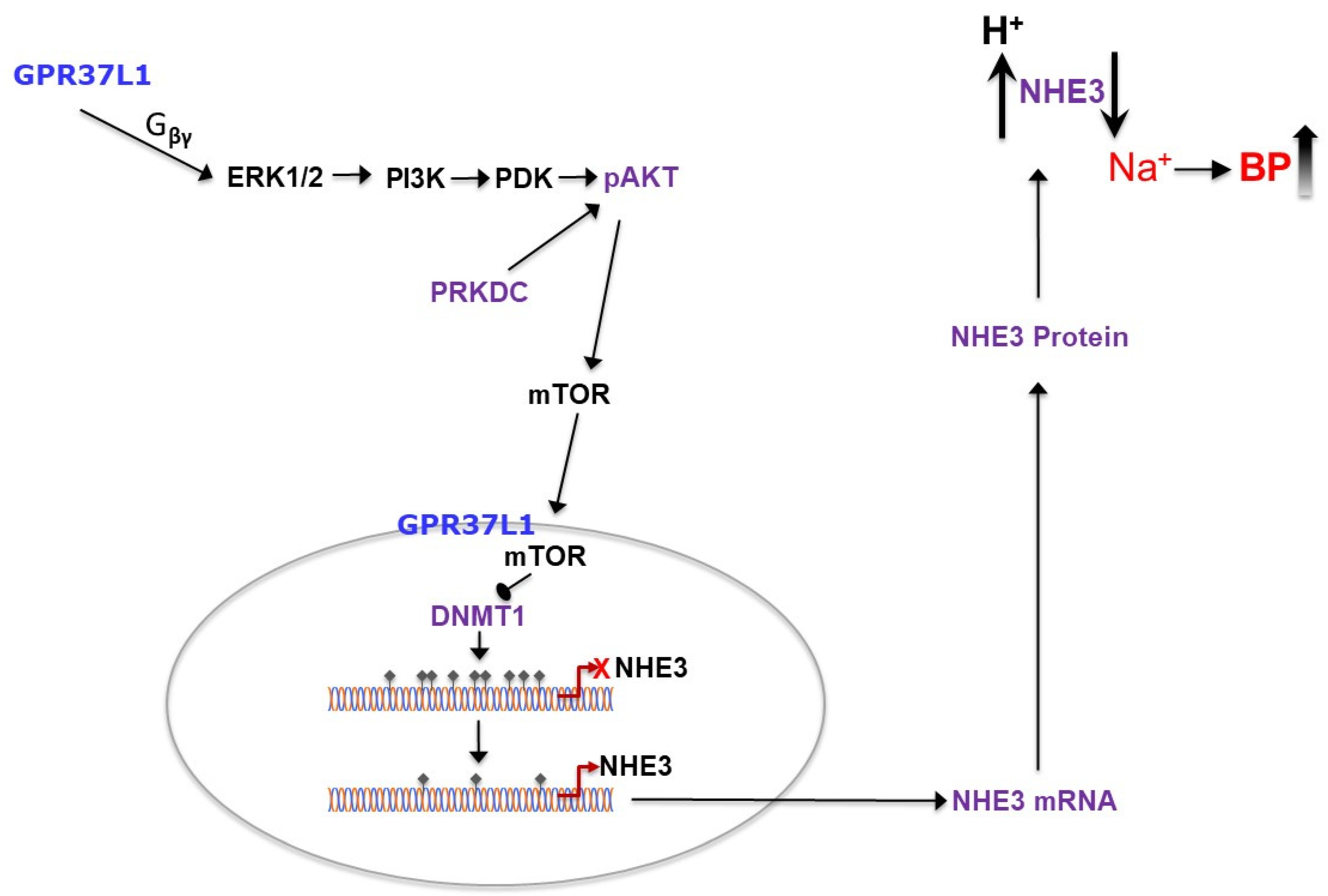
1. Introduction
2. Results
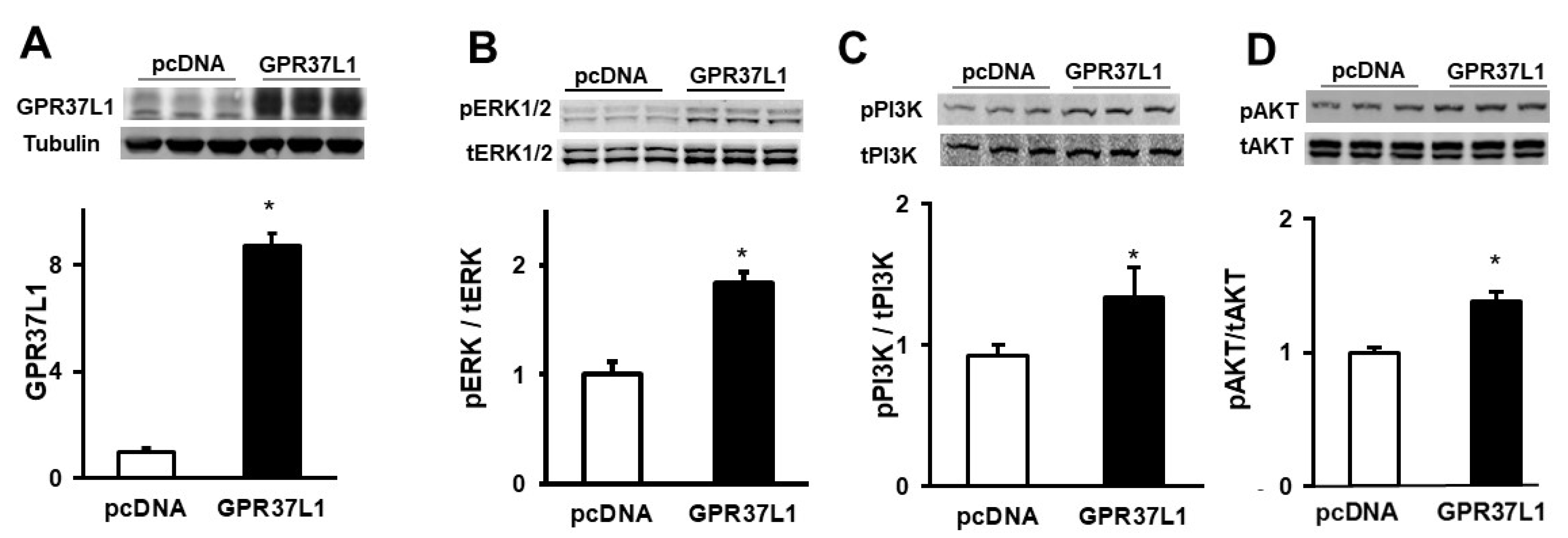
2.1. GPR37L1 Is Associated with Components of PI3K/AKT Signaling Pathway and Nuclear Transport Proteins
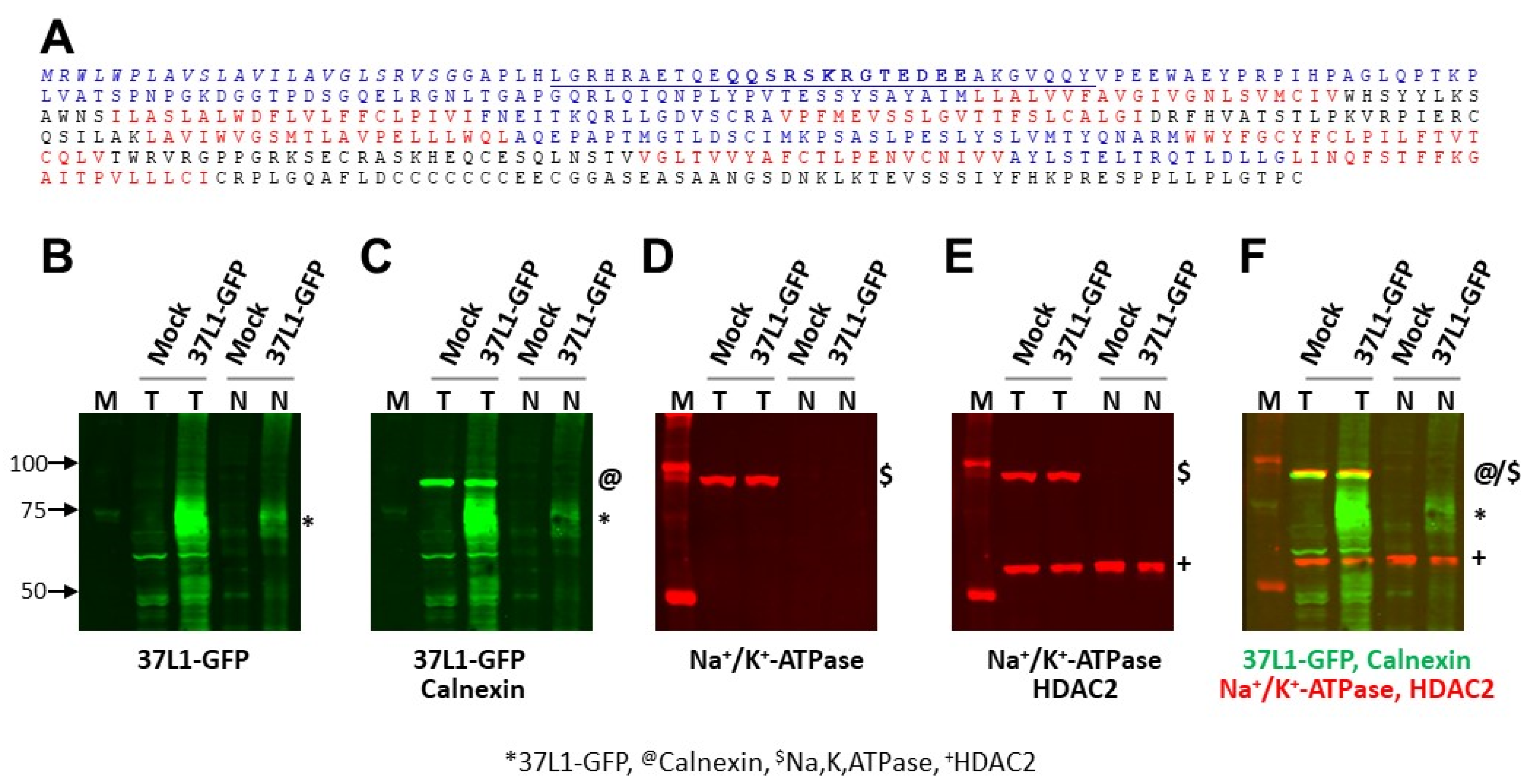
2.2. GPR37L1 Is Expressed on the Nuclear Membrane
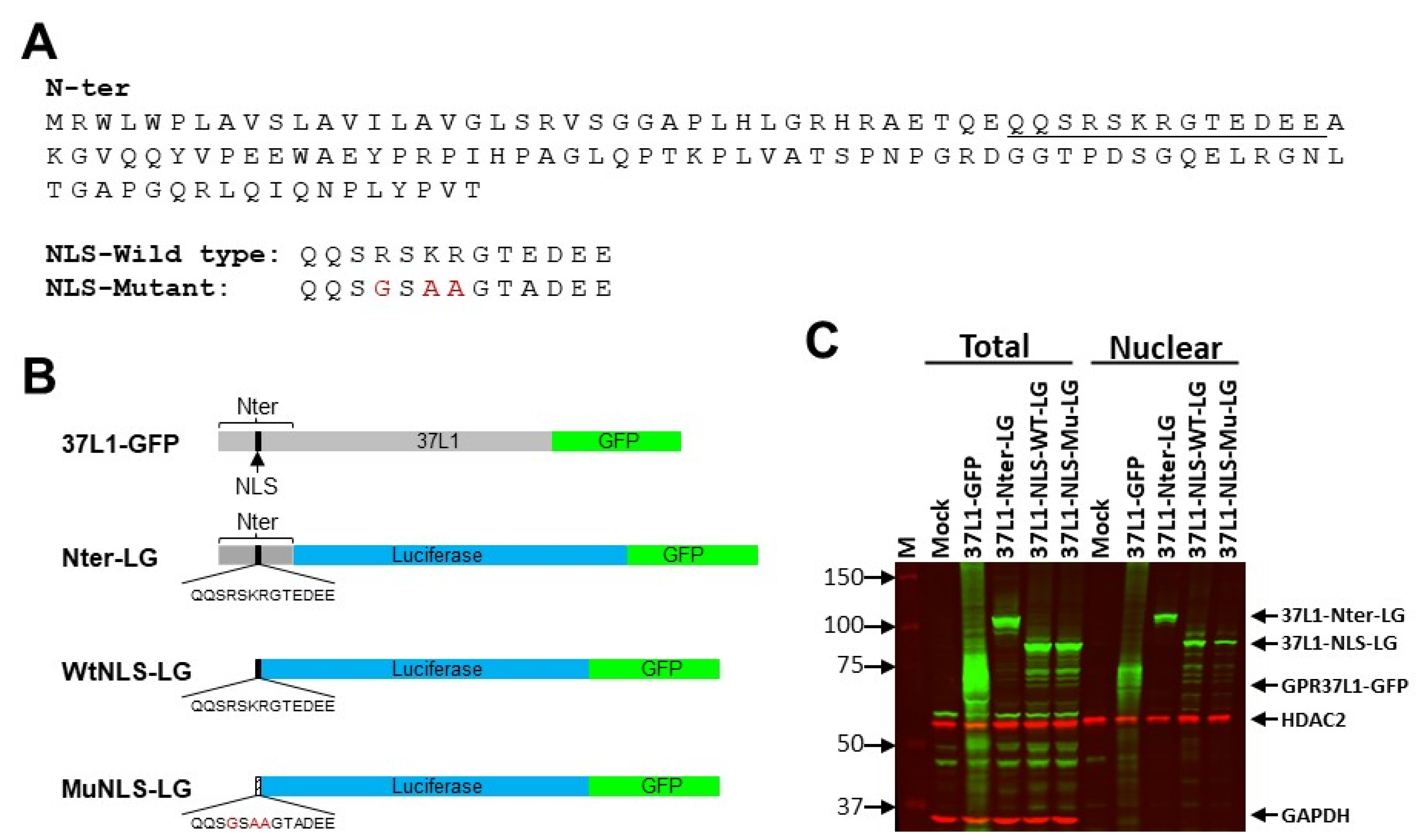
2.3. NLS Is on the N-Terminus of GPR37L1
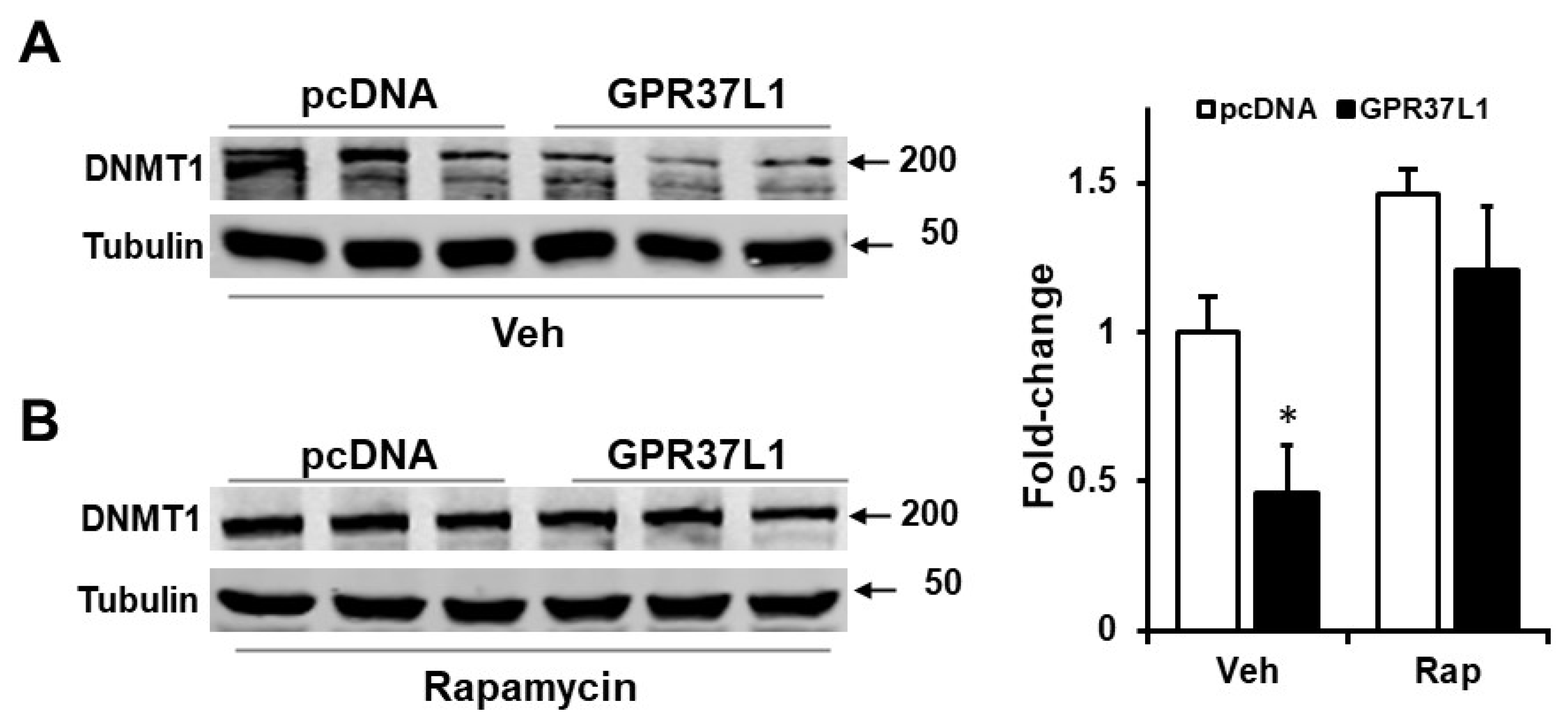
2.4. GPR37L1 Regulates the Expression of DNMT1 through ERK1/2/PI3K/AKT/mTOR Pathway
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Cell Culture
4.3. Plasmids
4.4. Glycerol Gradient Fractionation and Mass Spectrometry
4.5. Mass Spectrometry Analyses
4.6. Total and Nuclear Protein Preparation
4.7. In Silico Analyses
4.8. Immunoblotting
4.9. Immunofluorescence
4.10. Quantitative Real-Time PCR
4.11. Global DNA Methylation Assay
4.12. Methylated DNA Immunoprecipitation Assay
4.13. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E.; Collins, K.J.; Dennison Himmelfarb, C.; De Palma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA /ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2018, 71, e127–e248. [Google Scholar] [PubMed]
- Russo, A.; Di Gaetano, C.; Cugliari, G.; Matullo, G. Advances in the Genetics of Hypertension: The Effect of Rare Variants. Int. J. Mol. Sci. 2018, 19, 688. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, J. ‘Polygenic’ analyses may sharpen disease risk predictions. Science 2019, 366, 1431. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.E.; Granger, J.P.; do Carmo, J.M.; da Silva, A.A.; Dubinion, J.; George, E.; Hamza, S.; Speed, J.; Hall, M.E. Hypertension: Physiology and pathophysiology. Compr. Physiol. 2012, 4, 2393–2442. [Google Scholar]
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef]
- Kroeze, W.K.; Sheffler, D.J.; Roth, B.L. G-protein-coupled receptors at a glance. J. Cell Sci. 2003, 116, 4867–4869. [Google Scholar] [CrossRef]
- Zhuo, J.L.; Li, X.C. Proximal nephron. Compr Physiol. 2013, 3, 1079–1123. [Google Scholar]
- Vassilatis, D.K.; Hohmann, J.G.; Zeng, H.; Li, F.; Ranchalis, J.E.; Mortrud, M.T.; Brown, A.; Rodriguez, S.S.; Weller, J.R.; Wright, A.C.; et al. The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. USA 2003, 100, 4903–4908. [Google Scholar] [CrossRef] [PubMed]
- Regard, J.B.; Sato, I.T.; Coughlin, S.R. Anatomical profiling of G protein-coupled receptor expression. Cell 2008, 135, 561–571. [Google Scholar] [CrossRef]
- Rajkumar, P.; Aisenberg, W.H.; Acres, O.W.; Protzko, R.J.; Pluznick, J.L. Identification and characterization of novel renal sensory receptors. PLoS ONE 2014, 9, e111053. [Google Scholar] [CrossRef]
- Rapp, J.P.; Wang, S.M.; Dene, H. A genetic polymorphism in the renin gene of Dahl rats cosegregates with blood pressure. Science 1989, 243, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Flister, M.J.; Hoffman, M.J.; Reddy, P.; Jacob, H.J.; Moreno, C. Congenic mapping and sequence analysis of the Renin locus. Hypertension 2013, 61, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Giddens, M.M.; Wong, J.C.; Schroeder, J.P.; Farrow, E.G.; Smith, B.M.; Owino, S.; Soden, S.E.; Meyer, R.C.; Saunders, C.; LePichon, J.B.; et al. GPR37L1 modulates seizure susceptibility: Evidence from mouse studies and analyses of a human GPR37L1 variant. Neurobiol. Dis. 2017, 106, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Jolly, S.; Bazargani, N.; Quiroga, A.C.; Pringle, N.P.; Attwell, D.; Richardson, W.D.; Li, H. G protein-coupled receptor 37-like 1 modulates astrocyte glutamate transporters and neuronal NMDA receptors and is neuroprotective in ischemia. Glia 2018, 66, 47–61. [Google Scholar] [CrossRef]
- Di Pietro, C.; La Sala, G.; Matteoni, R.; Marazziti, D.; Tocchini-Valentini, G.P. Genetic ablation of Gpr37l1 delays tumor occurrence in Ptch1+/- mouse models of medulloblastoma. Exp. Neurol. 2019, 312, 33–42. [Google Scholar] [CrossRef]
- Arking, D.E.; Reinier, K.; Post, W.; Jui, J.; Hilton, G.; O’Connor, A.; Prineas, R.J.; Boerwinkle, E.; Psaty, B.M.; Tomaselli, G.F.; et al. Genome-wide association study identifies GPC5 as a novel genetic locus protective against sudden cardiac arrest. PLoS ONE 2010, 5, e9879. [Google Scholar] [CrossRef]
- Min, K.D.; Asakura, M.; Liao, Y.; Nakamaru, K.; Okazaki, H.; Takahashi, T.; Fujimoto, K.; Ito, S.; Takahashi, A.; Asanuma, H.; et al. Identification of genes related to heart failure using global gene expression profiling of human failing myocardium. Biochem. Biophys. Res. Commun. 2010, 393, 55–60. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Jobe, S.M.; Markham, B.E. Alpha-myosin heavy chain gene regulation: Delineation and characterization of the cardiac muscle-specific enhancer and muscle-specific promoter. J. Mol. Cell Cardiol. 1996, 28, 1211–1225. [Google Scholar] [CrossRef]
- Coleman, J.L.J.; Mouat, M.A.; Wu, J.; Jancovski, N.; Bassi, J.K.; Chan, A.Y.; Humphreys, D.T.; Mrad, N.; Yu, Z.Y.; Ngo, T.; et al. Orphan receptor GPR37L1 contributes to the sexual dimorphism of central cardiovascular control. Biol. Sex Differ. 2018, 9, 14. [Google Scholar] [CrossRef]
- Mouat, M.A.; Jackson, K.L.; Coleman, J.L.J.; Paterson, M.R.; Graham, R.M.; Head, G.A.; Smith, N.J. Deletion of Orphan G Protein-Coupled Receptor GPR37L1 in Mice Alters Cardiovascular Homeostasis in a Sex-Specific Manner. Front. Pharmacol. 2021, 11, 600266. [Google Scholar] [CrossRef]
- Zheng, X.; Asico, L.D.; Ma, X.; Konkalmatt, P.R. G protein-coupled receptor 37L1 regulates renal sodium transport and blood pressure. Am. J. Physiol. Renal. Physiol. 2019, 316, F506–F516. [Google Scholar] [CrossRef]
- Liu, B.; Mosienko, V.; Vaccari Cardoso, B.; Prokudina, D.; Huentelman, M.; Teschemacher, A.G.; Kasparov, S. Glio- and neuro-protection by prosaposin is mediated by orphan G-protein coupled receptors GPR37L1 and GPR37. Glia 2018, 66, 2414–2426. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.L.; Ngo, T.; Schmidt, J.; Mrad, N.; Liew, C.K.; Jones, N.M.; Graham, R.M.; Smith, N.J. Metalloprotease cleavage of the N terminus of the orphan G protein-coupled receptor GPR37L1 reduces its constitutive activity. Sci. Signal. 2016, 9, ra36. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.C.; Giddens, M.M.; Schaefer, S.A.; Hall, R.A. GPR37, and GPR37L1 are receptors for the neuroprotective, and glioprotective factors prosaptide, and prosaposin. Proc. Natl. Acad. Sci. USA 2013, 110, 9529–9534. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.C.; Giddens, M.M.; Coleman, B.M.; Hall, R.A. The protective role of prosaposin, and its receptors in the nervous system. Brain Res. 2014, 1585, 1–12. [Google Scholar] [CrossRef]
- Jacoby, E.; Bouhelal, R.; Gerspacher, M.; Seuwen, K. The 7 TM G-protein-coupled receptor target family. Chem. Med. Chem. 2006, 1, 761–782. [Google Scholar] [CrossRef]
- Seyedabadi, M.; Gharghabi, M.; Gurevich, E.V.; Gurevich, V.V. Structural basis of GPCR coupling to distinct signal transducers: Implications for biased signaling. Structural basis of GPCR coupling to distinct signal transducers: Implications for biased signaling. Trends Biochem. Sci. 2022, 47, 570–581. [Google Scholar] [CrossRef]
- Crilly, S.E.; Puthenveedu, M.A. Compartmentalized GPCR Signaling from Intracellular Membranes. J. Membr. Biol. 2021, 254, 259–271. [Google Scholar] [CrossRef]
- Calebiro, D.; Nikolaev, V.O.; Gagliani, M.C.; De Filippis, T.; Dees, C.; Tacchetti, C.; Lohse, M.J. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 2009, 7, e1000172. [Google Scholar] [CrossRef]
- Sorkin, A.; von Zastrow, M. Endocytosis and signalling: Intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009, 10, 609–622. [Google Scholar] [CrossRef]
- Gobeil, F.; Fortier, A.; Zhu, T.; Bossolasco, M.; Leduc, M.; Grandbois, M.; Barbaz, D. G protein-coupled receptors signalling at the cell nucleus: An emerging paradigm. Can. J. Physiol. Pharmacol. 2006, 84, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Bhosle, V.K.; Rivera, J.C.; Chemtob, S. New insights into mechanisms of nuclear translocation of G protein-coupled receptors. Small GTPases. 2019, 10, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Liccardp, F.; Luini, A.; Di Martino, R. Endomembrane-Based Signaling by GPCRs and G-Proteins. Cells 2022, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Vaniotis, G.; Allen, B.G.; Hébert, T.E. Nuclear GPCRs in cardiomyocytes: An insider’s view of β-adrenergic receptor signaling. Am. J. Physiol. Heart. Circ. Physiol. 2011, 301, H1754–H1764. [Google Scholar] [CrossRef]
- Campden, R.; Audet, N.; Hébert, T.E. Nuclear G protein signaling: New tricks for old dogs. J. Cardiovasc. Pharmacol. 2015, 65, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Boivin, B.; Lavoie, C.; Vaniotis, G.; Baragli, A.; Villeneuve, L.R.; Ethier, N.; Trieu, P.; Allen, B.G.; Hébert, T.E. Functional beta-adrenergic receptor signaling on nuclear membranes in adult rat and mouse ventricular cardiomyocytes. Cardiovasc Res. 2006, 71, 69–78. [Google Scholar] [CrossRef]
- Patel, N.; Itakura, T.; Gonzalez, J.M., Jr.; Schwartz, S.G.; Fini, M.E. GPR158, an orphan member of G protein-coupled receptor Family C: Glucocorticoid-stimulated expression and novel nuclear role. PLoS ONE 2013, 8, e57843. [Google Scholar] [CrossRef]
- Lu, D.; Yang, H.; Shaw, G.; Raizada, M.K. Angiotensin II-induced nuclear targeting of the angiotensin type 1 (AT1) receptor in brain neurons. Endocrinology 1998, 139, 365–375. [Google Scholar] [CrossRef]
- Kinsey, C.G.; Bussolati, G.; Bosco, M.; Kimura, T.; Pizzorno, M.C.; Chernin, M.I.; Cassoni, P.; Novak, J.F. Constitutive and ligand-induced nuclear localization of oxytocin receptor. J. Cell Mol. Med. 2007, 11, 96–110. [Google Scholar] [CrossRef]
- Lee, D.K.; Lanca, A.J.; Cheng, R.; Nguyen, T.; Ji, X.D.; Gobeil, F., Jr.; Chemtob, S.; George, S.R.; O’Dowd, B.F. Agonist-independent nuclear localization of the apelin, angiotensin AT1, and bradykinin B2 receptors. J. Biol. Chem. 2004, 279, 7901–7908. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. Regulation of, m.T.O.R.C.1.; its impact on gene expression at a glance. J. Cell Sci. 2013, 126 Pt 8, 1713–1719. [Google Scholar] [PubMed]
- Huang, K.; Fingar, D.C. Growing knowledge of the mTOR signaling network. Semin. Cell Dev. Biol. 2014, 36, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.A. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol. Metab. 2013, 24, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, S.; Wang, S.; Xie, J.; Cui, D. mTOR signaling: A pivotal player in Treg cell dysfunction in systemic lupus erythematosus. Clin. Immunol. 2022, 245, 109153. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Hresko, R.C.; Mueckler, M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 2005, 280, 40406–40416. [Google Scholar] [CrossRef]
- Dragoi, A.M.; Fu, X.; Ivanov, S.; Zhang, P.; Sheng, L.; Wu, D.; Li, G.C.; Chu, W.M. DNA-PKcs, but not TLR9, is required for activation of Akt by CpG-DNA. EMBO J. 2015, 24, 779–789. [Google Scholar] [CrossRef]
- Feng, J.; Park, J.; Cron, P.; Hess, D.; Hemmings, B.A. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004, 279, 41189–41196. [Google Scholar] [CrossRef]
- Luttrell, L.M.; van Biesen, T.; Hawes, B.E.; Koch, W.J.; Krueger, K.M.; Touhara, K.; Lefkowitz, R.J. G-protein-coupled receptors and their regulation: Activation of the MAP kinase signaling pathway by G-protein-coupled receptors. Adv. Second. Messenger Phosphoprot. Res. 1997, 31, 263–277. [Google Scholar]
- Zhang, Y.P.; Huang, Y.T.; Huang, T.S.; Pang, W.; Zhu, J.J.; Liu, Y.F.; Tang, R.Z.; Zhao, C.R.; Yao, W.J.; Li, Y.S.; et al. The Mammalian Target of Rapamycin and DNA methyltransferase 1 axis mediates vascular endothelial dysfunction in response to disturbed flow. Sci. Rep. 2017, 7, 14996. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Su, Z.; Fei, H.; Liu, X.; Pan, Q. The novel mTOR inhibitor Torin-2 induces autophagy and downregulates the expression of UHRF1 to suppress hepatocarcinoma cell growth. Oncol. Rep. 2015, 34, 1708–1716. [Google Scholar] [CrossRef] [PubMed]
- Lagerstrom, M.C.; Schioth, H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, R.J.; Shenoy, S.K. Transduction of receptor signals by beta-arrestins. Science 2005, 308, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves-Monteiro, S.; Ribeiro-Oliveira, R.; Vieira-Rocha, M.S.; Vojtek, M.; Sousa, J.B.; Diniz, C. Insights into Nuclear G-Protein-Coupled Receptors as Therapeutic Targets in Non-Communicable Diseases. Pharmaceuticals 2021, 14, 439. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Low, D.; Mizoguchi, A.; Mizoguchi, E. DNA methylation in inflammatory bowel disease and beyond. World J. Gastroenterol. 2013, 19, 5238–5249. [Google Scholar] [CrossRef]
- Wang, X.; Armando, I.; Upadhyay, K.; Pascua, A.; Jose, P.A. The regulation of proximal tubular salt transport in hypertension: An update. Curr. Opin. Nephrol. Hypertens. 2009, 18, 412–420. [Google Scholar] [CrossRef]
- Bobulescu, I.A.; Moe, O.W. Luminal Na(+)/H(+) exchange in the proximal tubule. Pflügers Arch. 2009, 458, 5–21. [Google Scholar] [CrossRef]
- Bobulescu, I.A.; Di Sole, F.; Moe, O.W. Na+/H+ exchangers: Physiology and link to hypertension and organ ischemia. Curr. Opin. Nephrol. Hypertens. 2005, 14, 485–494. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, X.; Patel, A.; Potru, R.; Azizi-Ghannad, S.; Dolinger, M.; Cao, J.; Bartholomew, C.; Mazurkiewicz, J.; Conti, D.; et al. Rapamycin Inhibition of mTOR Reduces Levels of the Na+/H+ Exchanger 3 in Intestines of Mice and Humans, Leading to Diarrhea. Gastroenterology 2015, 149, 151–162. [Google Scholar] [CrossRef]
- Kumar, A.; Malhotra, P.; Coffing, H.; Priyamvada, S.; Anbazhagan, A.N.; Krishnan, H.R.; Gill, R.K.; Alrefai, W.A.; Gavin, D.P.; Pandey, S.C.; et al. Epigenetic modulation of intestinal Na+/H+ exchanger-3 expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 314, G309–G318. [Google Scholar] [CrossRef] [PubMed]
- Gildea, J.J.; McGrath, H.E.; Van Sciver, R.E.; Wang, D.B.; Felder, R.A. Isolation, growth, and characterization of human renal epithelial cells using traditional and 3D methods. Methods Mol. Biol. 2013, 945, 329–345. [Google Scholar]
- Panigrahi, A.K.; Allen, T.E.; Stuart, K.; Haynes, P.A.; Gygi, S.P. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J. Am. Soc. Mass Spectrom. 2003, 14, 728–735. [Google Scholar] [CrossRef]
- Free, R.B.; Hazelwood, L.A.; Sibley, D.R. Identifying novel protein-protein interactions using co-immunoprecipitation and mass spectroscopy. Curr. Protoc. Neurosci. 2009, 46, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Have, S.; Boulon, S.; Ahmad, Y.; Lamond, A.I. Mass spectrometry-based immuno-precipitation proteomics—The user’s guide. Proteomics 2011, 11, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Collins, M.; An, J.; Geiser, R.; Tegeler, T.; Tsantilas, K.; Garcia, K.; Pirrotte, P.; Bowser, R. Immunoprecipitation and mass spectrometry defines an extensive RBM45 protein-protein interaction network. Brain Res. 2016, 15, 79–93. [Google Scholar] [CrossRef]
- Jensen, O.N.; Wilm, M.; Shevchenko, A.; Mann, M. Sample preparation methods for mass spectrometry peptide mapping directly from 2-DE gels. In Methods in Molecular Biology; Link, A.J., Ed.; Humana Press: Totowa, NJ, USA, 1999; Volume 112, pp. 513–530. [Google Scholar]
- Wang, P.; Bouwman, F.G.; Mariman, E.C. Generally detected proteins in comparative proteomics—A matter of cellular stress response? Proteomics 2009, 9, 2955–2966. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Thu, K.L.; Vucic, E.A.; Kennett, J.Y.; Heryet, C.; Brown, C.J.; Lam, W.L.; Wilson, I.M. Methylated DNA immunoprecipitation. J. Vis. Exp. 2009, 23, 935. [Google Scholar] [CrossRef]
- Borgel, J.; Guibert, S.; Weber, M. Methylated DNA immunoprecipitation (MeDIP) from low amounts of cells. Methods Mol. Biol. 2012, 925, 149–158. [Google Scholar]






| Protein ID | Protein Names | Gene Names | |
|---|---|---|---|
| Nuclear transport proteins | |||
| O95373 | Importin-1 | IPO1 | |
| O15397 | Importin-7 | IPO7 | |
| O14980 | Exportin-1 | XPO1 | |
| Q9HAV4 | Exportin-5 | XPO5 | |
| Q14974 | Importin subunit beta-1 | KPNB1 | |
| P52292 | Importin subunit alpha-1 | KPNA2 | |
| Q8TEM1 | Nuclear pore membrane glycoprotein 210 | NUP210 | |
| Kinases | |||
| P42345 | Serine/threonine-protein kinase mTOR | MTOR | |
| O14654 | Insulin receptor substrate 4 | IRS4 | |
| P78527 | DNA-dependent protein kinase catalytic subunit | PRKDC | |
| DNA methylation | P26358 | DNA (cytosine-5)-methyltransferase 1 | DNMT1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Armando, I.; Cuevas, S.; Fan, C.; Kumar, M.; Izzi, Z.; Jose, P.A.; Konkalmatt, P.R. G Protein-Coupled Receptor 37L1 Modulates Epigenetic Changes in Human Renal Proximal Tubule Cells. Int. J. Mol. Sci. 2022, 23, 14456. https://doi.org/10.3390/ijms232214456
Armando I, Cuevas S, Fan C, Kumar M, Izzi Z, Jose PA, Konkalmatt PR. G Protein-Coupled Receptor 37L1 Modulates Epigenetic Changes in Human Renal Proximal Tubule Cells. International Journal of Molecular Sciences. 2022; 23(22):14456. https://doi.org/10.3390/ijms232214456
Chicago/Turabian StyleArmando, Ines, Santiago Cuevas, Caini Fan, Megha Kumar, Zahra Izzi, Pedro A. Jose, and Prasad R. Konkalmatt. 2022. "G Protein-Coupled Receptor 37L1 Modulates Epigenetic Changes in Human Renal Proximal Tubule Cells" International Journal of Molecular Sciences 23, no. 22: 14456. https://doi.org/10.3390/ijms232214456
APA StyleArmando, I., Cuevas, S., Fan, C., Kumar, M., Izzi, Z., Jose, P. A., & Konkalmatt, P. R. (2022). G Protein-Coupled Receptor 37L1 Modulates Epigenetic Changes in Human Renal Proximal Tubule Cells. International Journal of Molecular Sciences, 23(22), 14456. https://doi.org/10.3390/ijms232214456








