Antiproliferative Activity Predictor: A New Reliable In Silico Tool for Drug Response Prediction against NCI60 Panel
Abstract
1. Introduction
2. Results and Discussion
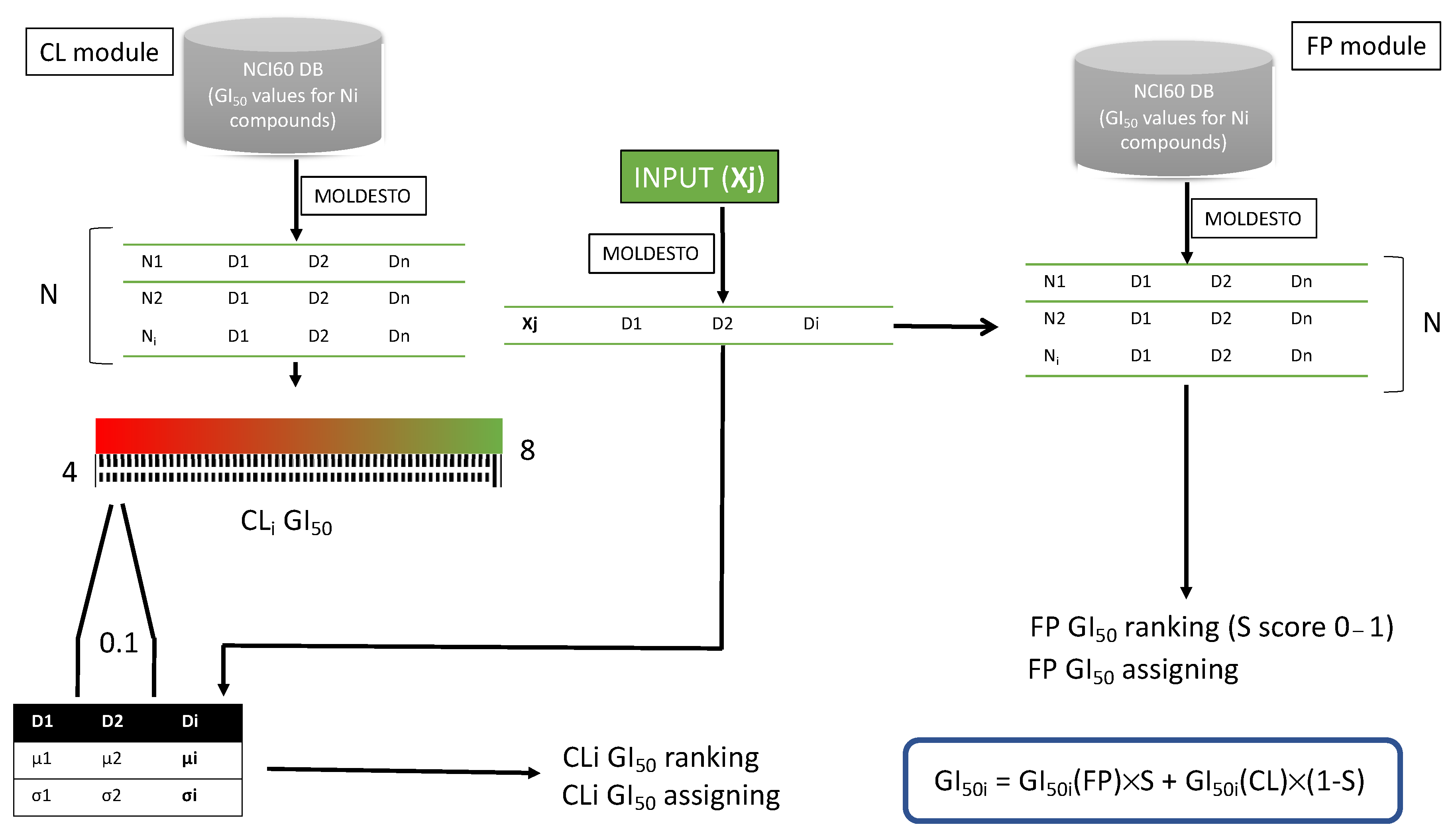
2.1. NCI60 Antiproliferative Activity Predictor Tool
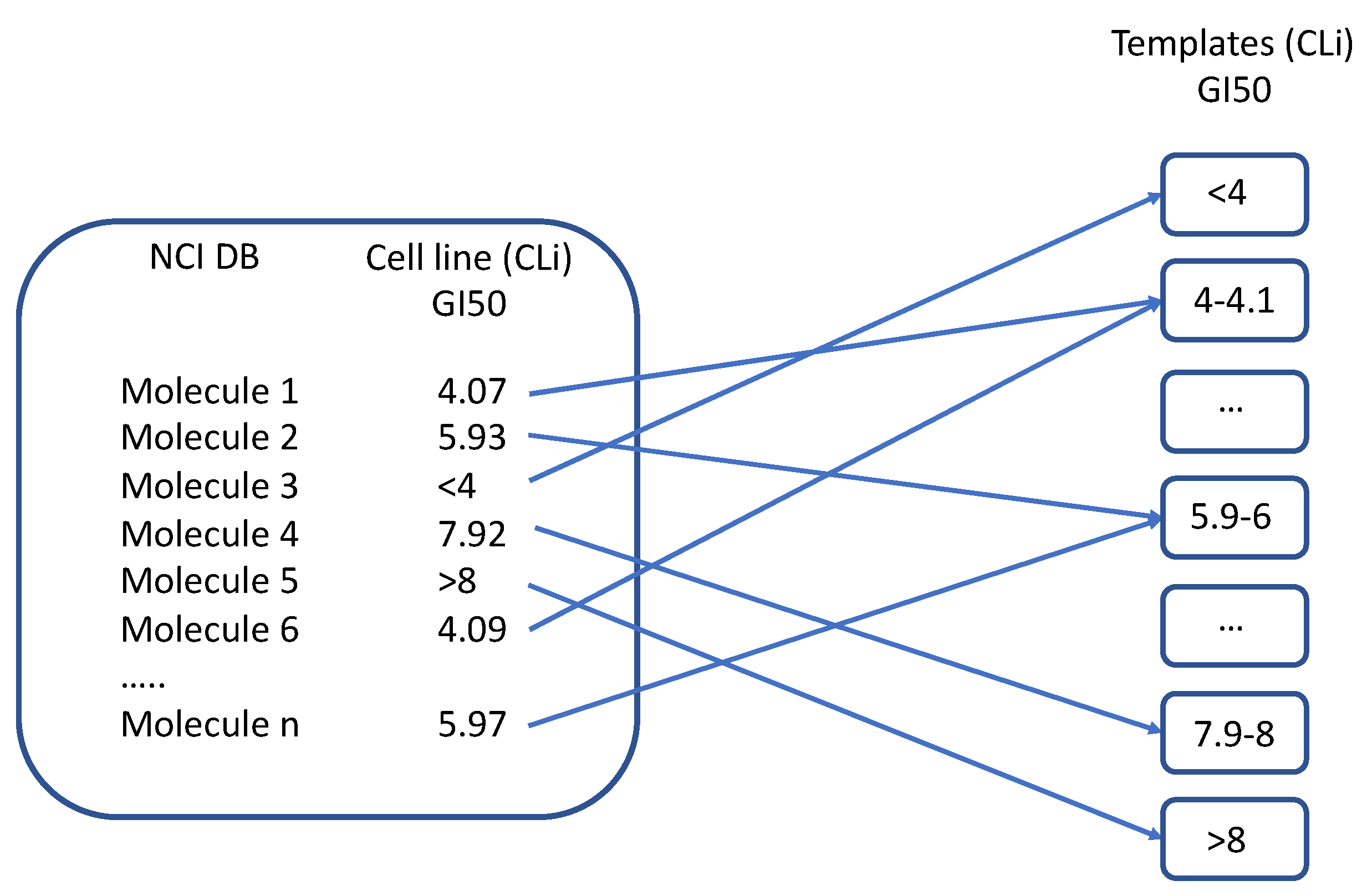
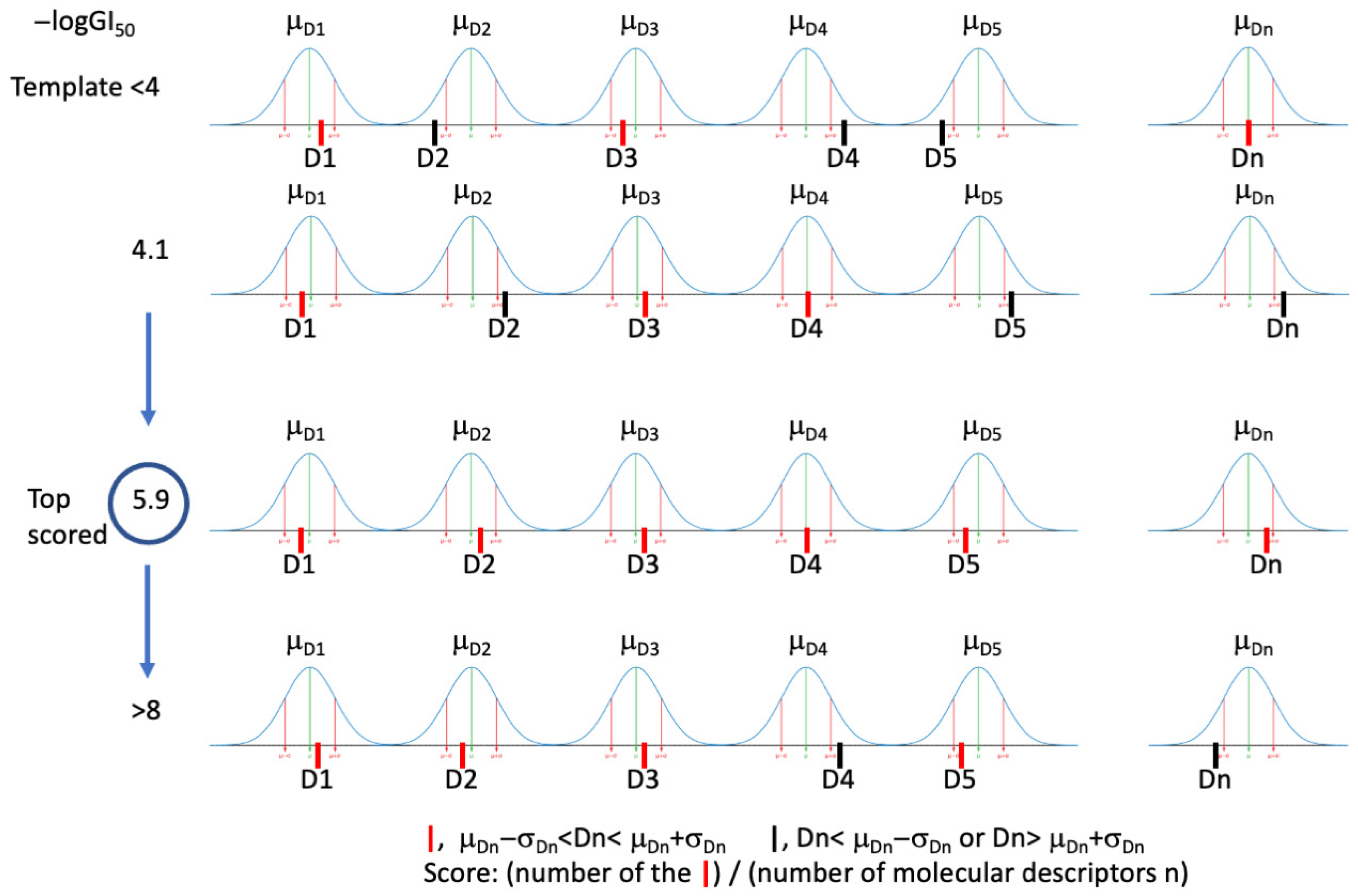
2.1.1. Description of the Tool Learning Process
2.1.2. Validation of the AAP Tool
2.1.3. Parameter Optimization for Cell Line/Subpanel Activity Prediction
2.1.4. Application of the AAP Tool for the Virtual Screening of an In-House Structure Database
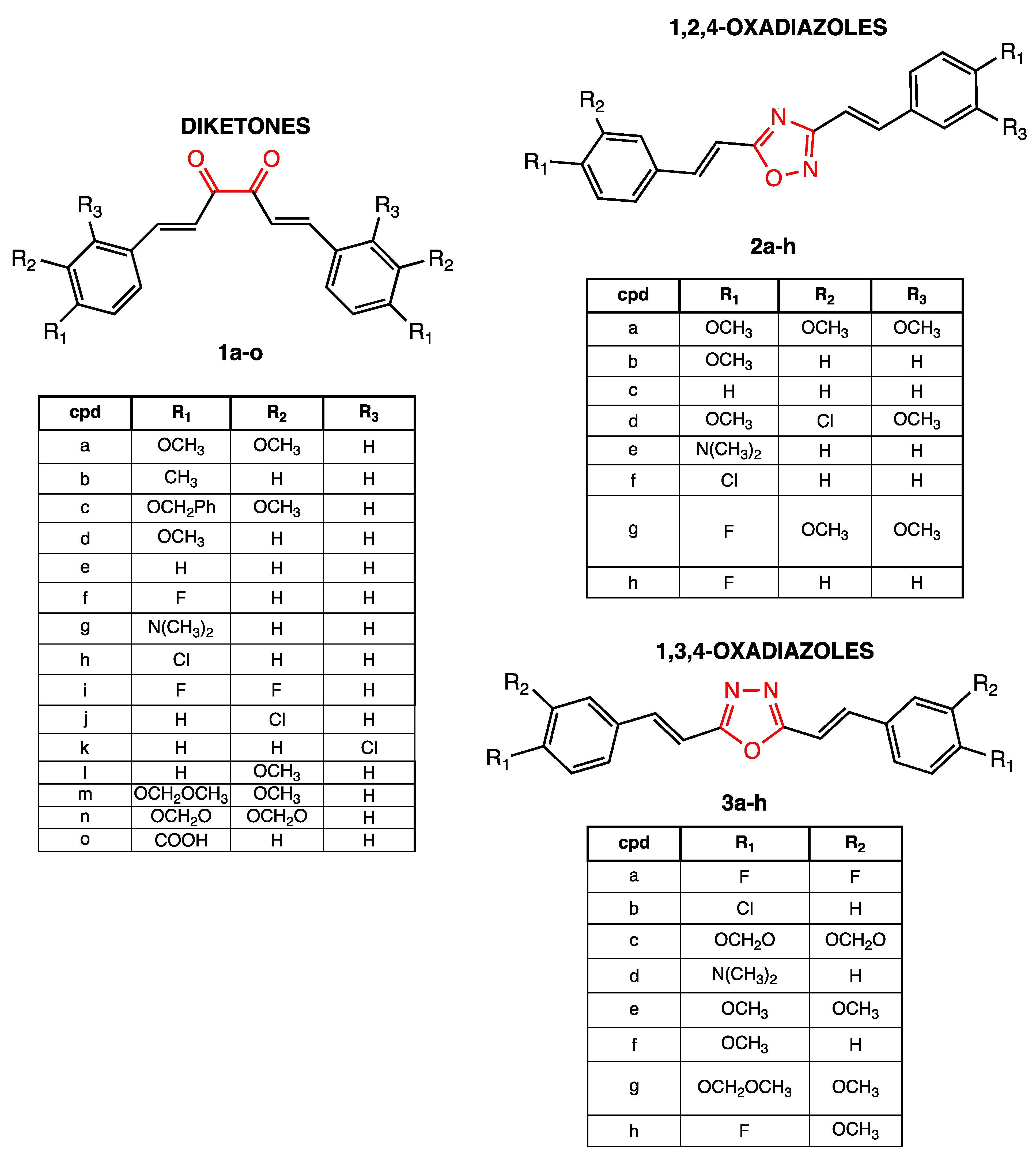
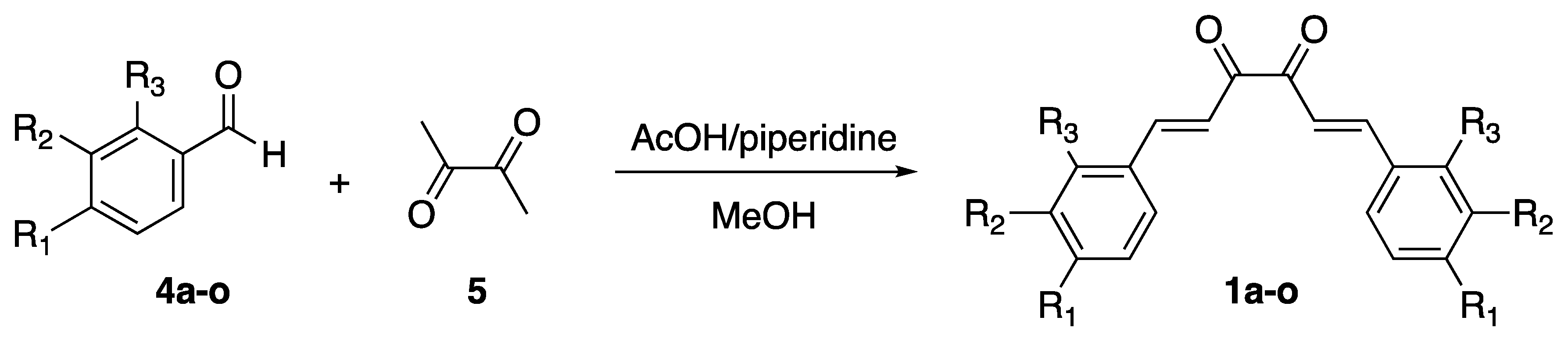
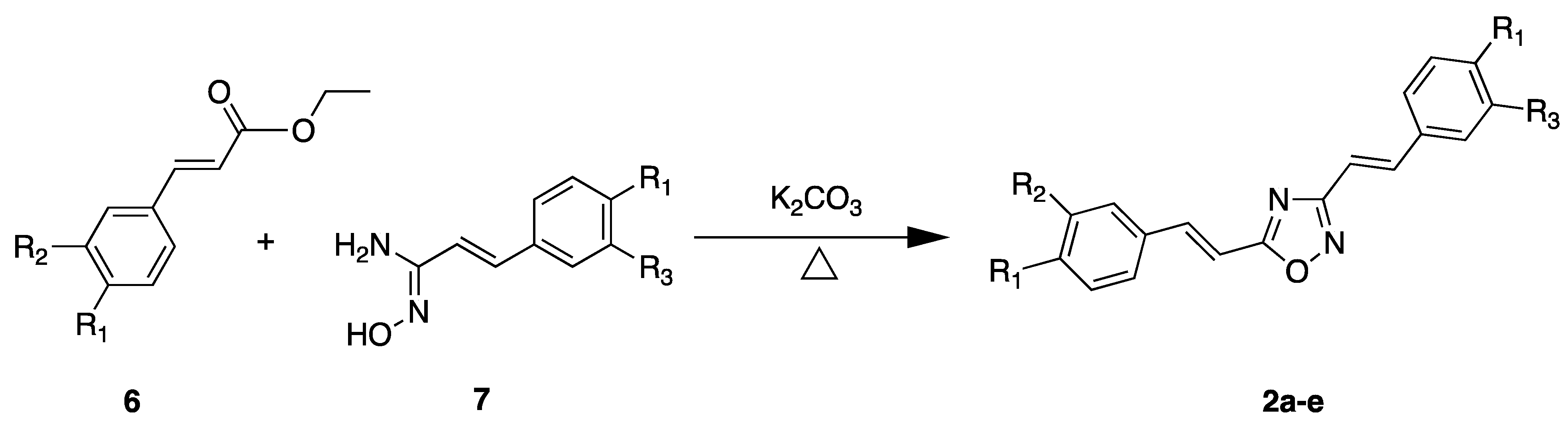
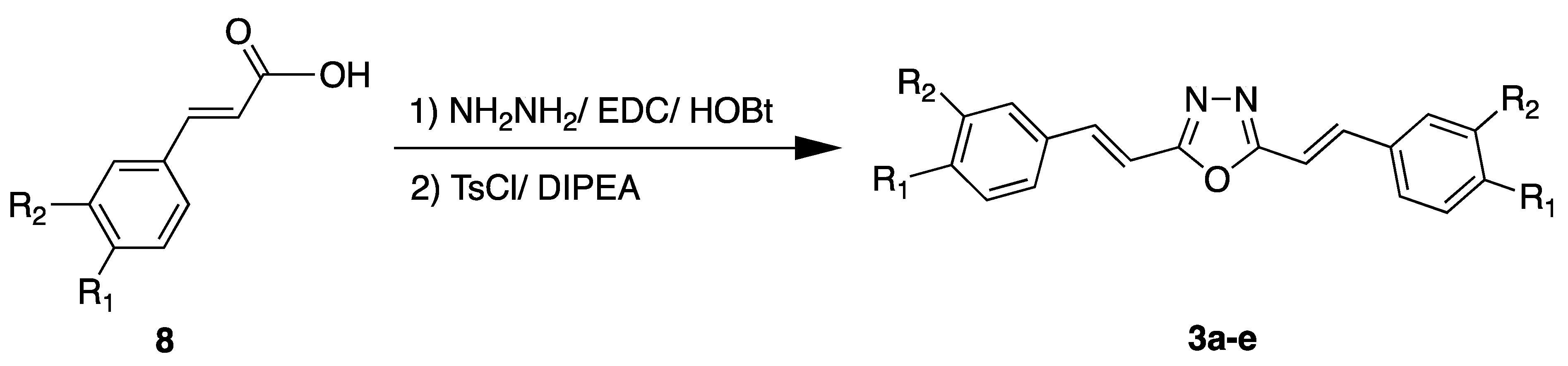
2.2. Chemistry
2.3. Biological Assays: NCI60 Human Tumor Cell Lines Screen Selected Compounds
2.3.1. One-Dose Antiproliferative Assay
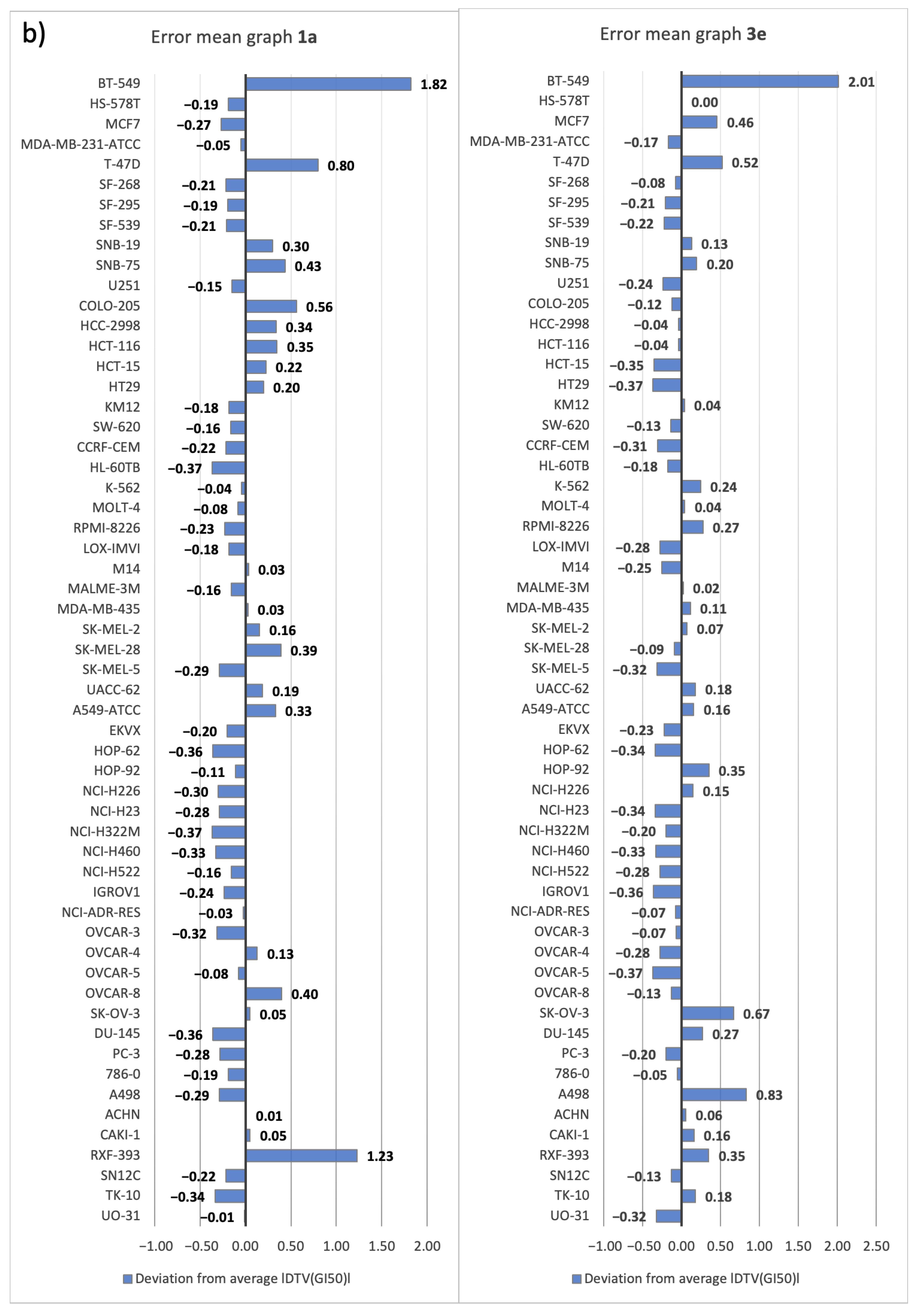
2.3.2. Five-Dose Antiproliferative Assay for the Most Active Derivatives, 1a and 3e
3. Materials and Methods
3.1. Computational Studies
3.1.1. Hardware
3.1.2. Software
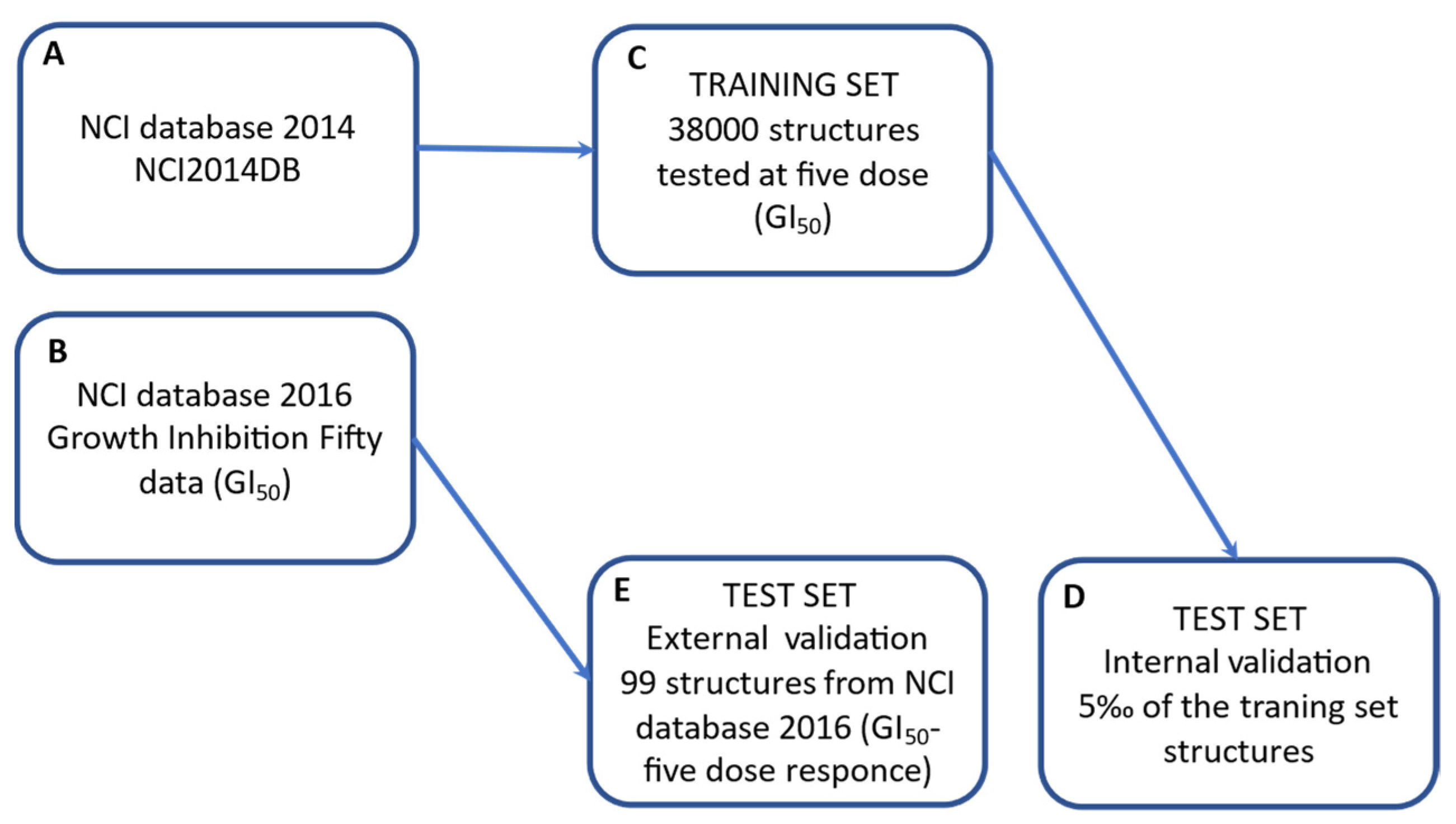
3.1.3. Database Selection and Dataset Building
3.1.4. MOLDESTO: A New Software for Molecular Descriptor Calculations
3.1.5. DRUDIT Settings for Antiproliferative Activity Predictor (AAP) Tool
3.2. Chemistry
3.3. NCI60 Antiproliferative Screenings
3.3.1. Compound Selection Guidelines
3.3.2. One-Dose Assay
3.3.3. Five-Dose Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Bodycombe, N.E.; Cheah, J.H.; Price, E.V.; Liu, K.; Schaefer, G.I.; Ebright, R.Y.; Stewart, M.L.; Ito, D.; Wang, S.; et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013, 154, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.P.; Varma, S.; Gottesman, M.M. The clinical relevance of cancer cell lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer cell lines are useful model systems for medical research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Goodspeed, A.; Heiser, L.M.; Gray, J.W.; Costello, J.C. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol. Cancer Res. 2016, 14, 3–13. [Google Scholar] [CrossRef]
- Takimoto, C.H. Anticancer drug development at the US National Cancer Institute. Cancer Chemother. Pharmacol. 2003, 52 (Suppl. 1), S29–S33. [Google Scholar] [CrossRef] [PubMed]
- NCI-60 Human Tumor Cell Lines Screen—Introduction. Available online: https://dtp.cancer.gov/discovery_development/nci-60/ (accessed on 15 November 2022).
- Molecular Characterization of the NCI-60. Available online: https://dtp.cancer.gov/discovery_development/nci-60/characterization.htm (accessed on 15 November 2022).
- Covell, D.G.; Huang, R.; Wallqvist, A. Anticancer medicines in development: Assessment of bioactivity profiles within the National Cancer Institute anticancer screening data. Mol. Cancer Ther. 2007, 6, 2261–2270. [Google Scholar] [CrossRef]
- NCI60 Human Timor Cell Lines Screen—Cell Lines in the In Vitro Screen. Available online: https://dtp.cancer.gov/discovery_development/nci-60/cell_list.htm (accessed on 15 November 2022).
- Firoozbakht, F.; Yousefi, B.; Schwikowski, B. An overview of machine learning methods for monotherapy drug response prediction. Brief. Bioinform. 2021, 23, bbab408. [Google Scholar] [CrossRef]
- Paull, K.D.; Shoemaker, R.H.; Hodes, L.; Monks, A.; Scudiero, D.A.; Rubinstein, L.; Plowman, J.; Boyd, M.R. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989, 81, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Zaharevitz, D.W.; Holbeck, S.L.; Bowerman, C.; Svetlik, P.A. COMPARE: A web accessible tool for investigating mechanisms of cell growth inhibition. J. Mol. Graph. Model. 2002, 20, 297–303. [Google Scholar] [CrossRef]
- NCI-60 Analysis Tools—CellMiner. Available online: http://discover.nci.nih.gov/cellminer/ (accessed on 15 November 2022).
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.; Elloumi, F.; Varma, S.; Wang, Y.; Rajapakse, V.N.; Aladjem, M.I.; Robert, J.; Sander, C.; Pommier, Y.; Reinhold, W.C. CellMiner Cross-Database (CellMinerCDB) version 1.2: Exploration of patient-derived cancer cell line pharmacogenomics. Nucleic Acids Res. 2021, 49, D1083–D1093. [Google Scholar] [CrossRef]
- Lind, A.P.; Anderson, P.C. Predicting drug activity against cancer cells by random forest models based on minimal genomic information and chemical properties. PLoS ONE 2019, 14, e0219774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, H.; Fang, Y.; Wang, J.; Zheng, X.; Liu, X.S. Predicting anticancer drug responses using a dual-layer integrated cell line-drug network model. PLoS Comput. Biol. 2015, 11, e1004498. [Google Scholar] [CrossRef] [PubMed]
- Cadow, J.; Born, J.; Manica, M.; Oskooei, A.; Rodríguez Martínez, M. PaccMann: A web service for interpretable anticancer compound sensitivity prediction. Nucleic Acids Res. 2020, 48, W502–W508. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Park, H.; Yang, H.J.; Lee, S.; Lee, K.Y.; Kim, T.S.; Jung, J.; Shin, J.M. Cancer Drug Response Profile scan (CDRscan): A deep learning model that predicts drug effectiveness from cancer genomic signature. Sci. Rep. 2018, 8, 8857. [Google Scholar] [CrossRef]
- Joo, M.; Park, A.; Kim, K.; Son, W.J.; Lee, H.S.; Lim, G.; Lee, J.; Lee, D.H.; An, J.; Kim, J.H.; et al. A deep learning model for cell growth inhibition IC50 prediction and its application for gastric cancer patients. Int. J. Mol. Sci. 2019, 20, 6276. [Google Scholar] [CrossRef]
- Al-Jarf, R.; de Sá, A.G.C.; Pires, D.E.V.; Ascher, D.B. pdCSM-cancer: Using graph-based signatures to identify small molecules with anticancer properties. J. Chem. Inf. Model. 2021, 61, 3314–3322. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Tutone, M.; Almerico, A.M. Virtual lock-and-key approach: The in silico revival of Fischer model by means of molecular descriptors. Eur. J. Med. Chem. 2011, 46, 4274–4280. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Tutone, M.; Barone, G.; Almerico, A.M. Multivariate analysis in the identification of biological targets for designed molecular structures: The BIOTA protocol. Eur. J. Med. Chem. 2014, 75, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Mannino, S.; Gentile, C.; Mannino, G.; Martorana, A.; Peri, D. DRUDIT: Web-based DRUgs DIscovery Tools to design small molecules as modulators of biological targets. Bioinformatics 2020, 36, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Abbate, I.; Patella, C.; Martorana, A.; Dattolo, G.; Almerico, A.M. New annelated thieno[2,3-e][1,2,3]triazolo[1,5-a]pyrimidines, with potent anticancer activity, designed through VLAK protocol. Eur. J. Med. Chem. 2013, 62, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Patella, C.; Abbate, I.; Martorana, A.; Almerico, A.M. Lead optimization through VLAK protocol: New annelated pyrrolo-pyrimidine derivatives as antitumor agents. Eur. J. Med. Chem. 2012, 55, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Abbate, I.; Gentile, C.; Angileri, F.; Martorana, A.; Almerico, A.M. Synthesis and biological activities of a new class of heat shock protein 90 inhibitors, designed by energy-based pharmacophore virtual screening. J. Med. Chem. 2013, 56, 3424–3428. [Google Scholar] [CrossRef] [PubMed]
- Diana, P.; Martorana, A.; Barraja, P.; Montalbano, A.; Carbone, A.; Cirrincione, G. Nucleophilic substitutions in the isoindole series as a valuable tool to synthesize derivatives with antitumor activity. Tetrahedron 2011, 67, 2072–2080. [Google Scholar] [CrossRef]
- Mingoia, F.; Di Sano, C.; Di Blasi, F.; Fazzari, M.; Martorana, A.; Almerico, A.M.; Lauria, A. Exploring the anticancer potential of pyrazolo[1,2-a]benzo[1,2,3,4]tetrazin-3-one derivatives: The effect on apoptosis induction, cell cycle and proliferation. Eur. J. Med. Chem. 2013, 64, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Lo Cascio, F.; Piccionello Palumbo, A.; Pace, A. Novel Small Molecules That Bind and/or Modulate Different Forms of Tau Oligomers. Patent WO2020/219714 A1, 23 April 2020. [Google Scholar]
- Battisti, A.; Palumbo Piccionello, A.; Sgarbossa, A.; Vilasi, S.; Ricci, C.; Ghetti, F.; Spinozzi, F.; Marino Gammazza, A.; Giacalone, V.; Martorana, A.; et al. Curcumin-like compounds designed to modify amyloid beta peptide aggregation patterns. RSC Adv. 2017, 7, 31714–31724. [Google Scholar] [CrossRef]
- Lo Cascio, F.; Puangmalai, N.; Ellsworth, A.; Bucchieri, F.; Pace, A.; Palumbo Piccionello, A.; Kayed, R. Toxic tau oligomers modulated by novel curcumin derivatives. Sci. Rep. 2019, 9, 19011. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Martorana, A.; La Monica, G.; Mannino, S.; Mannino, G.; Peri, D.; Gentile, C. In silico identification of small molecules as new Cdc25 inhibitors through the correlation between chemosensitivity and protein expression pattern. Int. J. Mol. Sci. 2021, 22, 3714. [Google Scholar] [CrossRef] [PubMed]
- NCI DTP Chemical Data. Available online: https://wiki.nci.nih.gov/display/NCIDTPdata/Chemical+Data (accessed on 15 November 2022).
- Sharma, R.A.; Gescher, A.J.; Steward, W.P. Curcumin: The story so far. Eur. J. Cancer 2005, 41, 1955–1968. [Google Scholar] [CrossRef] [PubMed]
- Adeluola, A.; Zulfiker, A.H.M.; Brazeau, D.; Amin, A.R.M.R. Perspectives for synthetic curcumins in chemoprevention and treatment of cancer: An update with promising analogues. Eur. J. Pharmacol. 2021, 906, 174266. [Google Scholar] [CrossRef]
- Ahsan, M.J.; Choudhary, K.; Jadav, S.S.; Yasmin, S.; Ansari, M.Y.; Sreenivasulu, R. Synthesis, antiproliferative activity, and molecular docking studies of curcumin analogues bearing pyrazole ring. Med. Chem. Res. 2015, 24, 4166–4180. [Google Scholar] [CrossRef]
- Ahsan, M.J. Evaluation of anticancer activity of curcumin analogues bearing a heterocyclic nucleus. Asian Pac. J. Cancer Prev. 2016, 17, 1739–1744. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Sabet, S.; Rashidinejad, A.; Melton, L.D.; McGillivray, D.J. Recent advances to improve curcumin oral bioavailability. Trends Food Sci. Technol. 2021, 110, 253–266. [Google Scholar] [CrossRef]
- Sanidad, K.Z.; Sukamtoh, E.; Xiao, H.; McClements, D.J.; Zhang, G. Curcumin: Recent advances in the development of strategies to improve oral bioavailability. Annu. Rev. Food Sci. Technol. 2019, 10, 597–617. [Google Scholar] [CrossRef]
- Kotha, R.R.; Luthria, D.L. Curcumin: Biological, pharmaceutical, nutraceutical, and analytical aspects. Molecules 2019, 24, 2930. [Google Scholar] [CrossRef]
- Nelson, K.M.; Dahlin, J.L.; Bisson, J.; Graham, J.; Pauli, G.F.; Walters, M.A. The essential medicinal chemistry of curcumin. J. Med. Chem. 2017, 60, 1620–1637. [Google Scholar] [CrossRef] [PubMed]
- Olotu, F.; Agoni, C.; Soremekun, O.; Soliman, M.E.S. An update on the pharmacological usage of curcumin: Has it failed in the drug discovery pipeline? Cell Biochem. Biophys. 2020, 78, 267–289. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.C.; Kumar, N.A.; Thakur, G. The potency of heterocyclic curcumin analogues: An evidence-based review. Pharmacol. Res. 2021, 166, 105489. [Google Scholar] [CrossRef] [PubMed]
- Lo Cascio, F.; Garcia, S.; Montalbano, M.; Puangmalai, N.; McAllen, S.; Pace, A.; Palumbo Piccionello, A.; Kayed, R. Modulating disease-relevant tau oligomeric strains by small molecules. J. Biol. Chem. 2020, 295, 14807–14825. [Google Scholar] [CrossRef] [PubMed]
- Sinu, C.R.; Padmaja, D.V.; Ranjini, U.P.; Seetha Lakshmi, K.C.; Suresh, E.; Nair, V. A cascade reaction actuated by nucleophilic heterocyclic carbene catalyzed intramolecular addition of enals via homoenolate to α,β-unsaturated esters: Efficient synthesis of coumarin derivatives. Org. Lett. 2013, 15, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Pace, A.; Buscemi, S.; Piccionello, A.P.; Pibiri, I. Recent advances in the chemistry of 1,2,4-oxadiazoles. Adv. Heterocycl. Chem. 2015, 116, 85–136. [Google Scholar] [CrossRef]
- NCI60 Growth Inhibition Data—Download NCI Cell Line Data. Available online: https://wiki.nci.nih.gov/display/NCIDTPdata/NCI-60+Growth+Inhibition+Data (accessed on 15 November 2022).
- Compound Submission for NCI60 Testing—Selection Guidelines. Available online: https://dtp.cancer.gov/organization/dscb/compoundSubmission/structureSelection.htm (accessed on 15 November 2022).
- NCI60 Screening Methodology—NCI60 Cell One/Five Doses Screen. Available online: https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm (accessed on 15 November 2022).
- NCI-60 Human Cancer Cell Line Screen—Standard Operating Procedures for Sample Preparation for NCI60 Screen. Available online: https://dtp.cancer.gov/discovery_development/nci-60/handling.htm (accessed on 15 November 2022).





| Z | N | G | ||
|---|---|---|---|---|
| a | b | c | ||
| 50 | 240 | 1.22 (1) | 1.23 (2) | 1.23 (3) |
| 500 | 1.22 (7) | 1.30 (8) | 1.31 (9) | |
| 760 | 1.32 (13) | 1.44 (14) | 1.42 (15) | |
| 100 | 240 | 1.31 (4) | 1.64(5) | 1.72 (6) |
| 500 | 1.23 (10) | 1.51 (11) | 1.53 (12) | |
| 760 | 1.28 (16) | 1.42 (17) | 1.44 (18) | |
| PANELS | CELL LINES | RUN | AVERAGE |DTV(GI50)| |
|---|---|---|---|
| Breast Cancer | BT-549 | 4 | 1.35 |
| HS-578T | 3 | 1.30 | |
| MCF7 | 1/7 | 1.30 | |
| MDA-MB-231-ATCC | 7 | 1.22 | |
| T-47D | 7 | 1.16 | |
| CNS Cancer | SF-268 | 7 | 1.17 |
| SF-295 | 1 | 1.25 | |
| SF-539 | 4 | 1.18 | |
| SNB-19 | 7 | 1.15 | |
| SNB-75 | 1 | 1.16 | |
| U251 | 1 | 1.24 | |
| Colon Cancer | COLO-205 | 10 | 1.13 |
| HCC-2998 | 1 | 1.09 | |
| HCT-116 | 2/7 | 1.13 | |
| HCT-15 | 2 | 1.21 | |
| HT29 | 1 | 1.14 | |
| KM12 | 1 | 1.19 | |
| SW-620 | 2 | 1.14 | |
| Leukemia | CCRF-CEM | 7 | 1.13 |
| HL-60TB | 7 | 1.22 | |
| K-562 | 2 | 1.27 | |
| MOLT-4 | 3 | 1.12 | |
| RPMI-8226 | 10 | 1.12 | |
| SR | 2 | 1.28 | |
| Melanoma | LOX-IMVI | 3 | 1.16 |
| M14 | 1/3 | 1.20 | |
| MALME-3M | 10 | 1.19 | |
| MDA-MB-435 | 3 | 1.22 | |
| SK-MEL-2 | 3 | 1.03 | |
| SK-MEL-28 | 2/3 | 0.97 | |
| SK-MEL-5 | 2 | 1.26 | |
| UACC-257 | 2 | 1.07 | |
| UACC-62 | 10 | 1.31 | |
| Non-Small-Cell Lung Cancer | A549-ATCC | 3 | 1.18 |
| EKVX | 1 | 1.02 | |
| HOP-62 | 1/8 | 1.19 | |
| HOP-92 | 10 | 1.21 | |
| NCI-H226 | 7 | 1.07 | |
| NCI-H23 | 4 | 1.16 | |
| NCI-H322M | 1 | 1.10 | |
| NCI-H460 | 2 | 1.26 | |
| NCI-H522 | 7 | 1.09 | |
| Ovarian Cancer | IGROV1 | 1/3 | 1.25 |
| NCI-ADR-RES | 4 | 1.31 | |
| OVCAR-3 | 1/4 | 1.22 | |
| OVCAR-4 | 7 | 1.00 | |
| OVCAR-5 | 16 | 1.02 | |
| OVCAR-8 | 1 | 1.14 | |
| SK-OV-3 | 7 | 1.18 | |
| Prostate Cancer | DU-145 | 10 | 1.19 |
| PC-3 | 2 | 1.19 | |
| Renal Cancer | 786-0 | 10 | 1.16 |
| A498 | 1 | 1.21 | |
| ACHN | 7 | 1.19 | |
| CAKI-1 | 1 | 1.11 | |
| RXF-393 | 10 | 1.12 | |
| SN12C | 1/10 | 1.16 | |
| TK-10 | 10 | 0.99 | |
| UO-31 | 2 | 1.16 |
| PANELS | RUN | AVERAGE|DTV(GI50)| |
|---|---|---|
| Breast Cancer | 1/3 | 1.37 |
| CNS Cancer | 1 | 1.23 |
| Colon Cancer | 1 | 1.19 |
| Leukemia | 2 | 1.23 |
| Melanoma | 3 | 1.18 |
| Non-Small-Cell Lung Cancer | 2/7 | 1.20 |
| Ovarian Cancer | 1 | 1.21 |
| Prostate Cancer | 1 | 1.23 |
| Renal Cancer | 10 | 1.15 |
| PANEL 1 | 1a | 1b | 1c | 2a | 3e |
|---|---|---|---|---|---|
| Leukemia | 14.84 | 78.71 | 96.37 | 77.47 | 18.53 |
| Non-Small-Cell Lung Cancer | 61.79 | 98.02 | 95.47 | 84.39 | 29.86 |
| Colon Cancer | −16.06 | 80.65 | 95.84 | 86.52 | 21.07 |
| CNS Cancer | 32.98 | 98.55 | 101.40 | 98.19 | 18.66 |
| Melanoma | 24.26 | 97.49 | 100.59 | 96.74 | 22.40 |
| Ovarian Cancer | 46.48 | 103.35 | 101.45 | 95.29 | 32.68 |
| Renal Cancer | 21.07 | 100.02 | 100.31 | 94.36 | 34.17 |
| Prostate Cancer | 33.06 | 102.04 | 101.62 | 88.89 | 40.34 |
| Breast Cancer | 18.21 | 86.77 | 99.62 | 88.93 | 24.71 |
| Overall average | 26.29 | 93.96 | 99.19 | 90.09 | 26.93 |
| PANEL | CELL LINE 1 | 1a | 3e | Curcumin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GI50 | TGI | LC50 | GI50 | TGI | LC50 | GI50 | TGI | LC50 | ||
| Leukemia | CCRF-CEM | 5.65 | 4.7 | 4 | 5.4 | 4 | 4 | 5.52 | 4.81 | 4 |
| HL-60(TB) | 5.63 | 4.77 | 4 | 5.63 | 5.07 | 4 | 5.14 | 4.60 | 4.04 | |
| K-562 | 5.97 | 4 | 4 | 5.74 | 4 | 4 | 5.51 | 4.26 | 4 | |
| MOLT-4 | 5.69 | 4.72 | 4 | 5.49 | 4 | 4 | 5.33 | 4.75 | 4.12 | |
| RPMI-8226 | 6.41 | 5.63 | 4 | 5.58 | 4 | 4 | 5.68 | 5.20 | 4 | |
| Panel average | 5.87 | 4.76 | 4 | 5.57 | 4.21 | 4 | 5.43 | 4.72 | 4.03 | |
| Non-Small-Cell Lung Cancer | A549/ATCC | 4 | 4 | 4 | 5.32 | 4 | 4 | 4.89 | 4.50 | 4.11 |
| EKVX | 5.3 | 4 | 4 | 5.21 | 4 | 4 | 4.82 | 4.45 | 4.10 | |
| HOP-62 | 5.17 | 4 | 4 | 5.28 | 4 | 4 | 5.44 | 4.72 | 4.24 | |
| HOP-92 | 4.82 | 4.08 | 4 | 5.6 | 4.63 | 4 | NT | NT | NT | |
| NCI-H226 | 5.63 | NT 1 | 4 | 4.83 | 4 | 4 | 4.73 | 4.27 | 4 | |
| NCI-H23 | 5.52 | 4 | 4 | 5.36 | 4 | 4 | 5.25 | 4.50 | 4 | |
| NCI-H322M | 5.17 | 4 | 4 | 4.84 | 4 | 4 | 4.78 | 4.49 | 4.21 | |
| NCI-H460 | 5.51 | 4.95 | 4 | 5.37 | 4 | 4 | 5.09 | 4.64 | 4.22 | |
| NCI-H522 | 5.43 | 4.72 | 4 | 5.72 | 5.17 | 4.02 | 5.27 | 4.78 | 4.07 | |
| Panel average | 5.17 | 4.22 | 4.00 | 5.28 | 4.20 | 4 | 5.03 | 4.54 | 4.12 | |
| Colon Cancer | COLO-205 | 5.72 | 5.31 | 4.4 | 5.08 | 4.01 | 4 | 4.87 | 4.54 | 4.21 |
| HCC-2998 | 5.77 | 5.49 | 5.22 | 4.71 | 4 | 4 | 5.52 | 5.09 | 4.53 | |
| HCT-116 | 6.5 | 5.88 | 5.39 | 5.59 | 4.75 | 4 | 5.53 | 5.03 | 4.28 | |
| HCT-15 | 6.11 | 5.21 | 4.25 | 5.56 | 4 | 4 | 5.39 | 4.73 | 4.14 | |
| HT-29 | 6.12 | 5.58 | 5.09 | 5.58 | 4.97 | 4 | 5.29 | 4.49 | 4 | |
| KM12 | 5.8 | 5.43 | 5.07 | 5.16 | 4 | 4 | 5.27 | 4.71 | 4.19 | |
| SW-620 | 5.97 | 5.48 | 4.97 | 5.48 | 4 | 4 | 5.38 | 4.67 | 4.07 | |
| Panel average | 6.00 | 5.48 | 4.91 | 5.31 | 4.25 | 4 | 5.32 | 4.75 | 4.20 | |
| CNS Cancer | SF-268 | 5.6 | 5.07 | 4 | 5.11 | 4 | 4 | 5.15 | 4.44 | 4 |
| SF-295 | 5.52 | 4.51 | 4 | 5.53 | 4.75 | 4 | 5.10 | 4.68 | 4.32 | |
| SF-539 | 5.67 | 5.29 | 4.22 | 5.57 | 5.03 | 4.09 | 5.55 | 5.05 | 4.48 | |
| SNB-19 | 5.51 | 4.63 | 4 | 5.39 | 4.04 | 4 | 5.05 | 4.61 | 4.20 | |
| SNB-75 | 5.6 | 4.47 | 4 | 5.38 | 4.34 | 4 | 5.17 | 4.74 | 4.35 | |
| U251 | 5.81 | 5.44 | 5.07 | 5.42 | 4.73 | 4 | 5.33 | 4.78 | 4.31 | |
| Panel average | 5.62 | 4.90 | 4.22 | 5.40 | 4.48 | 4.02 | 5.22 | 4.72 | 4.28 | |
| Melanoma | LOX IMVI | 5.84 | 5.49 | 5.15 | 5.51 | 4.54 | 4 | 5.57 | 5.07 | 4 |
| MALME-3M | 5.24 | 4.1 | 4 | 5.21 | 4 | 4 | 4.85 | 4.56 | 4.27 | |
| M14 | 5.77 | 5.36 | 4.24 | 5.51 | 4.58 | 4 | 5.42 | 4.80 | 4.35 | |
| MDA-MB-435 | 5.94 | 5.53 | 5.11 | 6.03 | 5.41 | 4.19 | 5.53 | 4.92 | 4.40 | |
| SK-MEL-2 | 4.51 | 4 | 4 | 5.54 | 4.73 | 4 | 4.78 | 4.39 | 4.06 | |
| SK-MEL-28 | 5.74 | 5.4 | NT | 5.3 | 4 | 4 | 5.35 | 4.80 | 4.30 | |
| SK-MEL-5 | 5.67 | 5.21 | 4 | 5.66 | 4.99 | 4 | 5.06 | 4.65 | 4.28 | |
| UACC-257 | 5.61 | 5.13 | 4 | 4.97 | 4 | 4 | 4.94 | 4.62 | 4.31 | |
| UACC-62 | 5.72 | 5.31 | 4.52 | 5.62 | 5 | 4 | 5.19 | 4.69 | 4.26 | |
| Panel average | 5.56 | 5.06 | 4.38 | 5.48 | 4.58 | 4.02 | 5.19 | 4.72 | 4.25 | |
| Ovarian Cancer | IGROV-1 | 5.57 | NT | 4 | 5.37 | 4 | 4 | 5.10 | 4.57 | 4.09 |
| OVCAR-3 | 5.55 | 5 | 4 | 5.3 | 4.09 | 4 | 5.18 | 4.61 | 4.17 | |
| OVCAR-4 | 5.39 | 4 | 4 | 5.04 | 4 | 4 | 5.03 | 4.44 | 4 | |
| OVCAR-5 | 5.6 | 5.09 | 4 | 5.31 | 4.27 | 4 | 4.78 | 4.45 | 4.12 | |
| OVCAR-8 | 5.53 | 4 | 4 | 5.06 | 4 | 4 | 5.13 | 4.55 | 4.08 | |
| NCI/ADR-RES | 5.38 | 4 | 4 | 5.45 | 4 | 4 | 5.14 | 4.12 | 4 | |
| SK-OV-3 | 4.44 | 4 | 4 | 5.06 | 4.03 | 4 | 5.05 | 4.68 | 4.33 | |
| Panel average | 5.35 | 4.35 | 4.00 | 5.23 | 4.06 | 4 | 5.06 | 4.49 | 4.11 | |
| Renal Cancer | 786-0 | 5.64 | 5.1 | 4 | 5.25 | 4 | 4 | 5.48 | 4.97 | 4.42 |
| A-498 | 5.65 | 4.7 | 4 | 6.62 | 5.04 | 4 | 4.80 | 4.48 | 4.16 | |
| ACHN | 5.69 | 5.27 | 4 | 4.86 | 4 | 4 | 4.91 | 4.54 | 4.17 | |
| CAKI-1 | 5.5 | 4 | 4 | 4.94 | 4 | 4 | 4.92 | 4.60 | 4.30 | |
| RXF-393 | 5.83 | 5.52 | 5.2 | 5.1 | 4.11 | 4 | 5.52 | 4.95 | 4.27 | |
| SN12C | 5.51 | 4.75 | 4 | 5.26 | 4 | 4 | 5.08 | 4.60 | 4.20 | |
| TK-10 | 5.42 | 4.6 | 4 | 5.42 | 4.56 | 4 | 4.85 | 4.51 | 4.18 | |
| UO-31 | 5.79 | 5.47 | 5.14 | 5.32 | 4 | 4 | 4.95 | 4.61 | 4.27 | |
| Panel average | 5.63 | 4.93 | 4.29 | 5.35 | 4.21 | 4 | 5.06 | 4.66 | 4.25 | |
| Prostate Cancer | PC-3 | 5.48 | 4 | 4 | 5.39 | 4 | 4 | 5.06 | 4.59 | 4.15 |
| DU-145 | 5.51 | 4.85 | 4 | 4.82 | 4 | 4 | 4.81 | 4.53 | 4.25 | |
| Panel average | 5.50 | 4.43 | 4.00 | 5.11 | 4.00 | 4 | 4.93 | 4.56 | 4.20 | |
| Breast Cancer | MCF7 | 6.29 | 5.02 | 4 | 5.46 | 4 | 4 | 5.48 | 4.46 | 4 |
| MDA-MB-231/ATCC | 5.65 | 5.22 | 4 | 5.44 | 4.36 | 4 | 4.75 | 4.25 | 4 | |
| HS 578T | 5.57 | 4 | 4 | 5.43 | 4.32 | 4 | 4.96 | 4.23 | 4 | |
| BT-549 | 5.74 | 5.35 | 4.66 | 5.54 | 4.45 | 4 | 5.30 | 4.86 | 4.37 | |
| T-47D | 5.65 | 4 | 4 | 5.46 | 4.09 | 4 | 5.08 | 4.33 | 4 | |
| MDA-MB-468 | 5.81 | 5.43 | 4 | 5.54 | 4.56 | 4 | NT | NT | NT | |
| Panel average | 5.79 | 4.84 | 4.11 | 5.48 | 4.30 | 4 | 5.11 | 4.43 | 4.07 | |
| Overall average | 5.59 | 4.81 | 4.24 | 5.37 | 4.28 | 4.01 | 5.16 | 4.63 | 4.17 | |
| Range | 4–6.5 | 4–5.88 | 4–5.39 | 4.71–6.62 | 4–5.41 | 4–4.19 | 4.73–5.68 | 4–5.20 | 4–4.53 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martorana, A.; La Monica, G.; Bono, A.; Mannino, S.; Buscemi, S.; Palumbo Piccionello, A.; Gentile, C.; Lauria, A.; Peri, D. Antiproliferative Activity Predictor: A New Reliable In Silico Tool for Drug Response Prediction against NCI60 Panel. Int. J. Mol. Sci. 2022, 23, 14374. https://doi.org/10.3390/ijms232214374
Martorana A, La Monica G, Bono A, Mannino S, Buscemi S, Palumbo Piccionello A, Gentile C, Lauria A, Peri D. Antiproliferative Activity Predictor: A New Reliable In Silico Tool for Drug Response Prediction against NCI60 Panel. International Journal of Molecular Sciences. 2022; 23(22):14374. https://doi.org/10.3390/ijms232214374
Chicago/Turabian StyleMartorana, Annamaria, Gabriele La Monica, Alessia Bono, Salvatore Mannino, Silvestre Buscemi, Antonio Palumbo Piccionello, Carla Gentile, Antonino Lauria, and Daniele Peri. 2022. "Antiproliferative Activity Predictor: A New Reliable In Silico Tool for Drug Response Prediction against NCI60 Panel" International Journal of Molecular Sciences 23, no. 22: 14374. https://doi.org/10.3390/ijms232214374
APA StyleMartorana, A., La Monica, G., Bono, A., Mannino, S., Buscemi, S., Palumbo Piccionello, A., Gentile, C., Lauria, A., & Peri, D. (2022). Antiproliferative Activity Predictor: A New Reliable In Silico Tool for Drug Response Prediction against NCI60 Panel. International Journal of Molecular Sciences, 23(22), 14374. https://doi.org/10.3390/ijms232214374









