Targeted NGS in Diagnostics of Genodermatosis Characterized by the Epidermolysis Bullosa Symptom Complex in 268 Russian Children
Abstract
1. Introduction
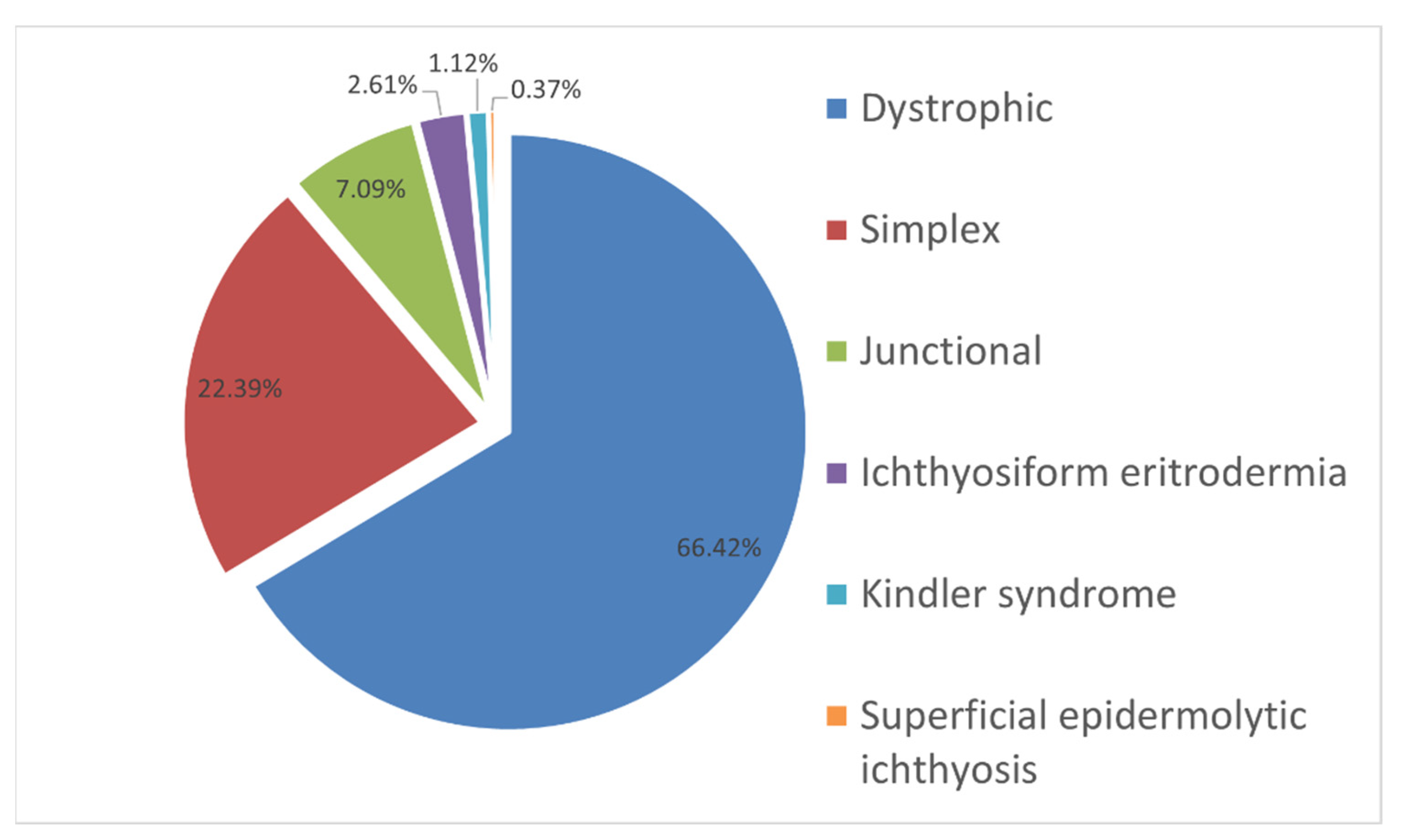
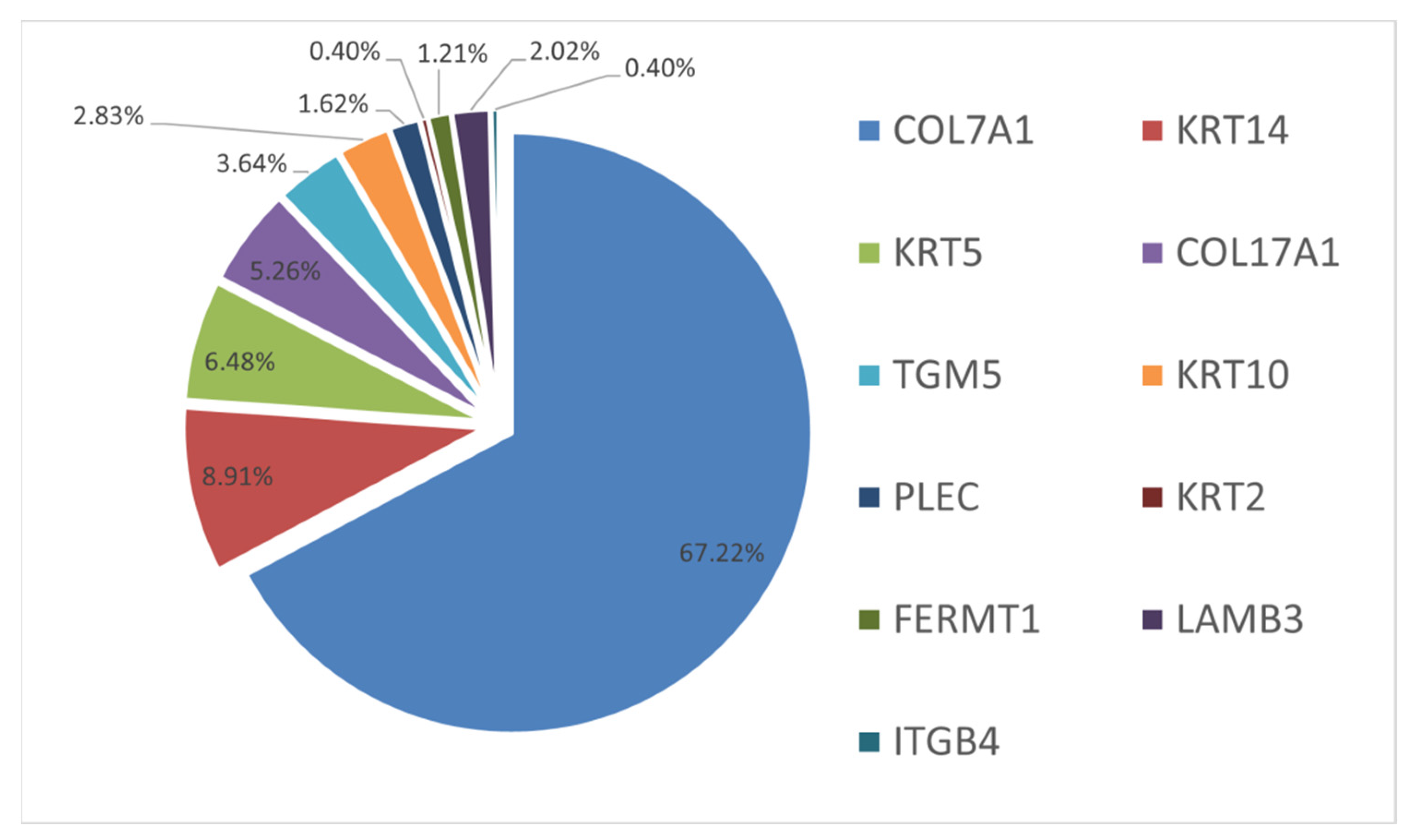
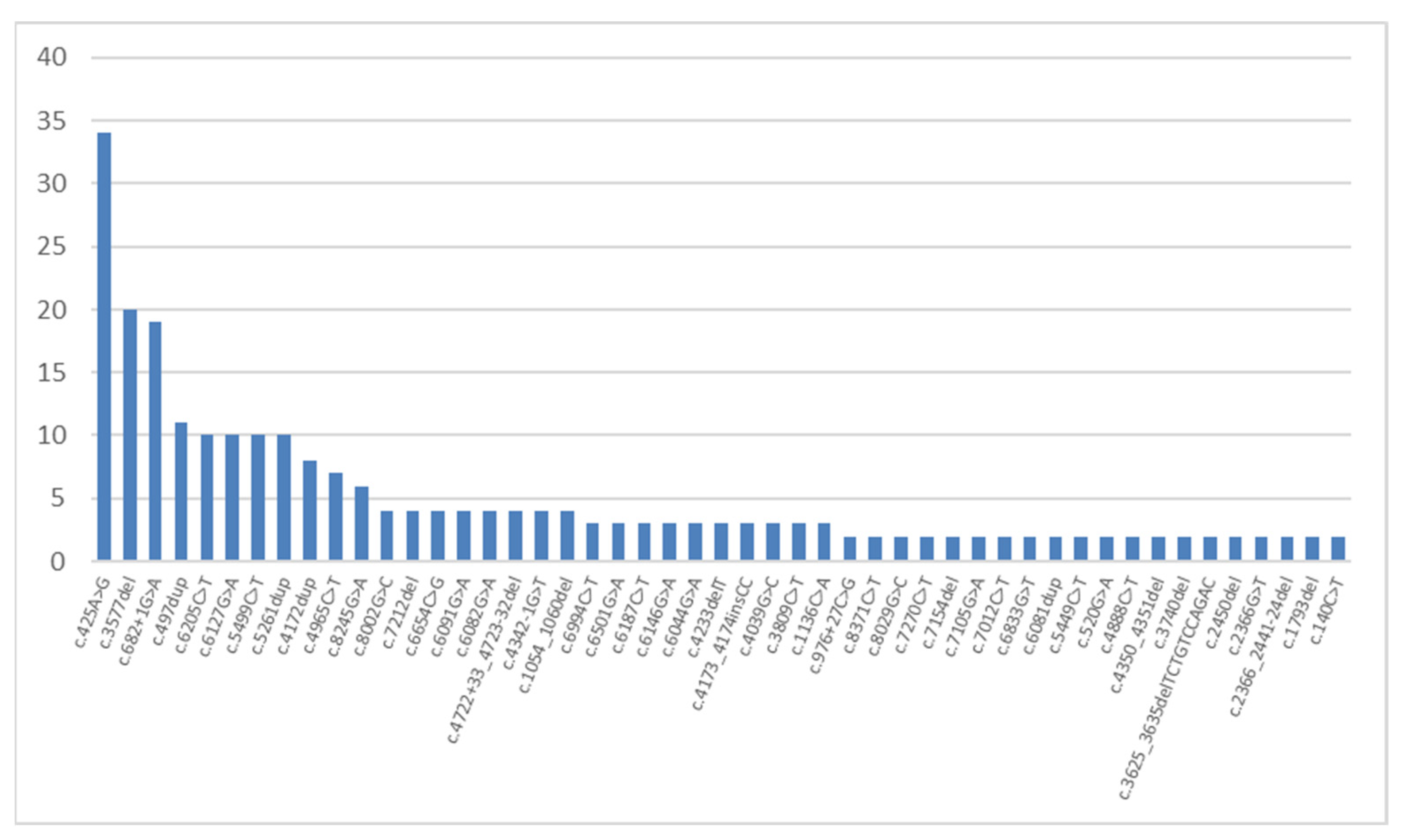
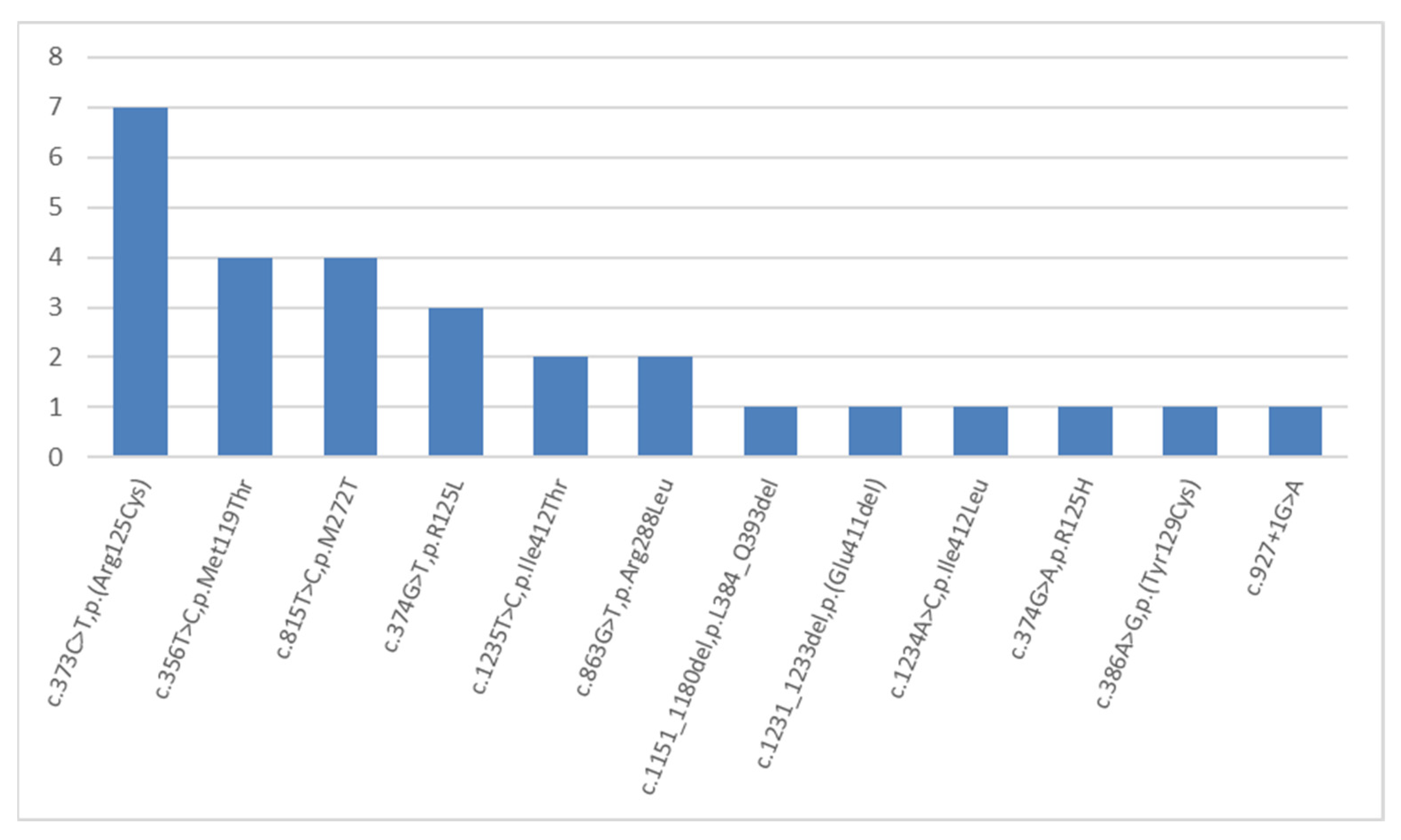
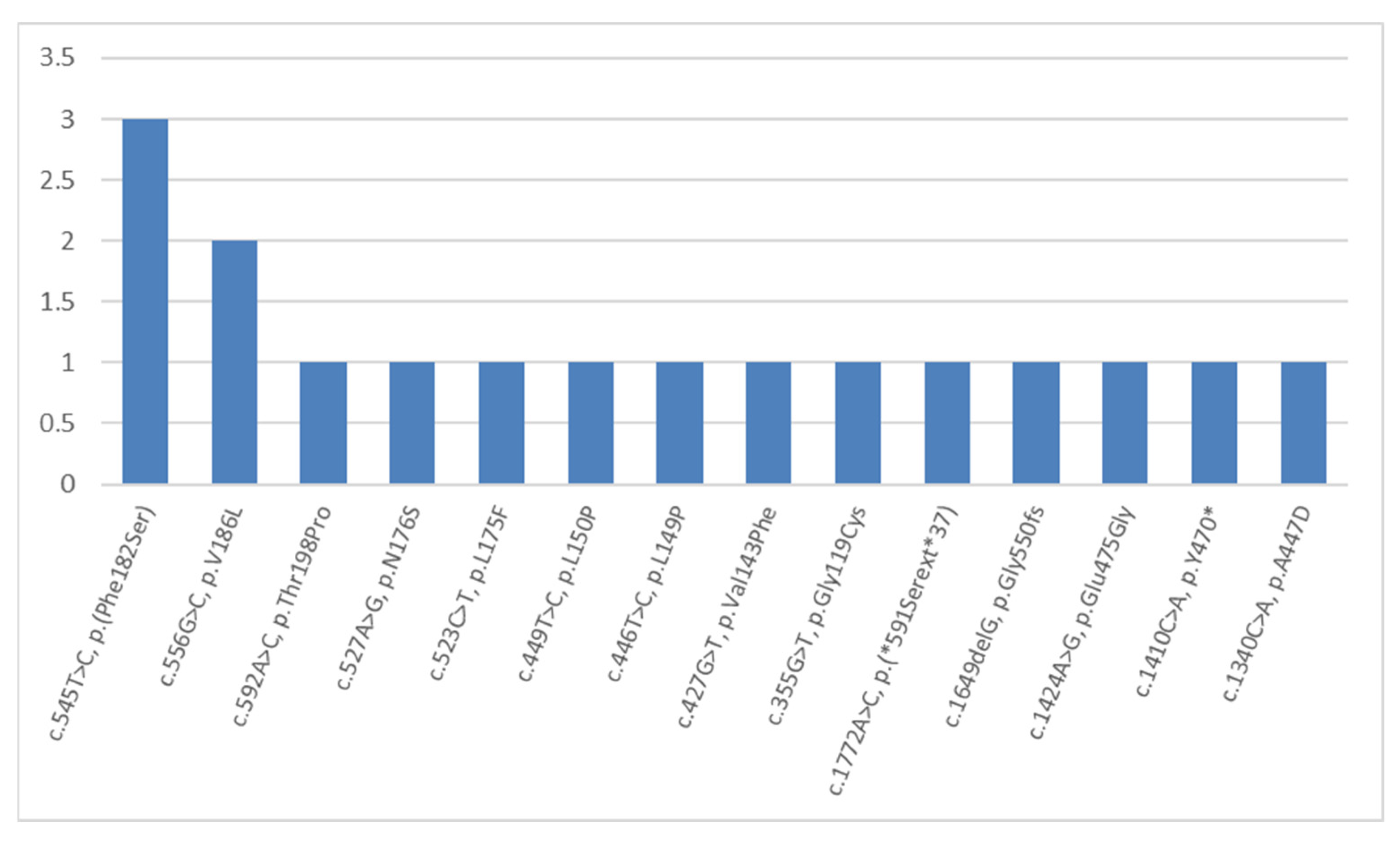
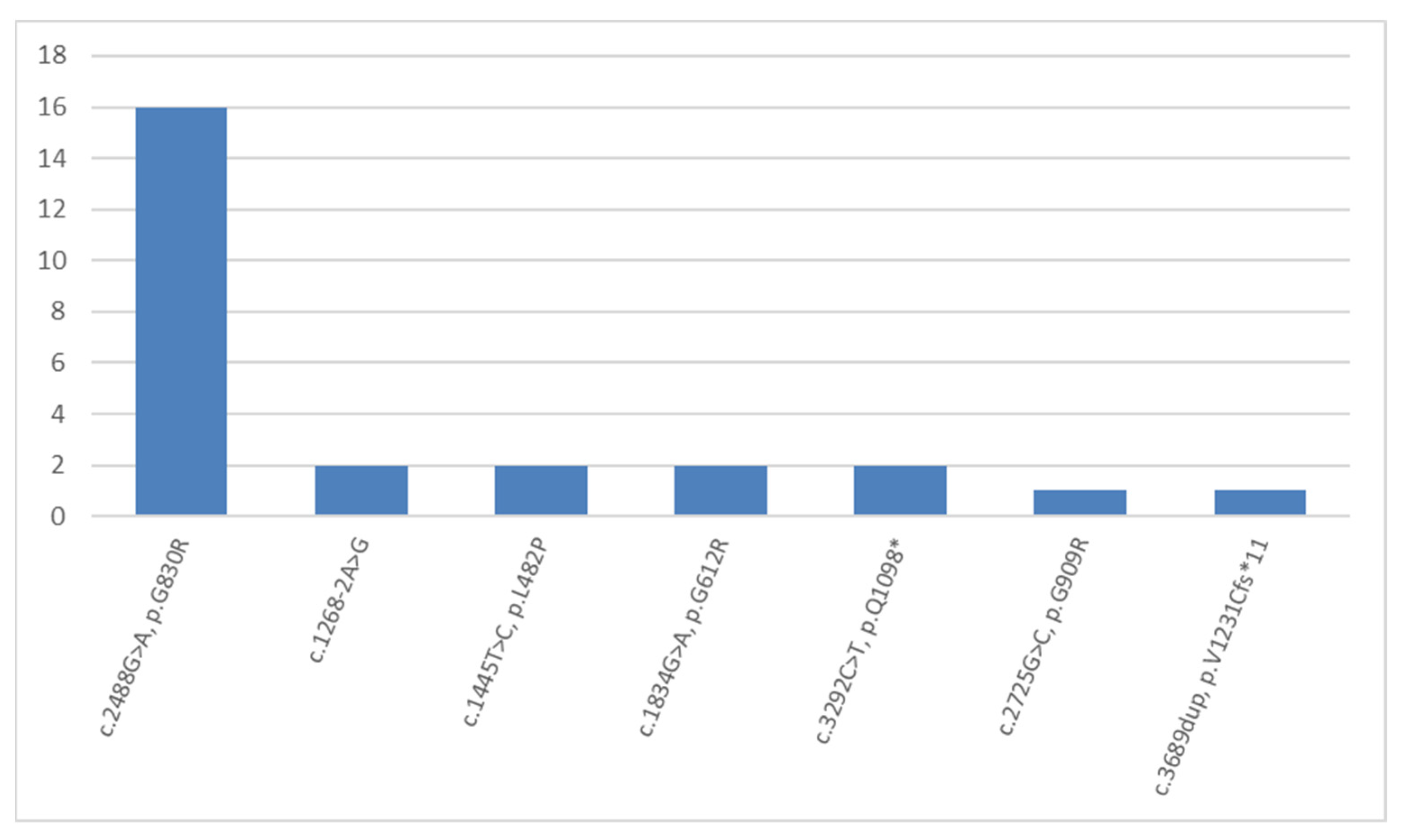
2. Results and Discussion
3. Materials and Methods
3.1. Molecular Diagnostics
3.2. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Patient’s ID | Sex | Genome Position (GRCh38) | Gene | Transcript ID | cDNA Variant | Protein Variant | Zygosity | Minor Allele Frequency ** | Inheritance *** |
|---|---|---|---|---|---|---|---|---|---|
| 36617 | M | 3:48575727 | COL7A1 | NM_000094 | c.5878G>A | p.V1960M | het | 0.001% | M |
| 36617 | M | 8:143922830 | PLEC | NM_201380 | c.7510A>C | p.T2504P | het | #N/A | M |
| 44039 | M | 8:143916607 | PLEC | NM_201384 | c.13214G>A | p.R4405H | het | <0.001% | M |
| 45268 | F | 8:143921585 | PLEC | NM_201384 | c.8236C>T | p.R2746W | het | <0.001% | F |
| 53826 | M | 8:143920574 | PLEC | NM_201384 | c.9247C>T | p.R3083C | het | 0.007% | F |
| 66124 | M | 12:52675434 | KRT1 | NM_006121 | c.1694G>C | p.S565T | het | #N/A | M |
| 66242 | F | 8:143931951 | PLEC | NM_201380 | c.2575G>A | p.A859T | het | 0.001% | M |
| 99424 | F | 3:48575692 | COL7A1 | NM_000094 | c.5913G>T | p.Leu1971L | het | 0.007% | F |
References
- Fine, J.-D.; Mellerio, J.E. Extracutaneous manifestations and complications of inherited epidermolysis bullosa: Part I. Epithelial associated tissues. J. Am. Acad. Dermatol. 2009, 61, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Fine, J.-D.; Mellerio, J.E. Extracutaneous manifestations and complications of inherited epidermolysis bullosa: Part II. Other organs. J. Am. Acad. Dermatol. 2009, 61, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Has, C.; Bruckner-Tuderman, L. The Genetics of Skin Fragility. Annu. Rev. Genom. Hum. Genet. 2014, 15, 245–268. [Google Scholar] [CrossRef]
- Ryynänen, M.; Knowlton, R.G.; Parente, M.G.; Chung, L.C.; Chu, M.L.; Uitto, J. Human type VII collagen: Genetic linkage of the gene (COL7A1) on chromosome 3 to dominant dystrophic epidermolysis bullosa. Am. J. Hum. Genet. 1991, 49, 797–803. [Google Scholar]
- Ryynänen, M.; Knowlton, R.G.; Uitto, J. Mapping of epidermolysis bullosa simplex mutation to chromosome 12. Am. J. Hum. Genet. 1991, 49, 978–984. [Google Scholar] [PubMed]
- Beilin, A.; Evtushenko, N.; Lukyanov, D.; Murashkin, N.; Ambarchian, E.; Pushkov, A.; Savostyanov, K.; Fisenko, A.; Rogovaya, O.; Vasiliev, A.; et al. Signatures of Dermal Fibroblasts from RDEB Pediatric Patients. Int. J. Mol. Sci. 2021, 22, 1792. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Savost’Anov, K.V.; Kuzenkova, L.M.; Gevorkyan, A.K.; Pushkov, A.A.; Nikitin, A.G.; Pakhomov, A.V.; Vashakmadze, N.D.; Zhurkova, N.V.; Podkletnova, T.V.; et al. Molecular characteristics of patients with glycosaminoglycan storage disorders in Russia. Clin. Chim. Acta 2014, 436, 112–120. [Google Scholar] [CrossRef]
- Savostyanov, K.; Pushkov, A.; Zhanin, I.; Mazanova, N.; Trufanov, S.; Pakhomov, A.; Alexeeva, A.; Sladkov, D.; Asanov, A.; Fisenko, A. The prevalence of Fabry disease among 1009 unrelated patients with hypertrophic cardiomyopathy: A Russian nationwide screening program using NGS technology. Orphanet J. Rare Dis. 2022, 17, 199. [Google Scholar] [CrossRef]
- Takeichi, T.; Liu, L.; Fong, K.; Ozoemena, L.; McMillan, J.; Salam, A.; Campbell, P.; Akiyama, M.; Mellerio, J.; McLean, W.; et al. Whole-exome sequencing improves mutation detection in a diagnostic epidermolysis bullosa laboratory. Br. J. Dermatol. 2014, 172, 94–100. [Google Scholar] [CrossRef]
- Fine, J.-D.; Bruckner-Tuderman, L.; Eady, R.A.; Bauer, E.A.; Bauer, J.W.; Has, C.; Heagerty, A.; Hintner, H.; Hovnanian, A.; Jonkman, M.F.; et al. Inherited epidermolysis bullosa: Updated recommendations on diagnosis and classification. J. Am. Acad. Dermatol. 2014, 70, 1103–1126. [Google Scholar] [CrossRef]
- El Hachem, M.; Giancristoforo, S.; Diociaiuti, A. Inherited epidermolysis bullosa. Orphanet J. Rare Dis. 2014, 149, 651–662. [Google Scholar]
- Kiritsi, D.; Cosgarea, I.; Franzke, C.W.; Schumann, H.; Oji, V.; Kohlhase, J.; Bruckner-Tuderman, L.; Has, C. Acral Peeling Skin Syndrome with TGM5 Gene Mutations May Resemble Epidermolysis Bullosa Simplex in Young Individuals. J. Investig. Dermatol. 2010, 130, 1741–1746. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.A. A Novel Genodermatosis Caused by Mutations in Plakophilin 1, A Structural Component of Desmosomes. J. Dermatol. 1999, 26, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Jonkman, M.F.; Pasmooij, A.M.; Pasmans, S.G.; Van Den Berg, M.; ter Horst, H.J.; Timmer, A.; Pas, H. Loss of Desmoplakin Tail Causes Lethal Acantholytic Epidermolysis Bullosa*. Am. J. Hum. Genet. 2005, 77, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Pigors, M.; Kiritsi, D.; Krümpelmann, S.; Wagner, N.; He, Y.; Podda, M.; Kohlhase, J.; Hausser, I.; Bruckner-Tuderman, L.; Has, C. Lack of plakoglobin leads to lethal congenital epidermolysis bullosa: A novel clinico-genetic entity. Hum. Mol. Genet. 2011, 20, 1811–1819. [Google Scholar] [CrossRef]
- Cabral, R.M.; Kurban, M.; Wajid, M.; Shimomura, Y.; Petukhova, L.; Christiano, A.M. Whole-exome sequencing in a single proband reveals a mutation in the CHST8 gene in autosomal recessive peeling skin syndrome. Genomics 2012, 99, 202–208. [Google Scholar] [CrossRef]
- Ishida-Yamamoto, A.; Furio, L.; Igawa, S.; Honma, M.; Tron, E.; Malan, V.; Murakami, M.; Hovnanian, A. Inflammatory peeling skin syndrome caused by homozygous genomic deletion in the PSORS1 region encompassing the CDSN gene. Exp. Dermatol. 2014, 23, 60–63. [Google Scholar] [CrossRef]
- Bolling, M.; Lemmink, H.; Jansen, G.; Jonkman, M. Mutations in KRT5 and KRT14 cause epidermolysis bullosa simplex in 75% of the patients. Br. J. Dermatol. 2011, 164, 637–644. [Google Scholar] [CrossRef]
- Groves, R.W.; Liu, L.; Dopping-Hepenstal, P.J.; Markus, H.S.; Lovell, P.A.; Ozoemena, L.; Lai-Cheong, J.E.; Gawler, J.; Owaribe, K.; Hashimoto, T.; et al. A Homozygous Nonsense Mutation within the Dystonin Gene Coding for the Coiled-Coil Domain of the Epithelial Isoform of BPAG1 Underlies a New Subtype of Autosomal Recessive Epidermolysis Bullosa Simplex. J. Investig. Dermatol. 2010, 130, 1551–1557. [Google Scholar] [CrossRef]
- Liu, L.; Mellerio, J.; Martinez, A.; McMillan, J.; Aristodemou, S.; Parsons, M.; McGrath, J. Mutations in EXPH5 result in autosomal recessive inherited skin fragility. Br. J. Dermatol. 2014, 170, 196–199. [Google Scholar] [CrossRef]
- Ruzzi, L.; Gagnoux-Palacios, L.; Pinola, M.; Belli, S.; Meneguzzi, G.; D’Alessio, M.; Zambruno, G. A homozygous mutation in the integrin alpha6 gene in junctional epidermolysis bullosa with pyloric atresia. J. Clin. Investig. 1997, 99, 2826–2831. [Google Scholar] [CrossRef] [PubMed]
- Vidal, F.; Aberdam, D.; Miquel, C.; Christiano, A.M.; Pulkkinen, L.; Uitto, J.; Ortonne, J.-P.; Meneguzzi, G.; Fr, D.A. Integrin β4 mutations associated with junctional epidermolysis bullosa with pyloric atresia. Nat. Genet. 1995, 10, 229–234. [Google Scholar] [CrossRef] [PubMed]
- McLean, W.H.; Pulkkinen, L.; Smith, F.J.; Rugg, E.L.; Lane, E.B.; Bullrich, F.; Burgeson, R.E.; Amano, S.; Hudson, D.L.; Owaribe, K.; et al. Loss of plectin causes epidermolysis bullosa with muscular dystrophy: cDNA cloning and genomic organization. Genes Dev. 1996, 10, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, S.; Feng, C.; Yang, S.; Wang, H.; Ma, D.; Zhang, J.; Gou, M.; Bu, D.; Zhang, T.; et al. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat. Genet. 2016, 48, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Kiritsi, D.; Has, C.; Bruckner-Tuderman, L. Laminin 332 in junctional epidermolysis bullosa. Cell Adhes. Migr. 2013, 7, 135–141. [Google Scholar] [CrossRef]
- Schumann, H.; Kiritsi, D.; Pigors, M.; Hausser, I.; Kohlhase, J.; Peters, J.; Ott, H.; Hyla-Klekot, L.; Gacka, E.; Sieron, A.; et al. Phenotypic spectrum of epidermolysis bullosa associated with α6β4 integrin mutations. Br. J. Dermatol. 2013, 169, 115–124. [Google Scholar] [CrossRef]
- Has, C.; Spartà, G.; Kiritsi, D.; Weibel, L.; Moeller, A.; Vega-Warner, V.; Waters, A.; He, Y.; Anikster, Y.; Esser, P.; et al. Integrin α3 Mutations with Kidney, Lung, and Skin Disease. New Engl. J. Med. 2012, 366, 1508–1514. [Google Scholar] [CrossRef]
- Pulkkinen, L.; Uitto, J.; Christiano, A.M. The molecular basis of the junctional forms of epidermolysis bullosa. In Epidermolysis Bullosa: Clinical, Epidemiologic, and Laboratory Advances, and the Findings of the National Epidermolysis Bullosa Registry; Johns Hopkins University Press: Baltimore, MD, USA, 1999. [Google Scholar]
- Crew, V.K.; Burton, N.; Kagan, A.; Green, C.A.; Levene, C.; Flinter, F.; Brady, R.L.; Daniels, G.; Anstee, D.J. CD151, the first member of the tetraspanin (TM4) superfamily detected on erythrocytes, is essential for the correct assembly of human basement membranes in kidney and skin. Blood 2004, 104, 2217–2223. [Google Scholar] [CrossRef]
- Yuen, W.Y.; Pas, H.; Sinke, R.J.; Jonkman, M.F. Junctional epidermolysis bullosa of late onset explained by mutations in COL17A1. Br. J. Dermatol. 2011, 164, 1280–1284. [Google Scholar] [CrossRef]
- Lai-Cheong, J.E.; McGrath, J.A. Kindler Syndrome. Dermatol. Clin. 2010, 28, 119–124. [Google Scholar] [CrossRef]
- McLean, W.I.; Eady, R.A.; Dopping-Hepenstal, P.J.; McMillan, J.R.; Leigh, I.M.; Navsaria, H.A.; Higgins, C.; Harper, J.I.; Paige, D.G.; Morley, S.M.; et al. Mutations in the Rod 1A Domain of Keratins 1 and 10 in Bullous Congenital Ichthyosiform Erythoderma (BCIE). J. Investig. Dermatol. 1994, 102, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Betlloch, I.; Costa, A.L.; Mataix, J.; Pérez-Crespo, M.; Ballester, I. Bullous Congenital Ichthyosiform Erythroderma: A Sporadic Case Produced by a NewKRT10Gene Mutation. Pediatr. Dermatol. 2009, 26, 489–491. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.J.; Maingi, C.; Covello, S.P.; Higgins, C.; Schmidt, M.; Lane, E.B.; Uitto, J.; Leigh, I.M.; McLean, W.I. Genomic Organization and Fine Mapping of the Keratin 2e Gene (KRT2E): K2e V1 Domain Polymorphism and Novel Mutations in Ichthyosis Bullosa of Siemens. J. Investig. Dermatol. 1998, 111, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, T.; Nanda, A.; Liu, L.; Salam, A.; Campbell, P.; Fong, K.; Akiyama, M.; Ozoemena, L.; Stone, K.L.; Al-Ajmi, H.; et al. Impact of next generation sequencing on diagnostics in a genetic skin disease clinic. Exp. Dermatol. 2013, 22, 825–831. [Google Scholar] [CrossRef]
- South, A.P.; Li, Q.; Uitto, J. Next-Generation Sequencing for Mutation Detection in Heritable Skin Diseases: The Paradigm of Pseudoxanthoma Elasticum. J. Investig. Dermatol. 2015, 135, 937–940. [Google Scholar] [CrossRef]
- Lucky, A.W.; Ms, N.D.; Lammers, K.; Husami, A.; Kissell, D.; Zhang, K.; Ms, K.L.; Bs, A.H.; Mhsa, D.K.; Zhang, M.K. A comprehensive next-generation sequencing assay for the diagnosis of epidermolysis bullosa. Pediatr. Dermatol. 2018, 35, 188–197. [Google Scholar] [CrossRef]
- Has, C.; Küsel, J.; Reimer, A.; Hoffmann, J.; Schauer, F.; Zimmer, A.; Fischer, J. The Position of Targeted Next-generation Sequencing in Epidermolysis Bullosa Diagnosis. Acta Derm. Venereol. 2018, 98, 437–440. [Google Scholar] [CrossRef]
- Hamada, T.; Tsuruta, D.; Fukuda, S.; Ishii, N.; Teye, K.; Numata, S.; Dainichi, T.; Karashima, T.; Ohata, C.; Furumura, M.; et al. How do keratinizing disorders and blistering disorders overlap? Exp. Dermatol. 2013, 22, 83–87. [Google Scholar] [CrossRef]
- Sproule, T.J.; Bubier, J.A.; Grandi, F.C.; Sun, V.Z.; Philip, V.M.; McPhee, C.G.; Adkins, E.B.; Sundberg, J.P.; Roopenian, D.C. Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice. PLoS Genet. 2014, 10, e1004068. [Google Scholar] [CrossRef]
- Vahidnezhad, H.; Youssefian, L.; Zeinali, S.; Saeidian, A.H.; Sotoudeh, S.; Mozafari, N.; Abiri, M.; Kajbafzadeh, A.-M.; Barzegar, M.; Ertel, A.; et al. Dystrophic Epidermolysis Bullosa: COL7A1 Mutation Landscape in a Multi-Ethnic Cohort of 152 Extended Families with High Degree of Customary Consanguineous Marriages. J. Investig. Dermatol. 2017, 137, 660–669. [Google Scholar] [CrossRef]
- Drera, B.; Ritelli, M.; Tadini, G.; Zoppi, N.; Venturini, M.; Calzavara-Pinton, P.; Barlati, S.; Colombi, M.; Calzavara-Pinton, P. The novel p.G150R missense mutation in the cartilage matrix protein subdomain of type VII collagen in compound heterozigosity with the c.682+1G>A COL7A1 splicing mutation leads to mild dystrophic epidermolysis bullosa. J. Dermatol. Sci. 2009, 53, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Mellerio, J.E.; Salas-Alanis, J.C.; Talamantes, M.L.; Horn, H.; Tidman, M.J.; Ashton, G.H.; Eady, R.A.; McGrath, J.A. A recurrent glycine substitution mutation, G2043R, in the type VII collagen gene (COL7A1) in dominant dystrophic epidermolysis bullosa. Br. J. Dermatol. 1998, 139, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Varki, R.; Sadowski, S.; Uitto, J.; Pfendner, E. Epidermolysis bullosa. II. Type VII collagen mutations and phenotype-genotype correlations in the dystrophic subtypes. J. Med. Genet. 2007, 44, 181–192. [Google Scholar] [CrossRef]
- Xu, K.; Nowak, I.; Kirchner, M.; Xu, Y. Recombinant Collagen Studies Link the Severe Conformational Changes Induced by Osteogenesis Imperfecta Mutations to the Disruption of a Set of Interchain Salt Bridges. J. Biol. Chem. 2008, 283, 34337–34344. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.; Almaani, N.; Liu, L.; Dopping-Hepenstal, P.; Lai-Cheong, J.; Wong, A.; Nanda, A.; Moss, C.; Martinéz, A.; Mellerio, J. Identical Glycine Substitution Mutations in Type VII Collagen May Underlie Both Dominant and Recessive Forms of Dystrophic Epidermolysis Bullosa. Acta Derm. Venereol. 2011, 91, 262–266. [Google Scholar] [CrossRef]
- Cummins, R.E.; Klingberg, S.; Wesley, J.; Rogers, M.; Zhao, Y.; Murrell, D.F. Keratin 14 Point Mutations at Codon 119 of Helix 1A Resulting in Different Epidermolysis Bullosa Simplex Phenotypes. J. Investig. Dermatol. 2001, 117, 1103–1107. [Google Scholar] [CrossRef]
- Rugg, E.L.; Horn, H.M.; Smith, F.J.; Wilson, N.J.; Hill, A.J.; Magee, G.J.; Shemanko, C.S.; Baty, D.U.; Tidman, M.J.; Lane, E.B. Epidermolysis Bullosa Simplex in Scotland Caused by a Spectrum of Keratin Mutations. J. Investig. Dermatol. 2007, 127, 574–580. [Google Scholar] [CrossRef]
- Gostyńska, K.B.; Yan Yuen, W.; Pasmooij AM, G.; Stellingsma, C.; Pas, H.H.; Lemmink, H.; Jonkman, M.F. Carriers with functional null mutations in LAMA3 have localized enamel abnormalities due to haploinsufficiency. Eur. J. Hum. Genet. 2017, 25, 94–99. [Google Scholar] [CrossRef]
| Marker | D3S3559 | D3S3563 | COL7A1 | D3S1588 | D3S1289 | D3S3666 | |
|---|---|---|---|---|---|---|---|
| Coordinates (cM) | 67.49 | 68.47 | 70.61 | 70.61 | 71.41 | 72.21 | |
| Chromosome | |||||||
| 9.1 | 4 | 3 | c.3577del | 7 | 4 | 9 | |
| 8.1 | 4 | 3 | c.3577del | 7 | 4 | 9 | |
| 8.2 | 1 | 3 | c.3577del | 7 | 4 | 7 | |
| 6.1 | 5 | 4 | c.3577del | 7 | 4 | 9 | |
| 6.2 | 4 | 1 | c.3577del | 7 | 4 | 8 | |
| 9.2 | 4 | 3 | c.3577del | 6 | 0 | 5 | |
| 1.1 | 4 | 3 | c.3577del | 6 | 4 | 9 | |
| 1.2 | 4 | 3 | c.3577del | 6 | 4 | 8 | |
| 4.1 | 4 | 3 | c.3577del | 6 | 4 | 9 | |
| 2.1 | 4 | 3 | c.3577del | 1 | 4 | 9 | |
| 3.1 | 4 | 3 | c.3577del | 1 | 4 | 8 | |
| 5.1 | 6 | 3 | c.3577del | 7 | 5 | 4 | |
| 5.2 | 6 | 3 | c.3577del | 7 | 5 | 4 | |
| 2.2. | 2 | 3 | c.3577del | 1 | 4 | 8 | |
| 3.2 | 2 | 3 | c.3577del | 1 | 4 | 7 | |
| 4.2 | 4 | 6 | c.3577del | 7 | 8 | 8 | |
| Marker | Coordinates, cM | Allele | p-Value | δ ± 95 CI |
|---|---|---|---|---|
| D3S3559 | 67.49 | 4 | p < 0.05 | 0.61 ± 0.13 |
| D3S3563 | 68.47 | 3 | p < 0.05 | 0.77 ± 0.12 |
| D3S1588 | 70.61 | 7 | p < 0.05 | 0.44 ± 0.15 |
| D3S1289 | 71.41 | 4 | p < 0.05 | 0.67 ± 0.15 |
| D3S3666 | 72.21 | 9 | p < 0.05 | 0.30 ± 0.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savostyanov, K.; Murashkin, N.; Pushkov, A.; Zhanin, I.; Suleymanov, E.; Akhkiamova, M.; Shchagina, O.; Balanovska, E.; Epishev, R.; Polyakov, A.; et al. Targeted NGS in Diagnostics of Genodermatosis Characterized by the Epidermolysis Bullosa Symptom Complex in 268 Russian Children. Int. J. Mol. Sci. 2022, 23, 14343. https://doi.org/10.3390/ijms232214343
Savostyanov K, Murashkin N, Pushkov A, Zhanin I, Suleymanov E, Akhkiamova M, Shchagina O, Balanovska E, Epishev R, Polyakov A, et al. Targeted NGS in Diagnostics of Genodermatosis Characterized by the Epidermolysis Bullosa Symptom Complex in 268 Russian Children. International Journal of Molecular Sciences. 2022; 23(22):14343. https://doi.org/10.3390/ijms232214343
Chicago/Turabian StyleSavostyanov, Kirill, Nikolay Murashkin, Alexander Pushkov, Ilya Zhanin, Elkhan Suleymanov, Mariya Akhkiamova, Olga Shchagina, Elena Balanovska, Roman Epishev, Aleksander Polyakov, and et al. 2022. "Targeted NGS in Diagnostics of Genodermatosis Characterized by the Epidermolysis Bullosa Symptom Complex in 268 Russian Children" International Journal of Molecular Sciences 23, no. 22: 14343. https://doi.org/10.3390/ijms232214343
APA StyleSavostyanov, K., Murashkin, N., Pushkov, A., Zhanin, I., Suleymanov, E., Akhkiamova, M., Shchagina, O., Balanovska, E., Epishev, R., Polyakov, A., & Fisenko, A. (2022). Targeted NGS in Diagnostics of Genodermatosis Characterized by the Epidermolysis Bullosa Symptom Complex in 268 Russian Children. International Journal of Molecular Sciences, 23(22), 14343. https://doi.org/10.3390/ijms232214343








