Mallotucin D, a Clerodane Diterpenoid from Croton crassifolius, Suppresses HepG2 Cell Growth via Inducing Autophagic Cell Death and Pyroptosis
Abstract
1. Introduction
2. Results
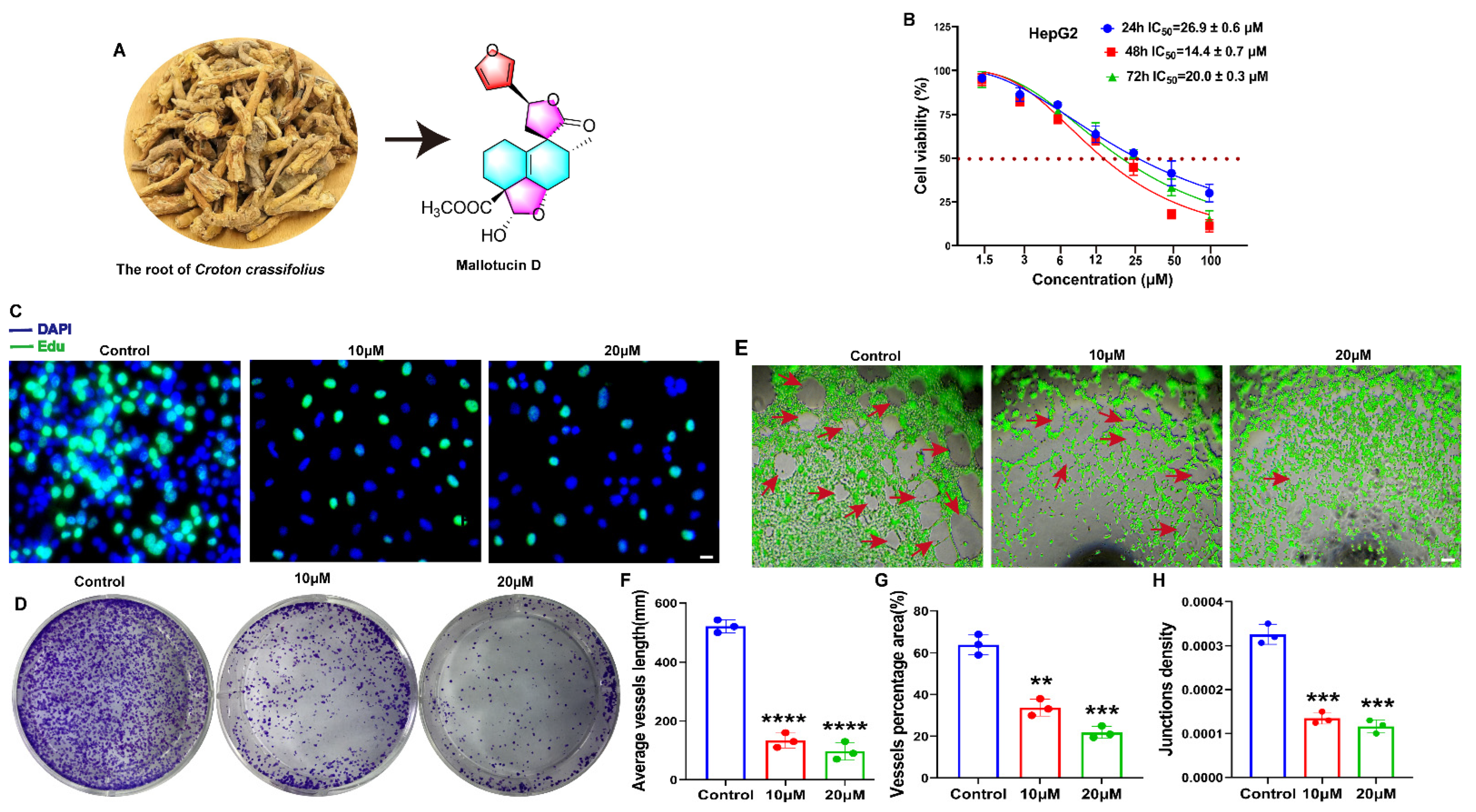
2.1. MLD Suppressed the Proliferation of HepG2 Cells and Angiogenesis of HUVECs
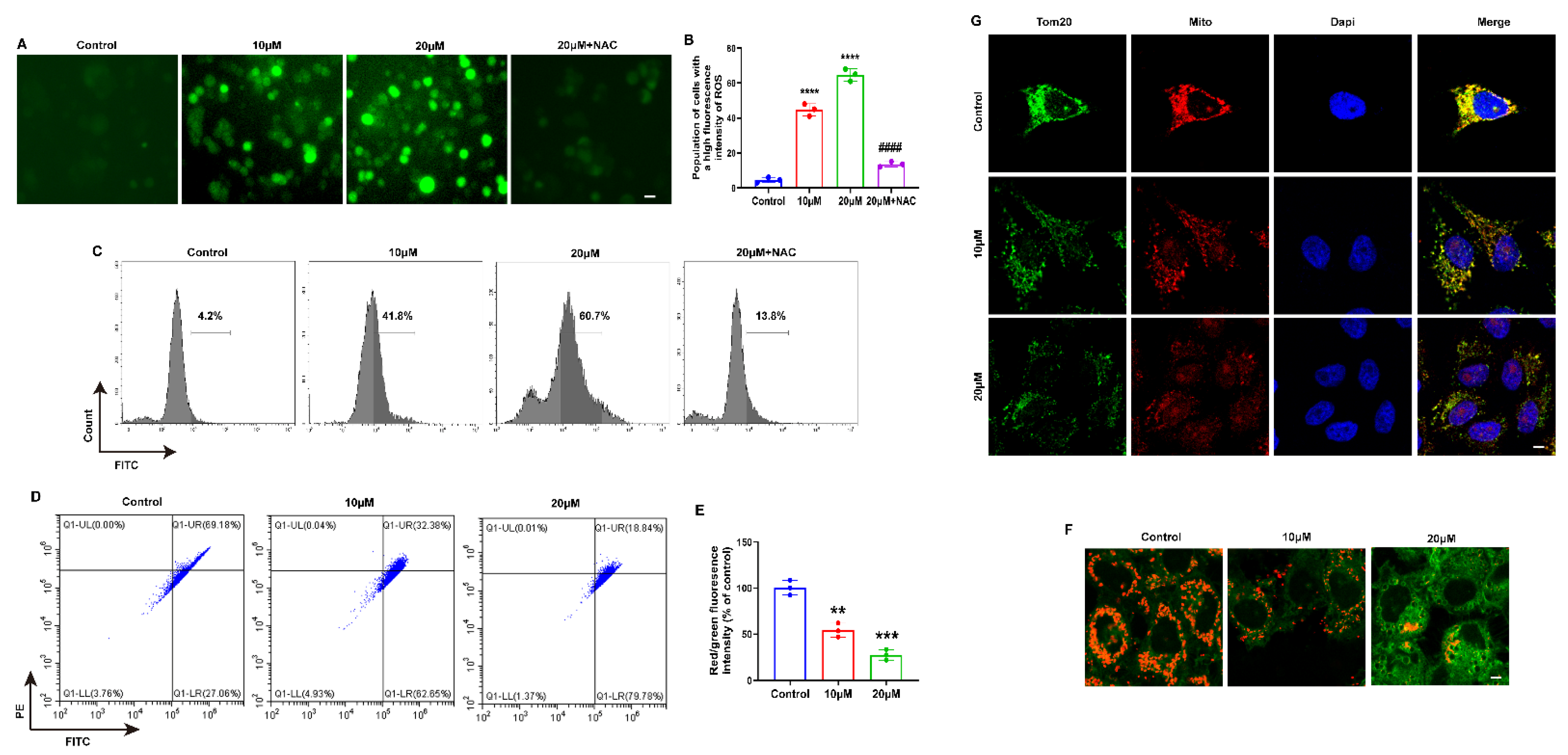
2.2. MLD Decreased the Mitochondrial Membrane Potential, Inducing ROS Production in HepG2 Cells
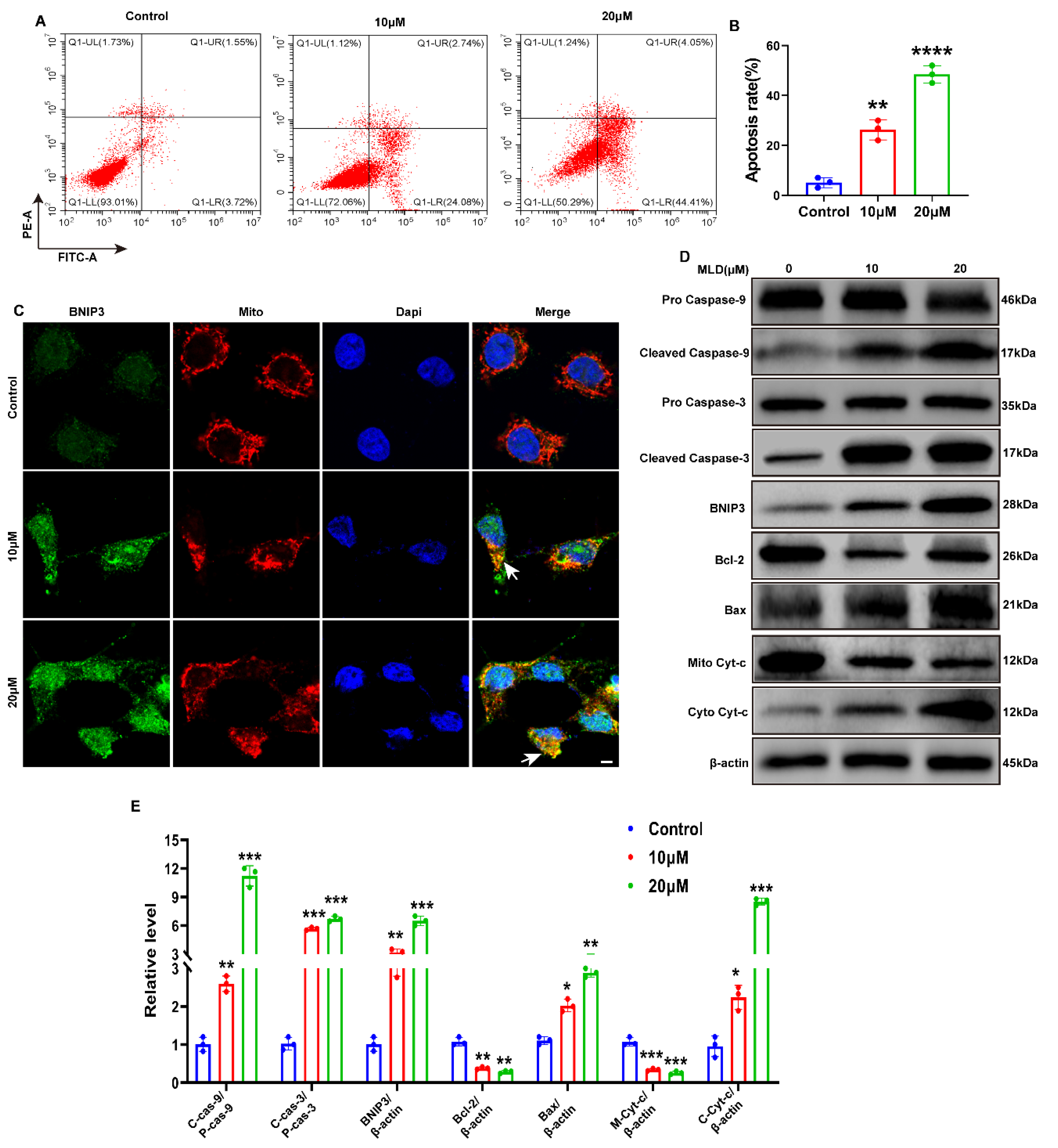
2.3. MLD Activated the Mitochondria Apoptosis of HepG2 Cells through Upregulating BNIP3
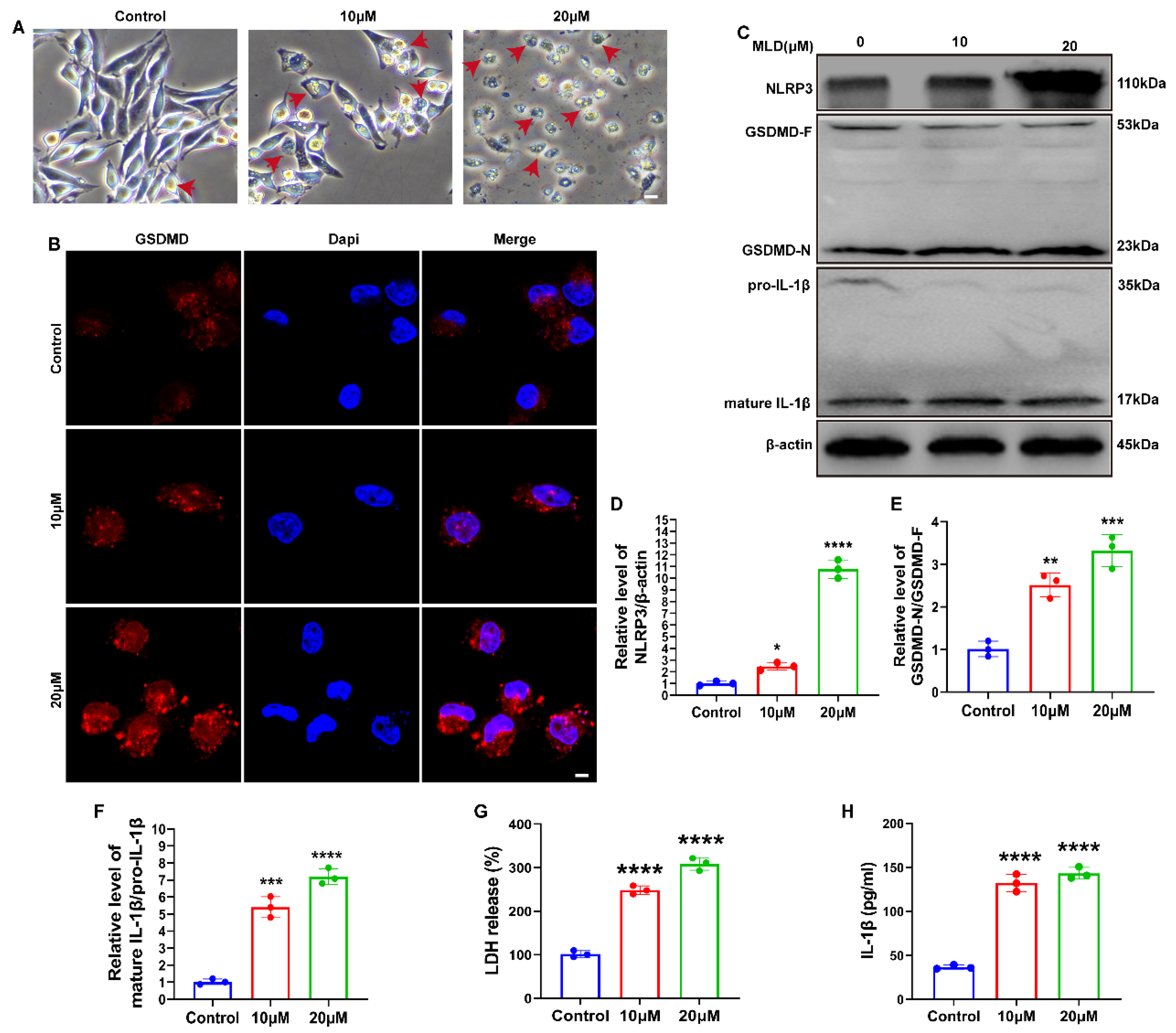
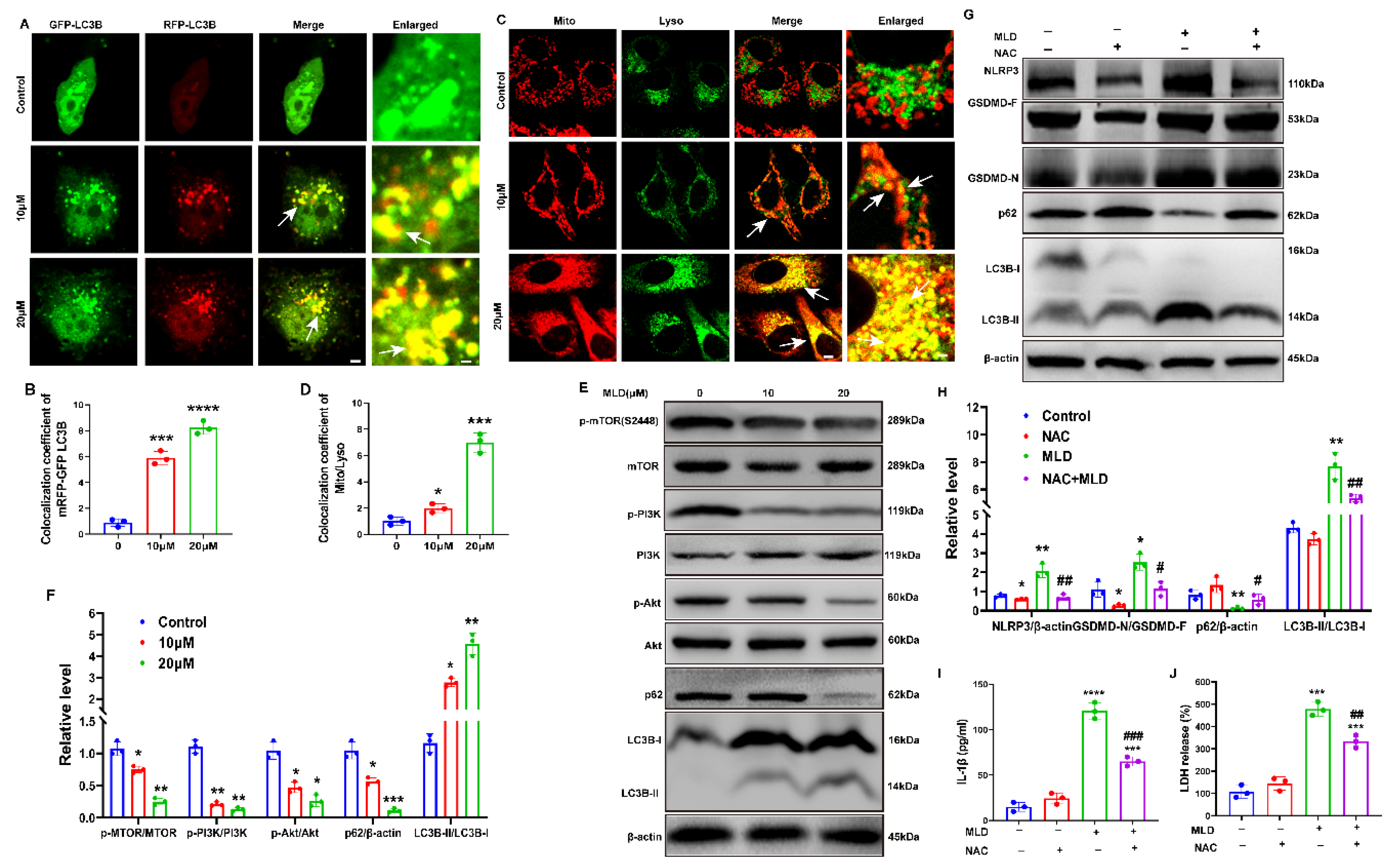
2.4. MLD Promoted HepG2 Cell Death via Activating GSDMD-Mediated Pyroptosis
2.5. MLD Induced HepG2 Cell Mitophagy by Inhibiting the PI3K/AKT/mTOR Pathway
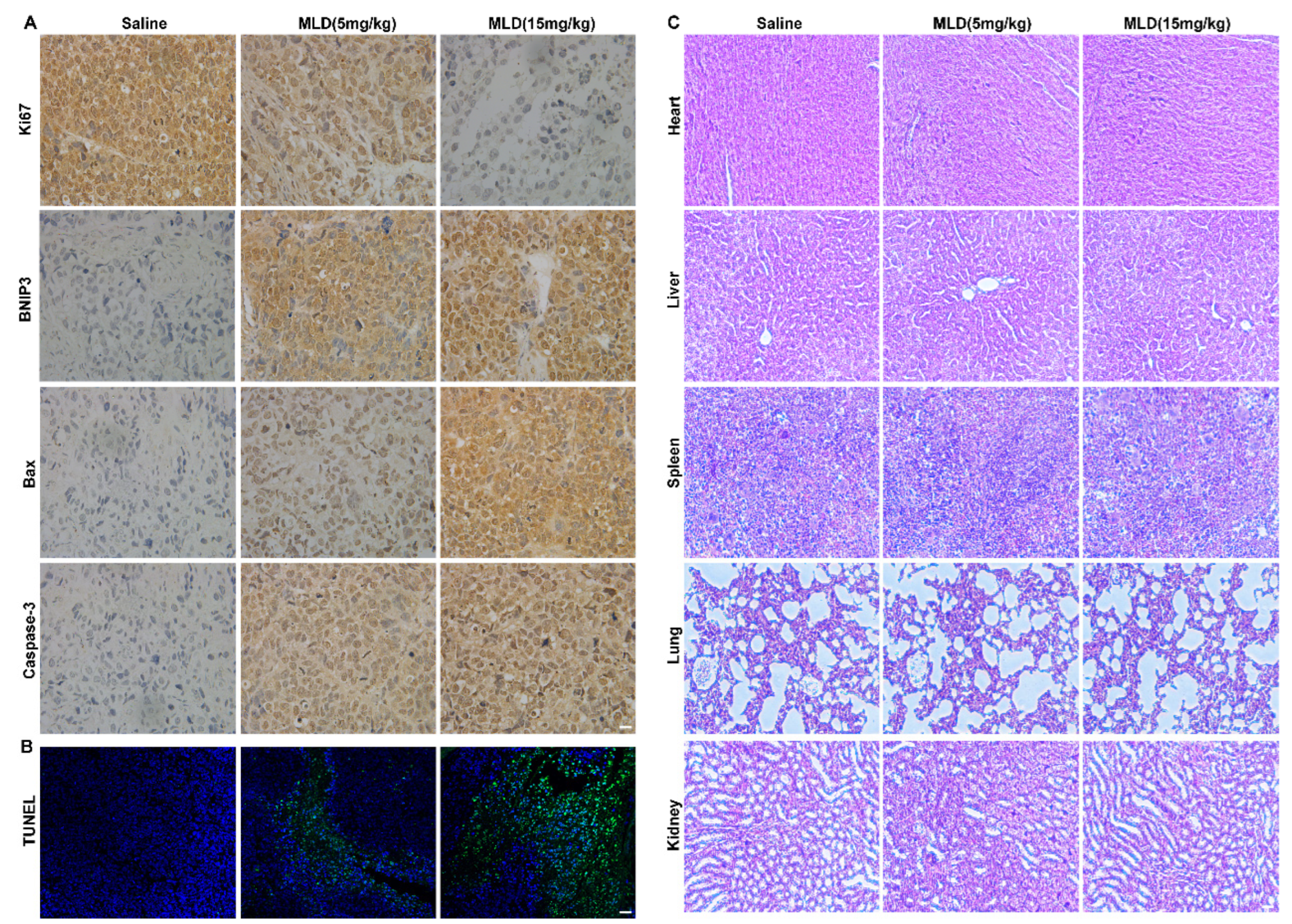
2.6. MLD Inhibited the HepG2 Cell Xenograft Tumor Growth In Vivo
3. Discussion
4. Materials and Methods
4.1. Extraction and Isolation
4.2. General Experimental Procedures
4.3. Cell Lines and Reagents
4.4. MTT Assay
4.5. ROS Assay
4.6. ELISA Assay
4.7. Immunofluorescence of Tom20, BNIP3, and GSDMD
4.8. Western Blot Assay
4.9. Colony Formation
4.10. Edu Staining Assay
4.11. Apoptosis Assay
4.12. Tube Formation Assay
4.13. JC-1 Staining Assay
4.14. MitoTracker Red CMXRos Staining and LysoTracker Green Staining
4.15. The HepG2 Cell Xenograft Tumor Study
4.16. HE Staining and Immunohistochemistry (IHC)
4.17. TUNEL Staining
4.18. The Transfection of shRNA
4.19. Serum Biochemistry and Hematological Assessments
4.20. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA A Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Sasaki-Tanaka, R.; Ray, R.; Moriyama, M. Molecular Changes in Relation to Alcohol Consumption and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 9679. [Google Scholar] [CrossRef] [PubMed]
- Mitten, E.K.; Baffy, G. Mechanotransduction in the pathogenesis of nonalcoholic fatty liver disease. J. Hepatol. 2022, 77, 1642–1656. [Google Scholar] [CrossRef] [PubMed]
- Laface, C.; Fedele, P.; Maselli, F.M.; Ambrogio, F.; Foti, C.; Molinari, P. Targeted Therapy for Hepatocellular Carcinoma: Old and New Opportunities. Cancers 2022, 14, 4028. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.B.; D’Angelica, M.I.; Abbott, D.E.; Anaya, D.A.; Anders, R.; Are, C.; Bachini, M.; Borad, M.; Brown, D.; Burgoyne, A.; et al. Hepatobiliary Cancers, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. JNCCN 2021, 19, 541–565. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Cao, L.; Zhou, R.; Yang, X.; Wu, M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 2019, 10, 1495. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhang, J.Y.; Liu, X.S.; Chen, H.Z.; Ai, Y.L.; Cheng, K.; Sun, R.Y.; Zhou, D.; Han, J.; Wu, Q. Tom20 senses iron-activated ROS signaling to promote melanoma cell pyroptosis. Cell Res. 2018, 28, 1171–1185. [Google Scholar] [CrossRef]
- Ashrafi, G.; Schwarz, T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013, 20, 31–42. [Google Scholar] [CrossRef]
- Panigrahi, D.P.; Praharaj, P.P.; Bhol, C.S.; Mahapatra, K.K.; Patra, S.; Behera, B.P.; Mishra, S.R.; Bhutia, S.K. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin. Cancer Biol. 2020, 66, 45–58. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Lu, S.; Wang, X.Z.; Wang, C.C.; Wang, L.; Liang, S.P.; Luo, T.F.; Wang, Z.C.; Piao, M.H.; Chi, G.F.; et al. FOXO3a protects glioma cells against temozolomide-induced DNA double strand breaks via promotion of BNIP3-mediated mitophagy. Acta Pharmacol. Sin. 2021, 42, 1324–1337. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Guo, Z.; Zhu, P.; Chen, J.; Huang, Y. Traditional Chinese medicine as a cancer treatment: Modern perspectives of ancient but advanced science. Cancer Med. 2019, 8, 1958–1975. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhong, Z.; Tan, H.Y.; Guo, W.; Zhang, C.; Tan, C.-W.; Li, S.; Wang, N.; Feng, Y. Uncovering the Anticancer Mechanisms of Chinese Herbal Medicine Formulas: Therapeutic Alternatives for Liver Cancer. Front. Pharmacol. 2020, 11, 293. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.C.; Li, J.G.; Li, G.Q.; Xu, J.J.; Wu, X.; Ye, W.C.; Li, Y.L. Clerodane diterpenoids from Croton crassifolius. J. Nat. Prod. 2012, 75, 2188–2192. [Google Scholar] [CrossRef]
- Jiang, S.; Shen, X.; Xuan, S.; Yang, B.; Ruan, Q.; Cui, H.; Zhao, Z.; Jin, J. Serum and colon metabolomics study reveals the anti-ulcerative colitis effect of Croton crassifolius Geisel. Phytomedicine Int. J. Phytother. Phytopharm. 2021, 87, 153570. [Google Scholar] [CrossRef]
- Huang, W.; Liang, Y.; Chung, H.Y.; Wang, G.; Huang, J.J.; Li, Y. Cyperenoic acid, a sesquiterpene derivative from Croton crassifolius, inhibits tumor growth through anti-angiogenesis by attenuating VEGFR2 signal pathway in breast cancer. Phytomedicine Int. J. Phytother. Phytopharm. 2020, 76, 153253. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, J.; Mao, L.; Guo, Y.; Hao, Y.; Deng, Y.; Han, X.; Li, Q.; Liao, W.; Yuan, M. Nobiletin Triggers Reactive Oxygen Species-Mediated Pyroptosis through Regulating Autophagy in Ovarian Cancer Cells. J. Agric. Food Chem. 2020, 68, 1326–1336. [Google Scholar] [CrossRef]
- Rapaport, D. Biogenesis of the mitochondrial TOM complex. Trends Biochem. Sci. 2002, 27, 191–197. [Google Scholar] [CrossRef]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Li, H.H.; Qi, F.M.; Dong, L.L.; Fan, G.X.; Che, J.M.; Guo, D.D.; Zhang, Z.X.; Fei, D.Q. Cytotoxic and antibacterial pyran-2-one derivatives from Croton crassifolius. Phytochem. Lett. 2014, 10, 304–308. [Google Scholar] [CrossRef]
- Zhang, D.B.; Tang, Z.S.; Xie, P.; Liang, Y.N.; Yu, J.G. A pair of new neo-clerodane diterpenoid epimers from the roots of Croton crassifolius and their anti-inflammatory. Nat. Product. Res. 2020, 34, 2945–2951. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Chung, H.Y.; Zhang, Y.B.; Li, G.Q.; Li, Y.L.; Huang, W.H.; Wang, G.C. Diterpenoids from the roots of Croton crassifolius and their anti-angiogenic activity. Phytochemistry 2016, 122, 270–275. [Google Scholar] [CrossRef]
- Li, W.; Hao, J.; Zhang, L.; Cheng, Z.; Deng, X.; Shu, G. Astragalin Reduces Hexokinase 2 through Increasing miR-125b to Inhibit the Proliferation of Hepatocellular Carcinoma Cells in Vitro and in Vivo. J. Agric. Food Chem. 2017, 65, 5961–5972. [Google Scholar] [CrossRef] [PubMed]
- Munakarmi, S.; Chand, L.; Shin, H.B.; Hussein, U.K.; Yun, B.S.; Park, H.R.; Jeong, Y.J. Anticancer effects of Poncirus fructus on hepatocellular carcinoma through regulation of apoptosis, migration, and invasion. Oncol. Rep. 2020, 44, 2537–2546. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.L.; Ho, C.T.; Chung, J.G.; Raghu, R.; Lo, Y.C.; Sheen, L.Y. Allicin induces anti-human liver cancer cells through the p53 gene modulating apoptosis and autophagy. J. Agric. Food Chem. 2013, 61, 9839–9848. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.J.; Shin, D.S.; Getzoff, E.D.; Tainer, J.A. The structural biochemistry of the superoxide dismutases. Biochim. Et Biophys. Acta 2010, 1804, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Kacso, T.P.; Zahu, R.; Tirpe, A. Reactive Oxygen Species and Long Non-Coding RNAs, an Unexpected Crossroad in Cancer Cells. Int. J. Mol. Sci. 2022, 23, 10133. [Google Scholar] [CrossRef]
- Liang, B.; Shao, W.; Zhu, C.; Wen, G.; Yue, X.; Wang, R.; Quan, J.; Du, J.; Bu, X. Mitochondria-Targeted Approach: Remarkably Enhanced Cellular Bioactivities of TPP2a as Selective Inhibitor and Probe toward TrxR. ACS Chem. Biol. 2016, 11, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Dewaele, M.; Maes, H.; Agostinis, P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy 2010, 6, 838–854. [Google Scholar] [CrossRef]
- Starkov, A.A. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N. Y. Acad. Sci. 2008, 1147, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Hamacher-Brady, A.; Brady, N.R.; Logue, S.E.; Sayen, M.R.; Jinno, M.; Kirshenbaum, L.A.; Gottlieb, R.A.; Gustafsson, A.B. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007, 14, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.W.; Li, W.; Zheng, X.J.; Liu, J.Y.; Yang, Y.H.; Li, S.; Zhang, S.; Fu, W.Q.; Xiao, B.; Wang, J.H.; et al. Benzimidazoles induce concurrent apoptosis and pyroptosis of human glioblastoma cells via arresting cell cycle. Acta Pharmacol. Sin. 2022, 43, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, P.; An, L.; Sun, N.; Peng, L.; Tang, W.; Ma, D.; Chen, J. Miltirone induces cell death in hepatocellular carcinoma cell through GSDME-dependent pyroptosis. Acta Pharm. Sin. B 2020, 10, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Huang, Z.; Chen, J.; Wang, J.; Wang, S. Protein phosphatase 2A activation mechanism contributes to JS-K induced caspase-dependent apoptosis in human hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. CR 2018, 37, 142. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Chiang, D.Y.; Newell, P.; Peix, J.; Thung, S.; Alsinet, C.; Tovar, V.; Roayaie, S.; Minguez, B.; Sole, M.; et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 2008, 135, 1972–1983. [Google Scholar] [CrossRef]
- Zhou, L.; Huang, Y.; Li, J.; Wang, Z. The mTOR pathway is associated with the poor prognosis of human hepatocellular carcinoma. Med. Oncol. 2010, 27, 255–261. [Google Scholar] [CrossRef]
- Chen, L.-M.; Song, T.-J.; Xiao, J.-H.; Huang, Z.-H.; Li, Y.; Lin, T.-Y. Tripchlorolide induces autophagy in lung cancer cells by inhibiting the PI3K/AKT/mTOR pathway and improves cisplatin sensitivity in A549/DDP cells. Oncotarget 2017, 8, 63911–63922. [Google Scholar] [CrossRef]
- Portal-Núñez, S.; Esbrit, P.; Alcaraz, M.J.; Largo, R. Oxidative stress, autophagy, epigenetic changes and regulation by miRNAs as potential therapeutic targets in osteoarthritis. Biochem. Pharmacol. 2016, 108, 1–10. [Google Scholar] [CrossRef]
- Russak, E.M.; Bednarczyk, E.M. Impact of Deuterium Substitution on the Pharmacokinetics of Pharmaceuticals. Ann. Pharmacother. 2019, 53, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tan, L.; Yu, Y.; Wang, B.; Chen, Z.; Han, J.; Li, M.; Chen, J.; Xiao, T.; Ambati, B.K.; et al. Placenta-specific drug delivery by trophoblast-targeted nanoparticles in mice. Theranostics 2018, 8, 2765–2781. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, X.; Sun, F.; Deng, K.; Lin, G.; Yin, W.; Chen, H.; Yang, D.; Liu, K.; Zhang, Y.; Huang, L. Mallotucin D, a Clerodane Diterpenoid from Croton crassifolius, Suppresses HepG2 Cell Growth via Inducing Autophagic Cell Death and Pyroptosis. Int. J. Mol. Sci. 2022, 23, 14217. https://doi.org/10.3390/ijms232214217
Dai X, Sun F, Deng K, Lin G, Yin W, Chen H, Yang D, Liu K, Zhang Y, Huang L. Mallotucin D, a Clerodane Diterpenoid from Croton crassifolius, Suppresses HepG2 Cell Growth via Inducing Autophagic Cell Death and Pyroptosis. International Journal of Molecular Sciences. 2022; 23(22):14217. https://doi.org/10.3390/ijms232214217
Chicago/Turabian StyleDai, Xiaoyong, Fen Sun, Kexin Deng, Gaoyang Lin, Wenjing Yin, Huaqing Chen, Dongye Yang, Kewei Liu, Yubo Zhang, and Laiqiang Huang. 2022. "Mallotucin D, a Clerodane Diterpenoid from Croton crassifolius, Suppresses HepG2 Cell Growth via Inducing Autophagic Cell Death and Pyroptosis" International Journal of Molecular Sciences 23, no. 22: 14217. https://doi.org/10.3390/ijms232214217
APA StyleDai, X., Sun, F., Deng, K., Lin, G., Yin, W., Chen, H., Yang, D., Liu, K., Zhang, Y., & Huang, L. (2022). Mallotucin D, a Clerodane Diterpenoid from Croton crassifolius, Suppresses HepG2 Cell Growth via Inducing Autophagic Cell Death and Pyroptosis. International Journal of Molecular Sciences, 23(22), 14217. https://doi.org/10.3390/ijms232214217






