Assessment of RACGAP1 as a Prognostic and Immunological Biomarker in Multiple Human Tumors: A Multiomics Analysis
Abstract
1. Introduction
2. Results
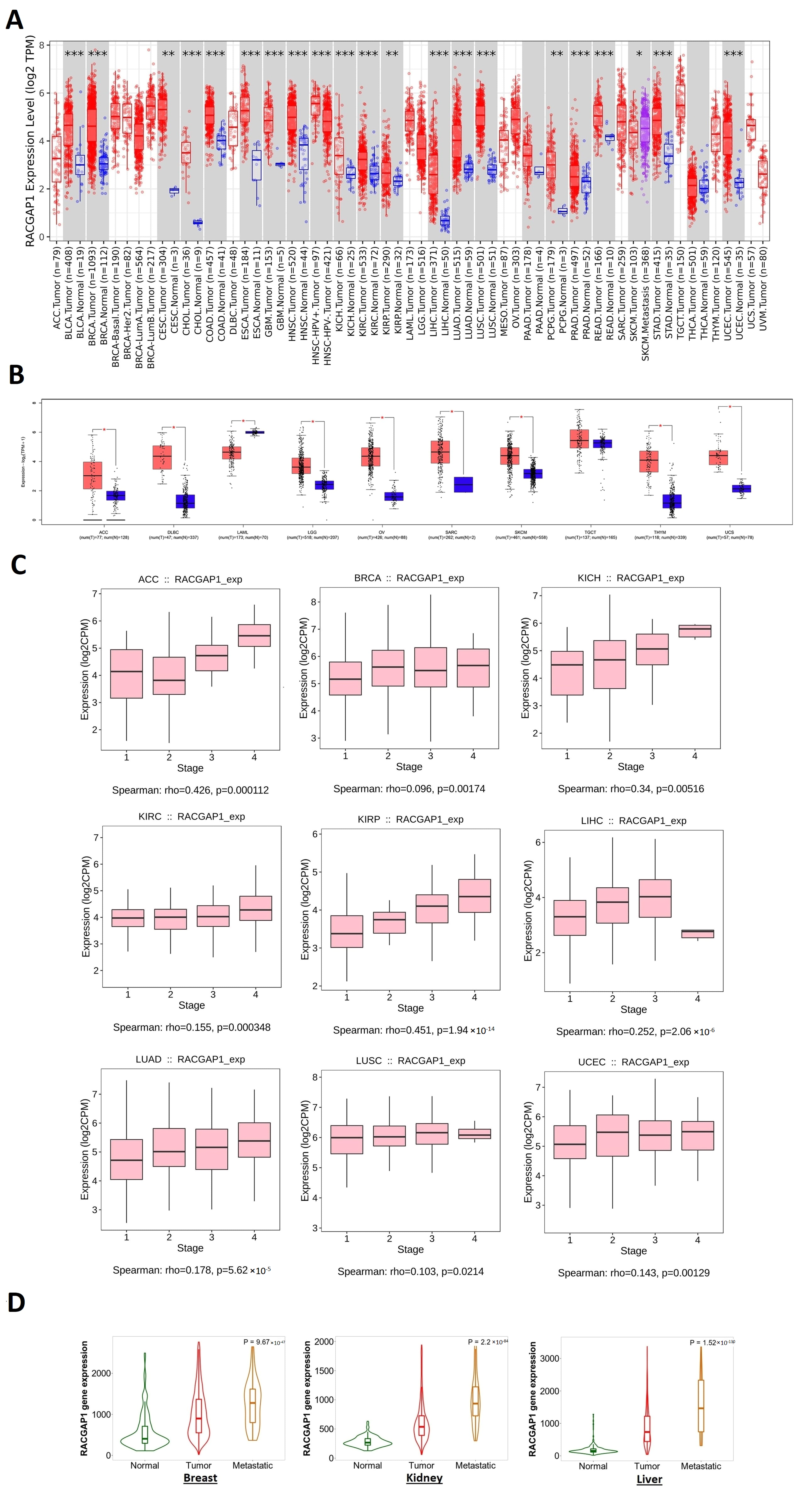
2.1. RACGAP1 Increased Expression in Many Tumor Types
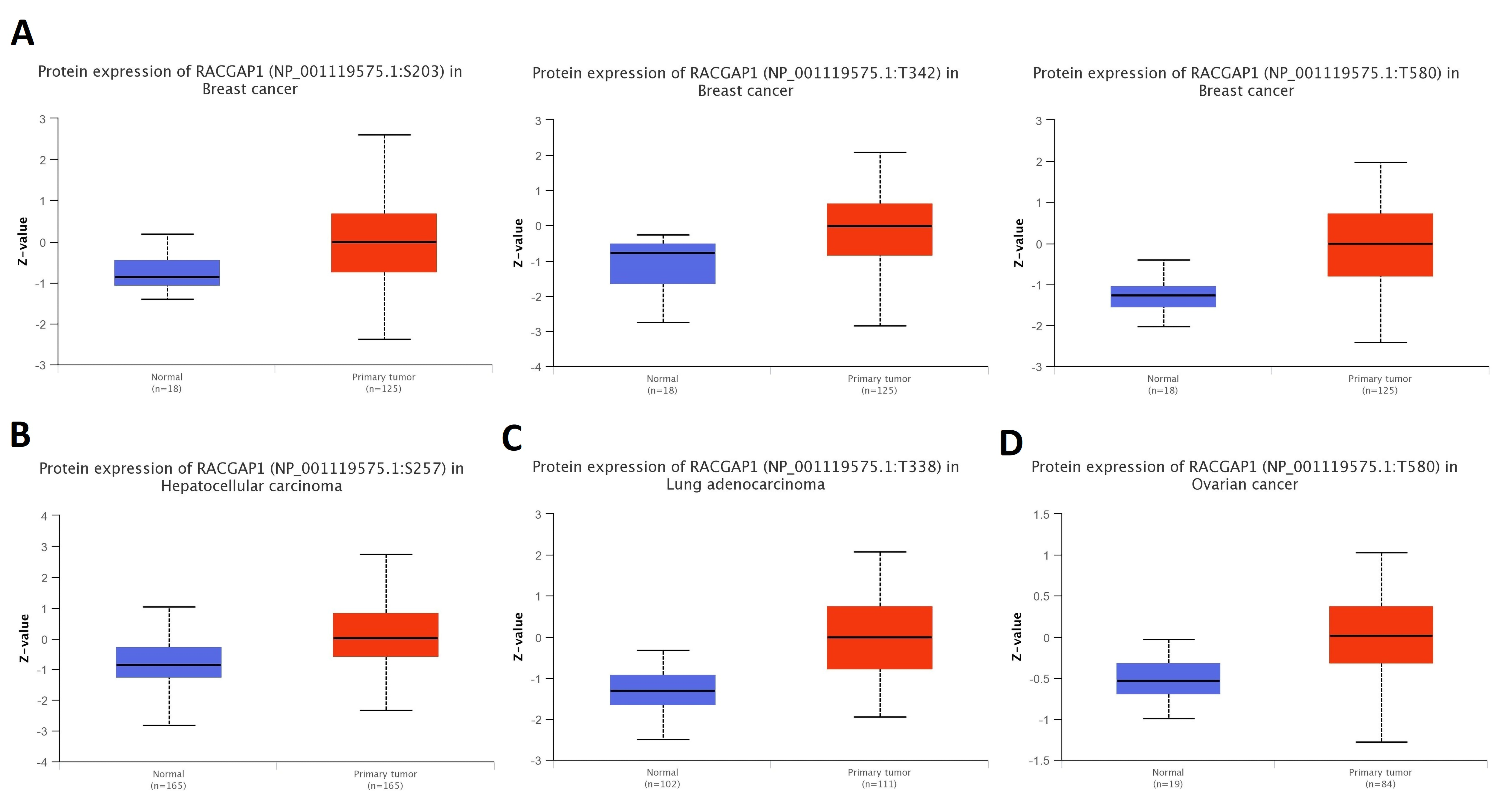
2.2. Differential Protein Expression
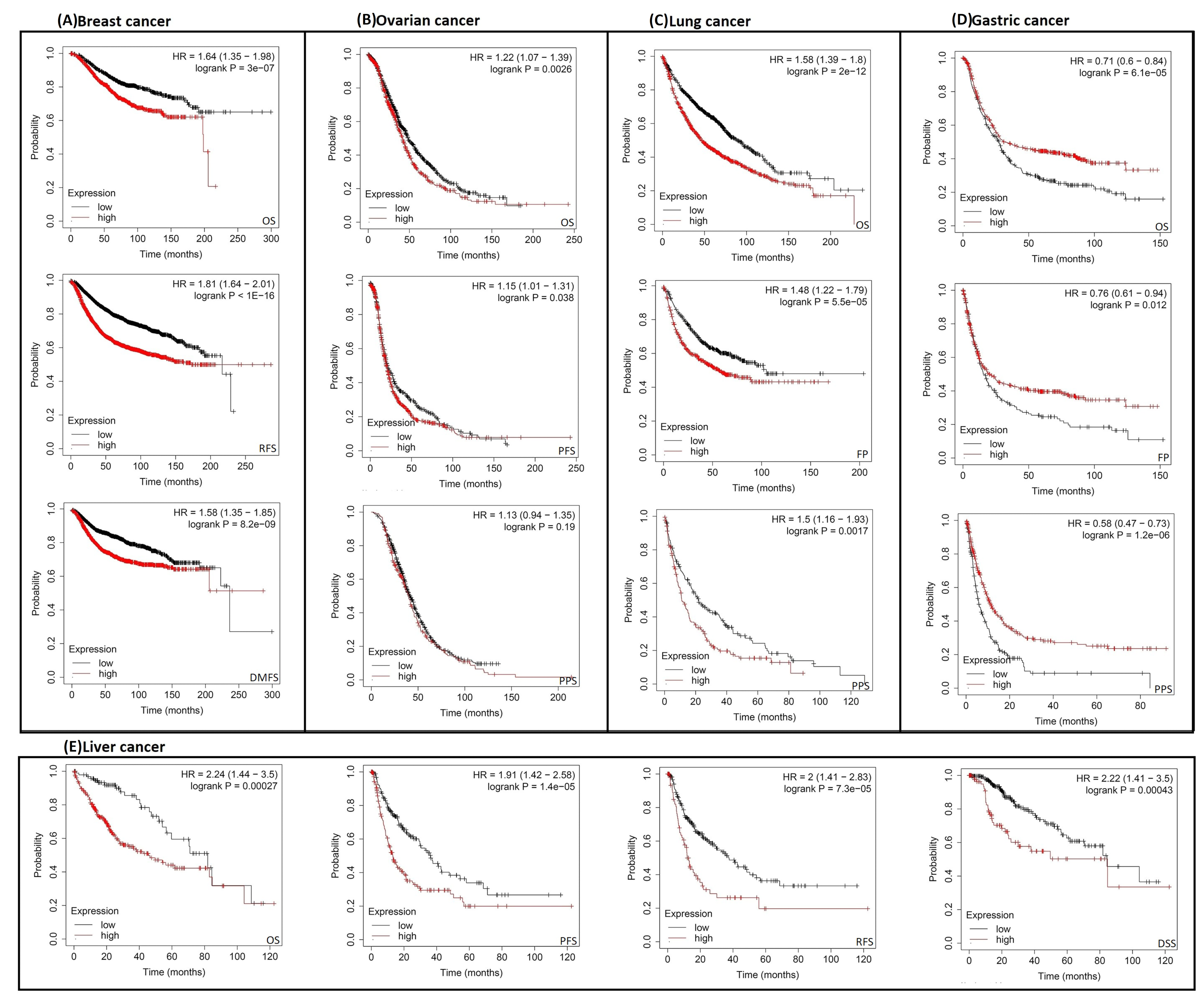
2.3. Increased RACGAP1 Level Estimates Poor Clinical Outcomes
2.4. RACGAP1 Mutation Analysis
2.5. RACGAP1 Phosphorylation Analysis
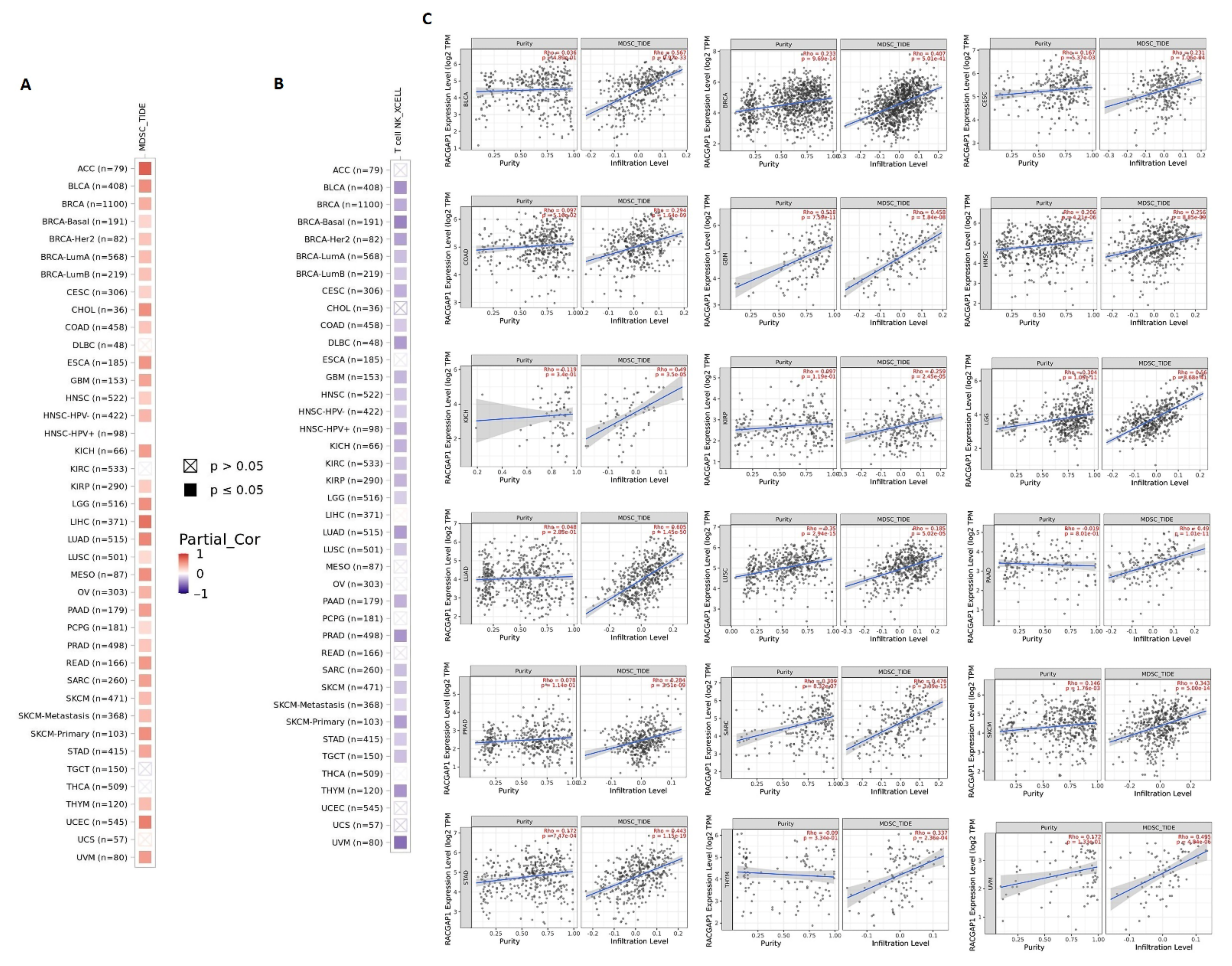
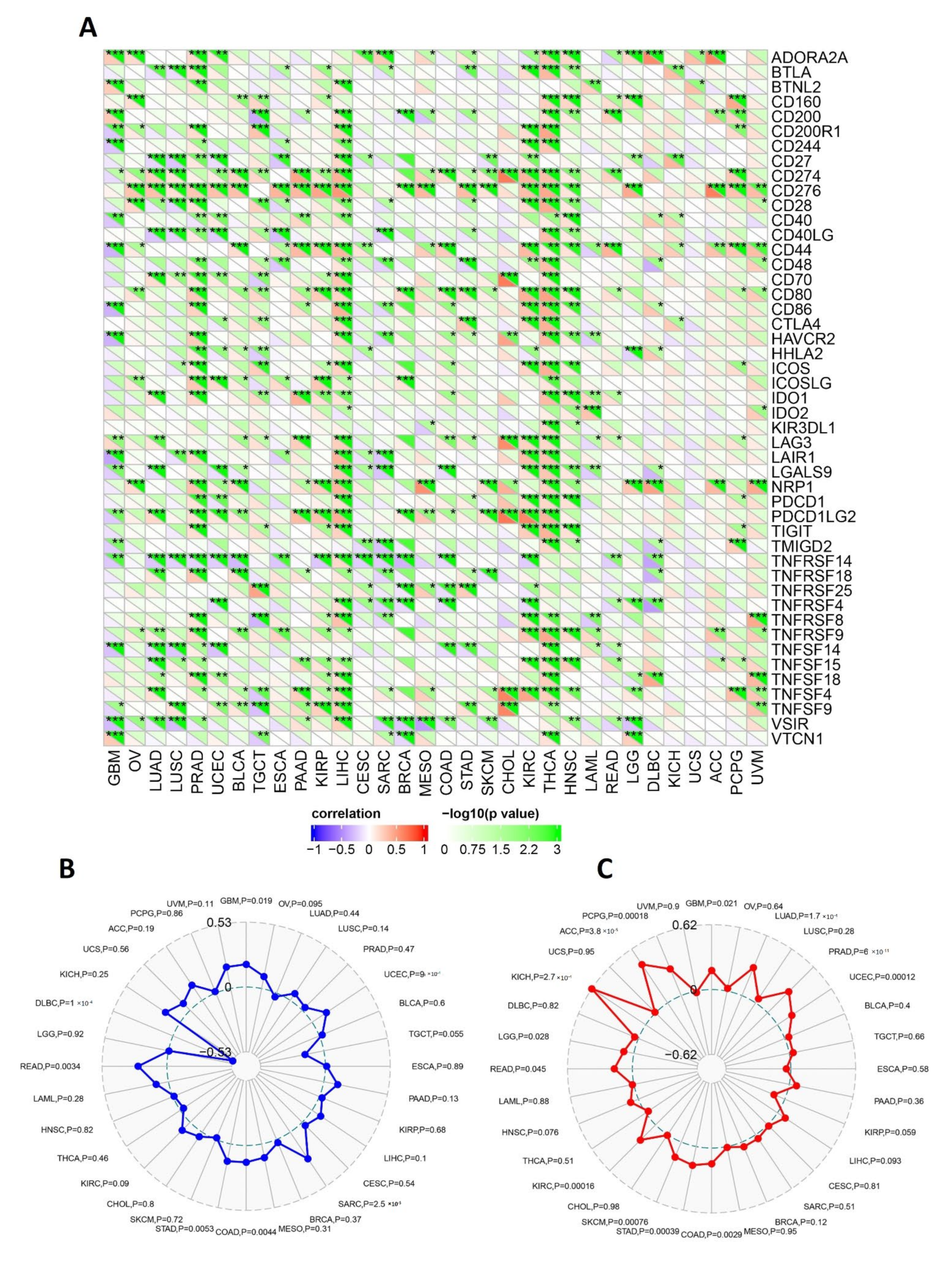
2.6. RACGAP1 Correlates with Immune Infiltration in Several Tumor Types
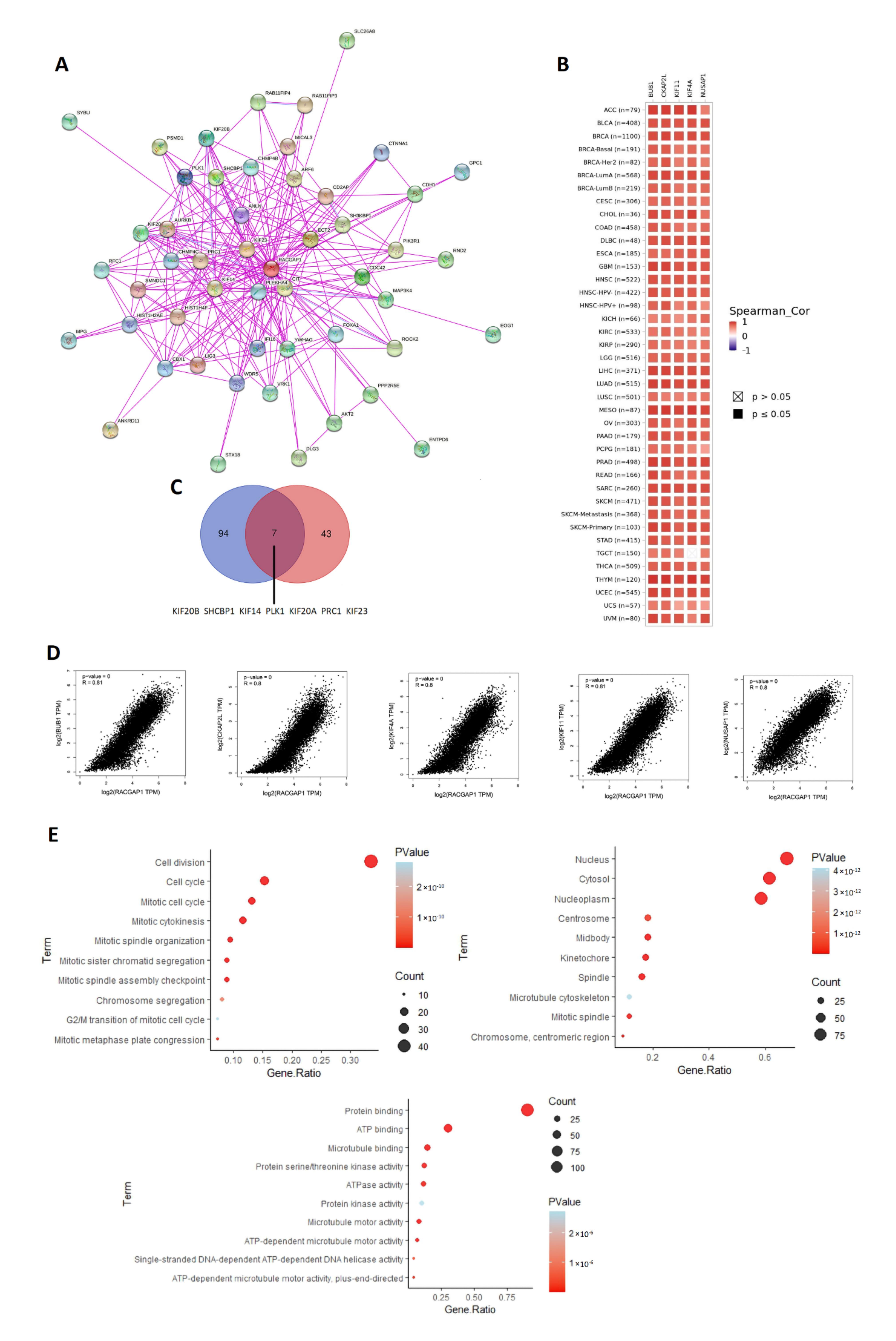
2.7. Analysis of Interacting and Correlated Proteins to RACGAP1
3. Discussion
4. Materials and Methods
4.1. Gene Expression Analysis
4.2. Protein Expression and Immunohistochemistry (IHC) Staining
4.3. Survival Prognosis Analysis
4.4. Gene Alteration Analysis
4.5. RACGAP1 Phosphorylation Analysis
4.6. Immune Reactivity Assessment
4.7. Protein–Protein Interaction and Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gharib, A.F.; Eldeen, M.A.; Khalifa, A.S.; Elsawy, W.H.; Eed, E.M.; El Askary, A.; Eid, R.A.; Soltan, M.A.; Raafat, N. Assessment of Glutathione Peroxidase-1 (GPX1) Gene Expression as a Specific Diagnostic and Prognostic Biomarker in Malignant Pleural Mesothelioma. Diagnostics 2021, 11, 2285. [Google Scholar] [CrossRef] [PubMed]
- Mahaboob Batcha, A.T.; Subramaniam, G.; Venkatachalam, K. Purified Banana lectin (BanLec) isolated from the ripen pulp of Musa Paradisiaca induces apoptosis in cancer cell lines: In vitro study. Adv. Tradit. Med. 2022, 3, 6373. [Google Scholar] [CrossRef]
- Hnisz, D.; Weintrau, A.S.; Day, D.S.; Valton, A.-L.; Bak, R.O.; Li, C.H.; Goldmann, J.; Lajoie, B.R.; Fan, Z.P.; Sigova, A.A.; et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 2016, 351, 1454–1458. [Google Scholar] [CrossRef] [PubMed]
- Blum, A.; Wang, P.; Zenklusen, J.C. SnapShot: TCGA-Analyzed Tumors. Cell 2018, 173, 530. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Wspolczesna Onkol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Lee, J.-S.; Kamijo, K.; Ohara, N.; Kitamura, T.; Miki, T. MgcRacGAP regulates cortical activity through RhoA during cytokinesis. Exp. Cell Res. 2003, 293, 275–282. [Google Scholar] [CrossRef]
- Zhao, W.-M.; Fang, G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. USA 2005, 102, 13158–13163. [Google Scholar] [CrossRef]
- Jantsch-Plunger, V.; Gönczy, P.; Romano, A.; Schnabel, H.; Hamill, D.; Schnabel, R.; Hyman, A.; Glotzer, M. CYK-4: A Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 2000, 149, 1391–1404. [Google Scholar] [CrossRef]
- Kawashima, T.; Bao, Y.C.; Minoshima, Y.; Nomura, Y.; Hatori, T.; Hori, T.; Fukagawa, T.; Fukada, T.; Takahashi, N.; Nosaka, T.; et al. A Rac GTPase-Activating Protein, MgcRacGAP, Is a Nuclear Localizing Signal-Containing Nuclear Chaperone in the Activation of STAT Transcription Factors. Mol. Cell. Biol. 2009, 29, 1796–1813. [Google Scholar] [CrossRef]
- Sahai, E. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 2005, 15, 87–96. [Google Scholar] [CrossRef]
- Mishima, M.; Kaitna, S.; Glotzer, M. Central Spindle Assembly and Cytokinesis Require a Kinesin-like Protein/RhoGAP Complex with Microtubule Bundling Activity. Dev. Cell 2002, 2, 41–54. [Google Scholar] [CrossRef]
- Touré, A.; Morin, L.; Pineau, C.; Becq, F.; Dorseuil, O.; Gacon, G. Tat1, a Novel Sulfate Transporter Specifically Expressed in Human Male Germ Cells and Potentially Linked to RhoGTPase Signaling. J. Biol. Chem. 2001, 276, 20309–20315. [Google Scholar] [CrossRef] [PubMed]
- Naud, N.; Toure, A.; Liu, J.; Pineau, C.; Morin, L.; Dorseuil, O.; Escalier, D.; Chardin, P.; Gacon, G. Rho family GTPase Rnd2 interacts and co-localizes with MgcRacGAP in male germ cells. Biochem. J. 2003, 372, 105–112. [Google Scholar] [CrossRef]
- Zhang, P.; Bai, H.; Fu, C.; Chen, F.; Zeng, P.; Wu, C.; Ye, Q.; Dong, C.; Song, Y.; Song, E. RacGAP1-driven focal adhesion formation promotes melanoma transendothelial migration through mediating adherens junction disassembly. Biochem. Biophys. Res. Commun. 2015, 459, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Strimpakos, A.; Sampaziotis, D.; Psyrri, A. RACGAP1 (Rac GTPase activating protein 1). Atlas Genet. Cytogenet. Oncol. Haematol. 2018, 20, 506. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, L.; Si, F.; Huang, L.; Zhao, Y.; Zhang, C.; Hoft, D.F.; Peng, G. NK and NKT cells have distinct properties and functions in cancer. Oncogene 2021, 40, 4521–4537. [Google Scholar] [CrossRef]
- Yamazaki, D.; Kurisu, S.; Takenawa, T. Involvement of Rac and Rho signaling in cancer cell motility in 3D substrates. Oncogene 2009, 28, 1570–1583. [Google Scholar] [CrossRef]
- Saigusa, S.; Tanaka, K.; Mohri, Y.; Ohi, M.; Shimura, T.; Kitajima, T.; Kondo, S.; Okugawa, Y.; Toiyama, Y.; Inoue, Y.; et al. Clinical significance of RacGAP1 expression at the invasive front of gastric cancer. Gastric Cancer 2015, 18, 84–92. [Google Scholar] [CrossRef]
- Sahin, S.; Ekinci, O.; Seckin, S.; Dursun, A. Proliferation markers RacGAP1 and Ki-67 in gastrointestinal stromal tumors by immunohistochemistry with respect to clinicopathological features and different risk stratification systems. Int. J. Clin. Exp. Pathol. 2017, 10, 11723–11736. [Google Scholar]
- Imaoka, H.; Toiyama, Y.; Saigusa, S.; Kawamura, M.; Kawamoto, A.; Okugawa, Y.; Hiro, J.; Tanaka, K.; Inoue, Y.; Mohri, Y.; et al. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis 2014, 36, 346–354. [Google Scholar] [CrossRef]
- Ren, K.; Zhou, D.; Wang, M.; Li, E.; Hou, C.; Su, Y.; Zou, Q.; Zhou, P.; Liu, X. RACGAP1 modulates ECT2-Dependent mitochondrial quality control to drive breast cancer metastasis. Exp. Cell Res. 2021, 400, 112493. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Toiyama, Y.; Okugawa, Y.; Shigemori, T.; Yamamoto, A.; Ide, S.; Kitajima, T.; Fujikawa, H.; Yasuda, H.; Okita, Y.; et al. Rac GTPase-Activating Protein 1 (RACGAP1) as an Oncogenic Enhancer in Esophageal Carcinoma. Oncology 2019, 97, 155–163. [Google Scholar] [CrossRef]
- Ruan, X.; Jiang, J. RACGAP1 promotes proliferation and cell cycle progression by regulating CDC25C in cervical cancer cells. Tissue Cell 2022, 76, 101804. [Google Scholar] [CrossRef]
- Hazar-Rethinam, M.; de Long, L.M.; Gannon, O.M.; Boros, S.; Vargas, A.C.; Dzienis, M.; Mukhopadhyay, P.; Saenz-Ponce, N.; Dantzic, D.D.; Simpson, F.; et al. RacGAP1 Is a Novel Downstream Effector of E2F7-Dependent Resistance to Doxorubicin and Is Prognostic for Overall Survival in Squamous Cell Carcinoma. Mol. Cancer Ther. 2015, 14, 1939–1950. [Google Scholar] [CrossRef]
- Wang, S.M.; Ooi, L.L.P.; Hui, K.M. Upregulation of Rac GTPase-Activating Protein 1 Is Significantly Associated with the Early Recurrence of Human Hepatocellular Carcinoma. Clin. Cancer Res. 2011, 17, 6040–6051. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-Y.; Chen, D.-P.; Qi, B.; Li, M.-Y.; Zhu, Y.-Y.; Yin, W.-J.; He, L.; Yu, Y.; Li, Z.-Y.; Lin, L.; et al. Pseudogene RACGAP1P activates RACGAP1/Rho/ERK signalling axis as a competing endogenous RNA to promote hepatocellular carcinoma early recurrence. Cell Death Dis. 2019, 10, 426. [Google Scholar] [CrossRef]
- Zabady, S.; Mahran, N.; Soltan, M.A.; Eldeen, M.A.; Eid, R.A.; Albogami, S.; Fayad, E.; Matboli, M.; Habib, E.K.; Hasanin, A.H.; et al. Cyanidin-3-Glucoside Modulates hsa_circ_0001345/miRNA106b/ATG16L1 Axis Expression as a Potential Protective Mechanism against Hepatocellular Carcinoma. Curr. Issues Mol. Biol. 2022, 44, 1677–1687. [Google Scholar] [CrossRef]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 153–168. [Google Scholar] [CrossRef]
- Shen, H.; Che, K.; Cong, L.; Dong, W.; Zhang, T.; Liu, Q.; Du, J. Diagnostic and prognostic value of blood samples for KRAS mutation identification in lung cancer: A meta-analysis. Oncotarget 2017, 8, 36812–36823. [Google Scholar] [CrossRef] [PubMed]
- Jakob, J.A.; Bassett, R.L.; Ng, C.S.; Curry, J.L.; Joseph, R.W.; Rohlfs, M.L.; Alvarado, G.C.; Richard, J.; Gershenwald, J.E.; Kim, K.B.; et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012, 118, 4014–4023. [Google Scholar] [CrossRef] [PubMed]
- Anglim, P.P.; A Alonzo, T.; A Laird-Offringa, I. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: An update. Mol. Cancer 2008, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Romero-Garcia, S.; Prado-Garcia, H.; Carlos-Reyes, A. Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front. Oncol. 2020, 10, 1152. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Sun, W.; Tan, M.; Glazer, C.A.; Bhan, S.; Zhong, X.; Fakhry, C.; Sharma, R.; Westra, W.H.; Hoque, M.O.; et al. Integrated, Genome-Wide Screening for Hypomethylated Oncogenes in Salivary Gland Adenoid Cystic Carcinoma. Clin. Cancer Res. 2011, 17, 4320–4330. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2013, 63, 635–646. [Google Scholar] [CrossRef]
- Søes, S.; Daugaard, I.L.; Sørensen, B.S.; Carus, A.; Mattheisen, M.; Alsner, J.; Overgaard, J.; Hager, H.; Hansen, L.L.; Kristensen, L.S. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience 2014, 1, 367–374. [Google Scholar] [CrossRef]
- Peng, M.; Mo, Y.; Wang, Y.; Wu, P.; Zhang, Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; Li, X.; et al. Neoantigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 2019, 18, 128. [Google Scholar] [CrossRef]
- Chang, E.; Pelosof, L.; Lemery, S.; Gong, Y.; Goldberg, K.B.; Farrell, A.T.; Keegan, P.; Veeraraghavan, J.; Wei, G.; Blumenthal, G.M.; et al. Systematic Review of PD-1/PD-L1 Inhibitors in Oncology: From Personalized Medicine to Public Health. Oncologist 2021, 26, e1786–e1799. [Google Scholar] [CrossRef]
- Condamine, T.; Ramachandran, I.; Youn, J.-I.; Gabrilovich, D.I. Regulation of Tumor Metastasis by Myeloid-Derived Suppressor Cells. Annu. Rev. Med. 2015, 66, 97–110. [Google Scholar] [CrossRef]
- Weber, R.; Fleming, V.; Hu, X.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018, 9, 1310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, X.; Zhu, C.; Liu, L.; Wang, G.; Yuan, X. The Role of Myeloid-Derived Suppressor Cells in Patients with Solid Tumors: A Meta-Analysis. PLoS ONE 2016, 11, e0164514. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Bae, E.-A.; Seo, H.; Kim, I.-K.; Jeon, I.; Kang, C.-Y. Roles of NKT cells in cancer immunotherapy. Arch. Pharm. Res. 2019, 42, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B.J.; Choi, J.E.; Exley, M.A. Novel Approaches to Exploiting Invariant NKT Cells in Cancer Immunotherapy. Front. Immunol. 2018, 9, 384. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, W.; Zhang, X.; Ge, M.; Song, C. Correlation between TMB and MSI in patients with solid tumors. J. Clin. Oncol. 2020, 38, e15169. [Google Scholar] [CrossRef]
- Diaz, L.A.; Marabelle, A.; Delord, J.-P.; Shapira-Frommer, R.; Geva, R.; Peled, N.; Kim, T.W.; Andre, T.; Van Cutsem, E.; Guimbaud, R.; et al. Pembrolizumab therapy for microsatellite instability high (MSI-H) colorectal cancer (CRC) and non-CRC. J. Clin. Oncol. 2017, 35, 3071. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Schiewer, M.J.; Goodwin, J.F.; Han, S.; Brenner, J.C.; Augello, M.A.; Dean, J.L.; Liu, F.; Planck, J.L.; Ravindranathan, P.; Chinnaiyan, A.M.; et al. Dual Roles of PARP-1 Promote Cancer Growth and Progression. Cancer Discov. 2012, 2, 1134–1149. [Google Scholar] [CrossRef]
- Byers, L.A.; Wang, J.; Nilsson, M.B.; Fujimoto, J.; Saintigny, P.; Yordy, J.; Giri, U.; Peyton, M.; Fan, Y.H.; Diao, L.; et al. Proteomic Profiling Identifies Dysregulated Pathways in Small Cell Lung Cancer and Novel Therapeutic Targets Including PARP. Cancer Discov. 2012, 2, 798–811. [Google Scholar] [CrossRef]
- Mazzotta, A.; Partipilo, G.; De Summa, S.; Giotta, F.; Simone, G.; Mangia, A. Nuclear PARP1 expression and its prognostic significance in breast cancer patients. Tumor Biol. 2015, 37, 6143–6153. [Google Scholar] [CrossRef]
- Liang, H.; Tan, A.R. Iniparib, a PARP1 inhibitor for the potential treatment of cancer, including triple-negative breast cancer. IDrugs Investig. Drugs J. 2010, 13, 646–656. [Google Scholar]
- Lazaro-Camp, V.J.; Salari, K.; Meng, X.; Yang, S. SETDB1 in cancer: Overexpression and its therapeutic implications. Am. J. Cancer Res. 2021, 11, 1803–1827. [Google Scholar] [PubMed]
- Fei, Q.; Shang, K.; Zhang, J.; Chuai, S.; Kong, D.; Zhou, T.; Fu, S.; Liang, Y.; Li, C.; Chen, Z.; et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of P53. Nat. Commun. 2015, 6, 8651. [Google Scholar] [CrossRef]
- Sun, Q.-Y.; Ding, L.-W.; Xiao, J.-F.; Chien, W.; Lim, S.-L.; Hattori, N.; Goodglick, L.; Chia, D.; Mah, V.; Alavi, M.; et al. SETDB1 accelerates tumourigenesis by regulating the WNT signalling pathway. J. Pathol. 2014, 235, 559–570. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor–immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Severson, E.; Pignon, J.-C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eid, R.A.; Soltan, M.A.; Eldeen, M.A.; Shati, A.A.; Dawood, S.A.; Eissa, M.; Zaki, M.S.A.; Algahtani, M.; Theyab, A.; Abdel-Daim, M.M.; et al. Assessment of RACGAP1 as a Prognostic and Immunological Biomarker in Multiple Human Tumors: A Multiomics Analysis. Int. J. Mol. Sci. 2022, 23, 14102. https://doi.org/10.3390/ijms232214102
Eid RA, Soltan MA, Eldeen MA, Shati AA, Dawood SA, Eissa M, Zaki MSA, Algahtani M, Theyab A, Abdel-Daim MM, et al. Assessment of RACGAP1 as a Prognostic and Immunological Biomarker in Multiple Human Tumors: A Multiomics Analysis. International Journal of Molecular Sciences. 2022; 23(22):14102. https://doi.org/10.3390/ijms232214102
Chicago/Turabian StyleEid, Refaat A., Mohamed A. Soltan, Muhammad Alaa Eldeen, Ayed A. Shati, Samy A. Dawood, Mohamed Eissa, Mohamed Samir A. Zaki, Mohammad Algahtani, Abdulrahman Theyab, Mohamed M. Abdel-Daim, and et al. 2022. "Assessment of RACGAP1 as a Prognostic and Immunological Biomarker in Multiple Human Tumors: A Multiomics Analysis" International Journal of Molecular Sciences 23, no. 22: 14102. https://doi.org/10.3390/ijms232214102
APA StyleEid, R. A., Soltan, M. A., Eldeen, M. A., Shati, A. A., Dawood, S. A., Eissa, M., Zaki, M. S. A., Algahtani, M., Theyab, A., Abdel-Daim, M. M., & Kim, B. (2022). Assessment of RACGAP1 as a Prognostic and Immunological Biomarker in Multiple Human Tumors: A Multiomics Analysis. International Journal of Molecular Sciences, 23(22), 14102. https://doi.org/10.3390/ijms232214102






