A R2R3 MYB Transcription Factor, TaMYB391, Is Positively Involved in Wheat Resistance to Puccinia striiformis f. sp. tritici
Abstract
1. Introduction
2. Results
2.1. Identification and Sequence Analysis of TaMYB391
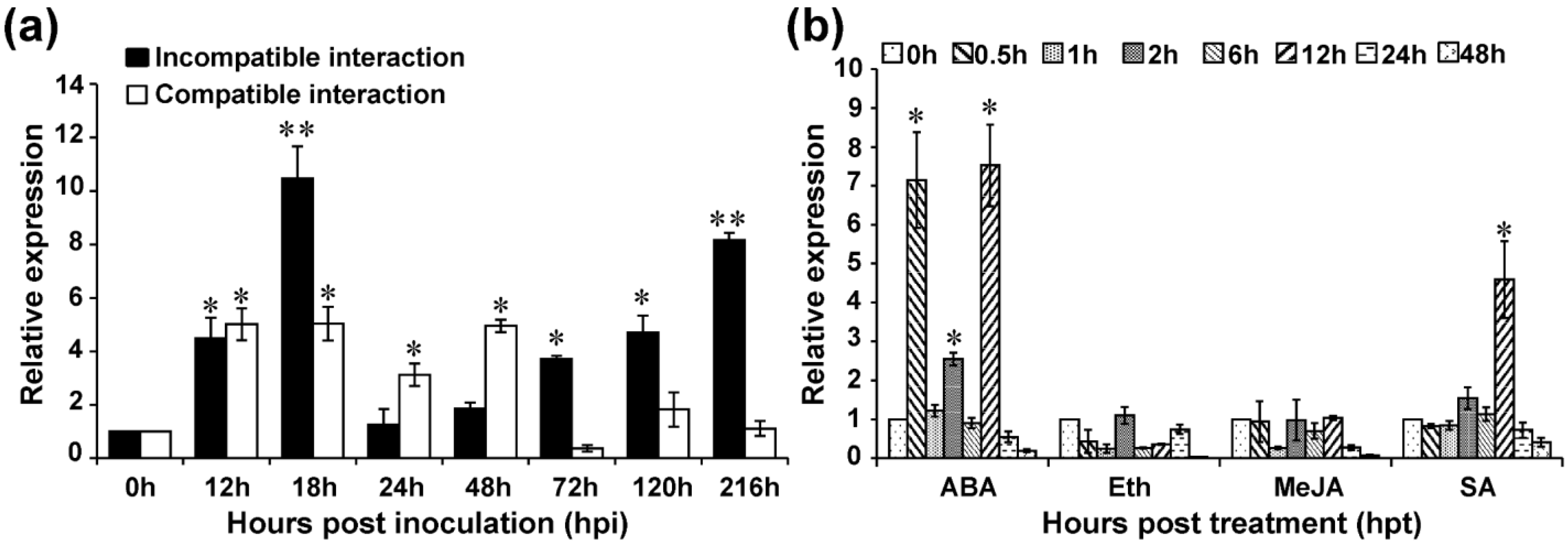
2.2. Transcriptional Responses of TaMYB391 to Pst
2.3. Transcriptional Response of TaMYB391 to Exogeneous Hormones
2.4. TaMYB391 Is Localized in the Nucleus
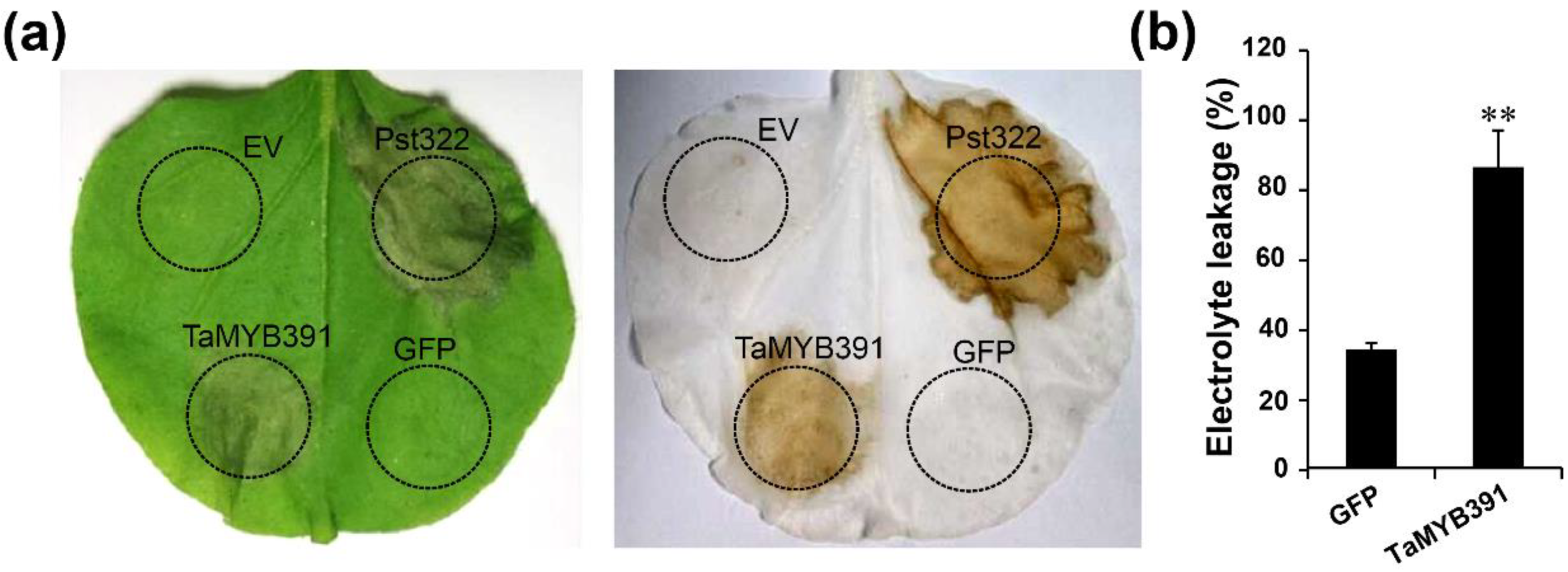
2.5. Transient Overexpression of TaMYB391 in N. benthamiana Triggered HR-Related Programmed Cell Death
2.6. TaMYB391 Activates Plant Immunity Responses
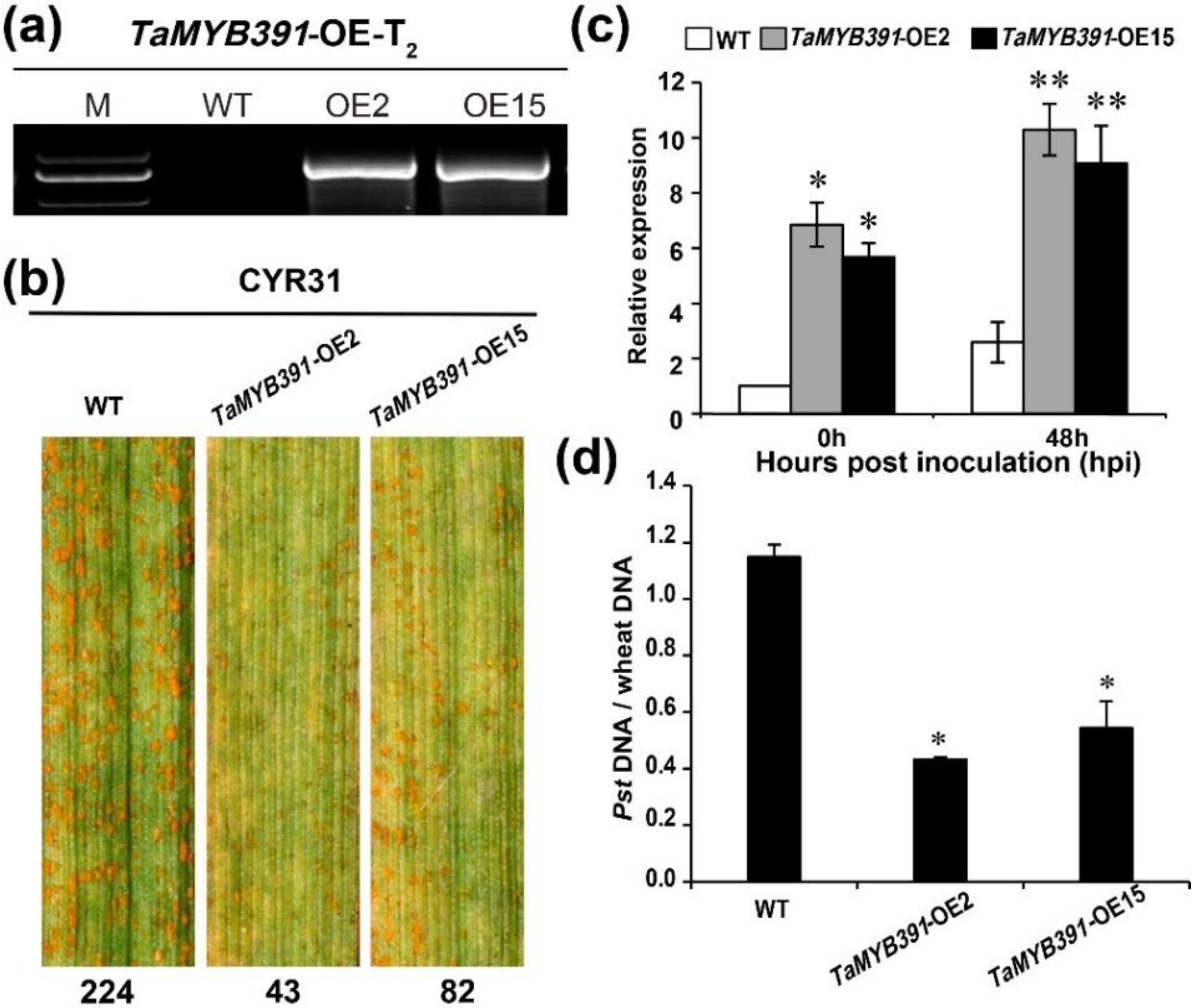
2.7. Overexpression of TaMYB391 in Wheat Enhances Wheat Resistance to Pst Infection
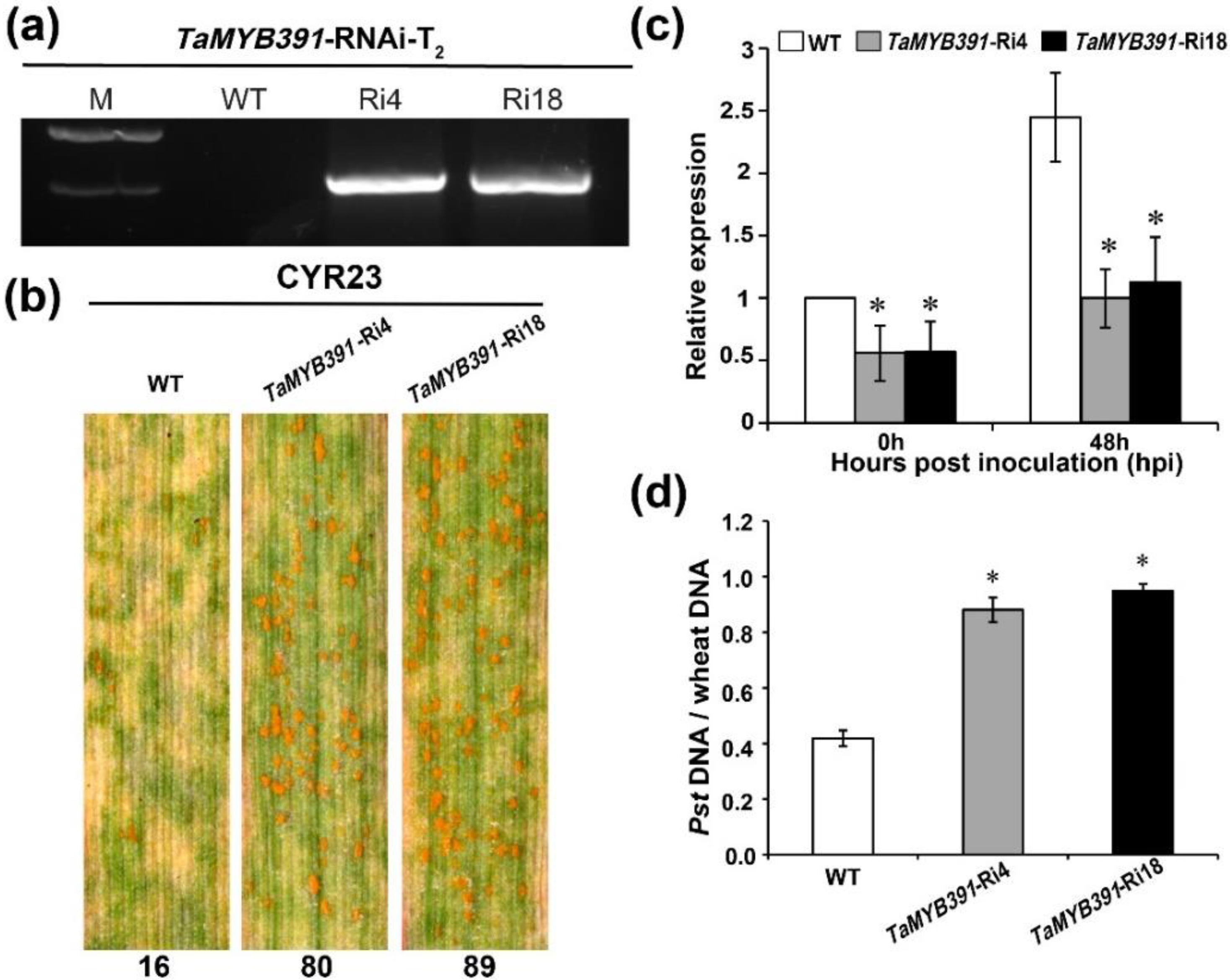
2.8. RNAi-Mediated Silencing of TaMYB391 Impairs Wheat Resistance to Pst Infection
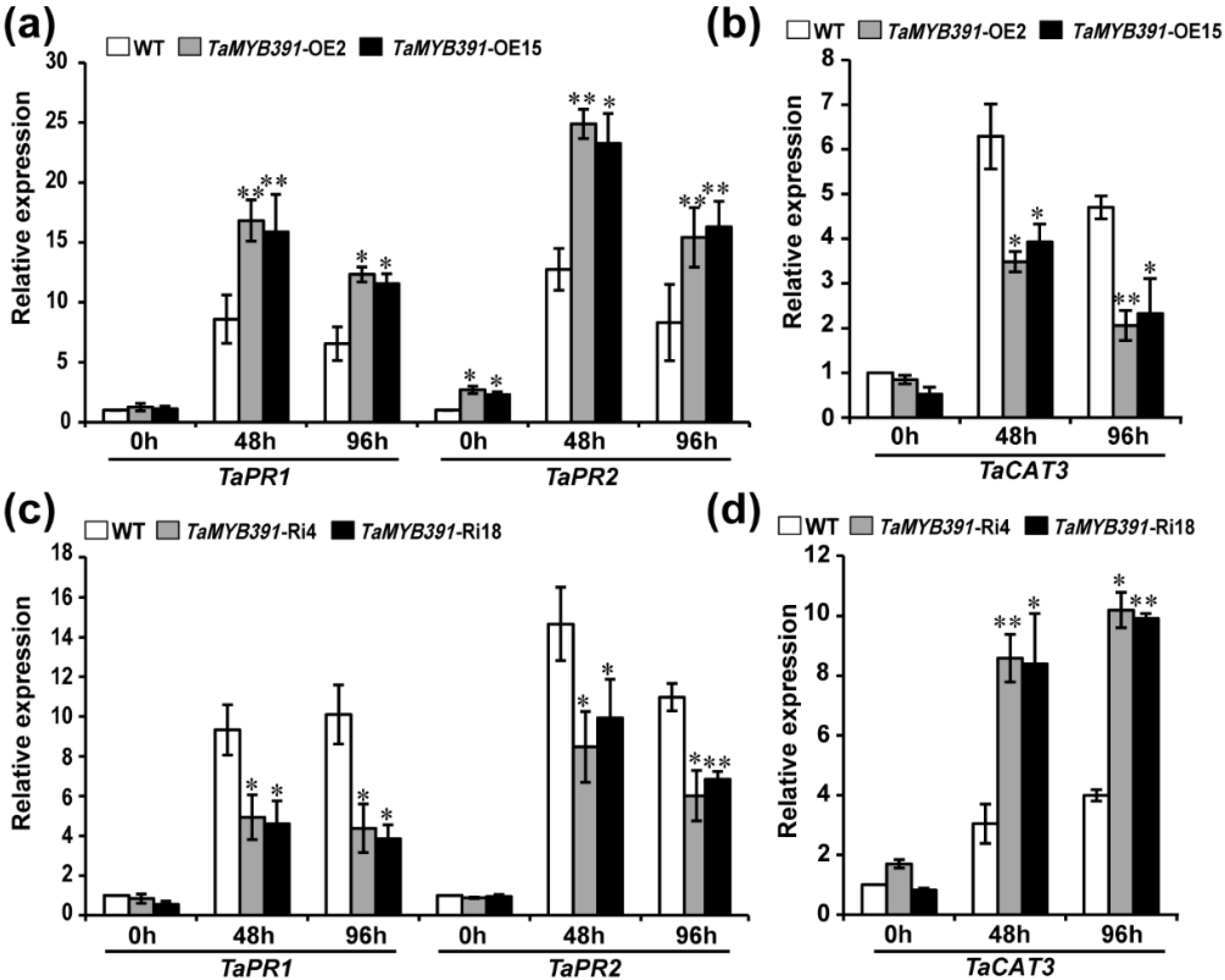
2.9. TaMYB391 Positively Regulates the Expression of Pathogenesis-Related (PR) and ROS-Scavenging Genes during Pst Infection
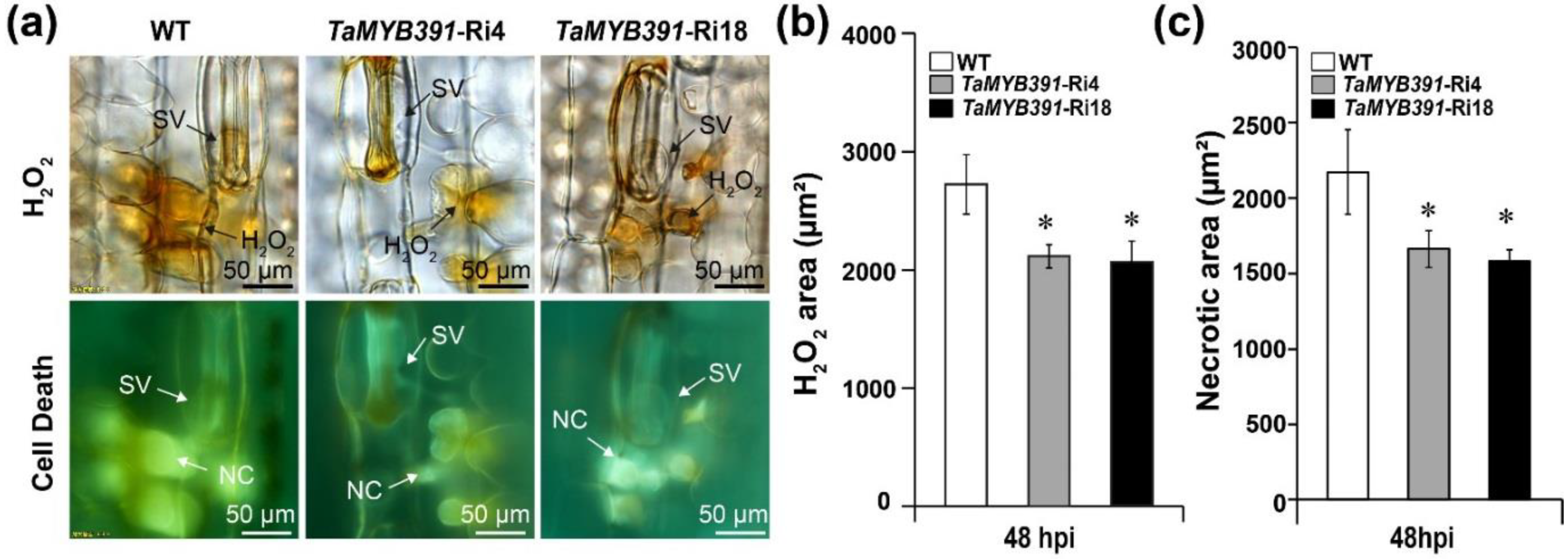
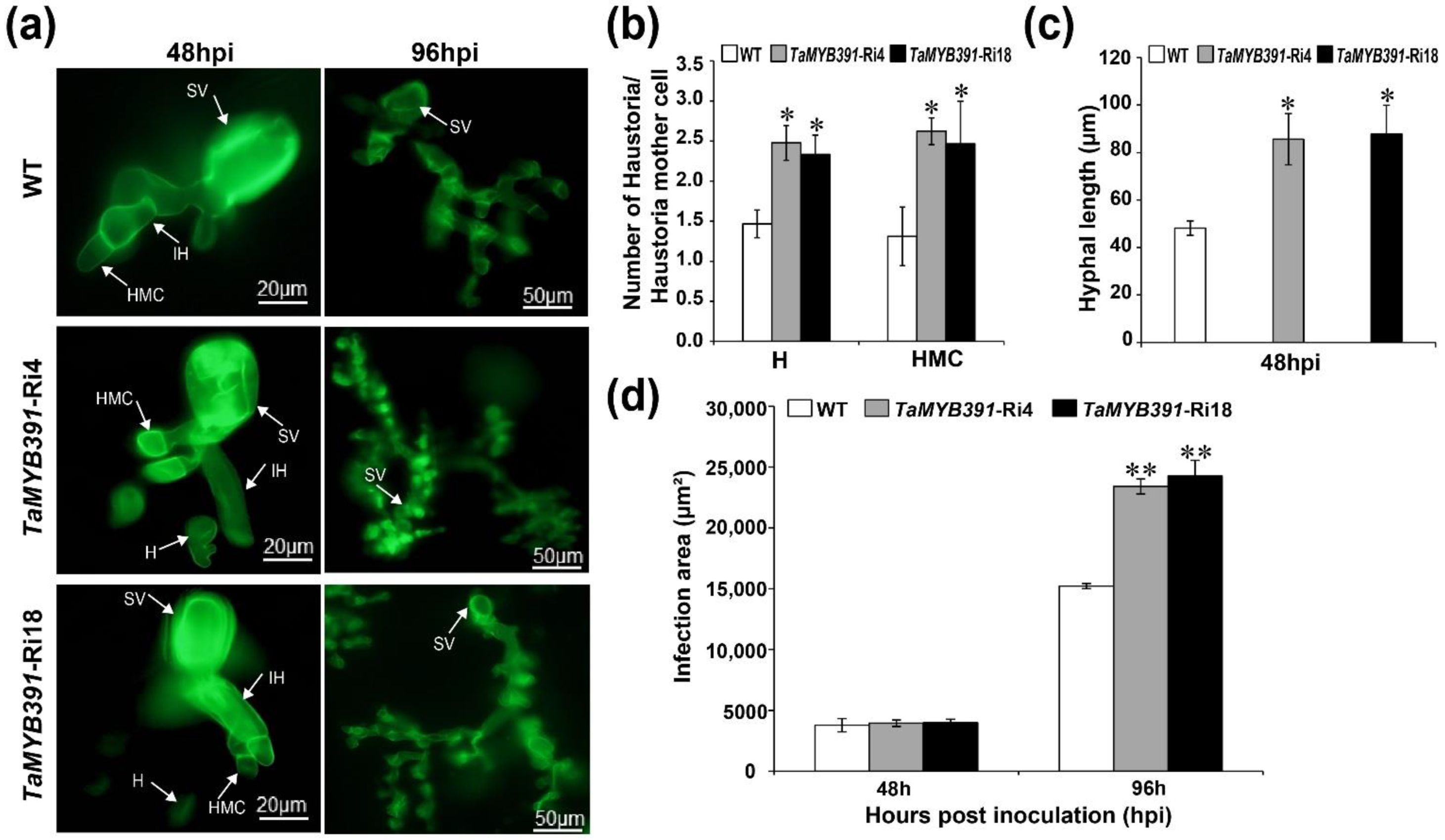
2.10. Pst Growth and H2O2 Accumulation Were Significantly Affected in TaMYB391-Overexpressing and TaMYB391-RNAi Transgenic Lines
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Fungal Isolates and Inoculation/Treatments
4.2. RNA Extraction, cDNA Synthesis and Gene Expression Analysis
4.3. Identification, Cloning and Sequence Analysis of TaMYB391
4.4. Subcellular Localization of TaMYB391 in Wheat and N. benthamiana
4.5. Transient Overexpression of TaMYB391 in N. benthamiana
4.6. Measurement of Electrolyte Leakage
4.7. Generation of Transgenic Lines
4.8. H2O2 Accumulation and Histological Observation of Fungal Growth
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X. Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Secur. 2020, 12, 239–251. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Yang, C.; Dolatabadian, A.; Fernando, W.D. The wonderful world of intrinsic and intricate immunity responses in plants against pathogens. Can. J. Plant Pathol. 2022, 44, 1–20. [Google Scholar] [CrossRef]
- Ng, D.W.K.; Abeysinghe, J.K.; Kamali, M. Regulatingthe regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef]
- Bai, X.; Zhan, G.; Tian, S.; Peng, H.; Cui, X.; Islam, M.A.; Goher, F.; Ma, Y.; Kang, Z.; Xu, Z.S. Transcription factor BZR2 activates chitinase Cht20.2 transcription to confer resistance to wheat stripe rust. Plant Physiol. 2021, 187, 2749–2762. [Google Scholar] [CrossRef]
- Qi, T.; Guo, J.; Liu, P.; He, F.; Wan, C.; Islam, M.A.; Tyler, B.M.; Kang, Z.; Guo, J. Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 2019, 12, 1624–1638. [Google Scholar] [CrossRef]
- Wang, F.; Lin, R.; Li, Y.; Wang, P.; Feng, J.; Chen, W.; Xu, S. TabZIP74 acts as a positive regulator in wheat stripe rust resistance and involves root development by mRNA splicing. Front. Plant. Sci. 2019, 10, 1551. [Google Scholar] [CrossRef]
- Hawku, M.D.; Goher, F.; Islam, M.A.; Guo, J.; He, F.; Bai, X.; Yuan, P.; Kang, Z.; Guo, J. TaAP2-15, an AP2/ERF transcription factor, is positively involved in wheat resistance to Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2021, 22, 2080. [Google Scholar] [CrossRef]
- Zhu, C.; Li, Z.; Tang, Y.; Zhang, L.; Wen, J.; Wang, Z.; Su, Y.; Zhang, H. TaWRKY10 plays a key role in the upstream of circadian gene TaLHY in wheat. Plant Sci. 2021, 310, 110973. [Google Scholar] [CrossRef]
- Wang, N.; Tang, C.; Fan, X.; He, M.; Gan, P.; Zhang, S.; Hu, Z.; Wang, X.; Yan, T.; Shu, W. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 2022, 185, 2961–2974. [Google Scholar] [CrossRef]
- Wang, J.; Tao, F.; Tian, W.; Guo, Z.; Chen, X.; Xu, X.; Shang, H.; Hu, X. The wheat WRKY transcription factors TaWRKY49 and TaWRKY62 confer differential high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. PLoS ONE 2017, 12, e0181963. [Google Scholar] [CrossRef]
- Wang, J.; Tao, F.; An, F.; Zou, Y.; Tian, W.; Chen, X.; Xu, X.; Hu, X. Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2017, 18, 649–661. [Google Scholar] [CrossRef]
- Ma, R.; Liu, B.; Geng, X.; Ding, X.; Yan, N.; Sun, X.; Wang, W.; Sun, X.; Zheng, C. Biological function and stress response mechanism of MYB transcription factor family genes. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Wei, Q.; Chen, R.; Wei, X.; Liu, Y.; Zhao, S.; Yin, X.; Xie, T. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genom. 2020, 21, 792. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef]
- Kim, S.H.; Lam, P.Y.; Lee, M.-H.; Jeon, H.S.; Tobimatsu, Y.; Park, O.K. The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity. Front. Plant Sci. 2020, 11, 583153. [Google Scholar] [CrossRef]
- Wang, L.; Ran, L.; Hou, Y.; Tian, Q.; Li, C.; Liu, R.; Fan, D.; Luo, K. The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytol. 2017, 215, 351–367. [Google Scholar] [CrossRef]
- Ren, H.; Bai, M.; Sun, J.; Liu, J.; Ren, M.; Dong, Y.; Wang, N.; Ning, G.; Wang, C. RcMYB84 and RcMYB123 mediate jasmonate-induced defense responses against Botrytis cinerea in rose (Rosa chinensis). Plant J. 2020, 103, 1839–1849. [Google Scholar] [CrossRef]
- Liu, X.; Yang, L.; Zhou, X.; Zhou, M.; Lu, Y.; Ma, L.; Ma, H.; Zhang, Z. Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. J. Exp. Bot. 2013, 64, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Dong, L.; Han, X.; Jin, H.; Yin, C.; Han, Y.; Li, B.; Qin, H.; Zhang, J.; Shen, Q. The TuMYB46L-TuACO3 module regulates ethylene biosynthesis in einkorn wheat defense to powdery mildew. New Phytol. 2020, 225, 2526–2541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, X.; Wang, X.; Zhou, M.; Zhou, X.; Ye, X.; Wei, X. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 2012, 196, 1155–1170. [Google Scholar] [CrossRef]
- Wei, X.; Shan, T.; Hong, Y.; Xu, H.; Liu, X.; Zhang, Z. TaPIMP2, a pathogen-induced MYB protein in wheat, contributes to host resistance to common root rot caused by Bipolaris sorokinian. Sci. Rep. 2017, 7, 1754. [Google Scholar] [CrossRef]
- Shan, T.; Rong, W.; Xu, H.; Du, L.; Liu, X.; Zhang, Z. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci. Rep. 2016, 6, 28777. [Google Scholar] [CrossRef]
- Al-Attala, M.N.; Wang, X.; Abou-Attia, M.A.; Duan, X.; Kang, Z. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp tritici and abiotic stresses. Plant Mol. Biol. 2014, 84, 589–603. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, J.; Su, Y.; Liu, H.; Chen, Y.; Luo, P.; Du, X.; Wang, D.; Zhang, H. TaLHY, a 1R-MYB transcription factor, plays an important role in disease resistance against stripe rust fungus and ear heading in wheat. PLoS ONE 2015, 10, e0127723. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; He, Q.; Guo, D.; Liu, C.; Cao, J.; Wu, Z.; Kang, Z.; Wang, X. TaMYB29: A novel R2R3-MYB transcription factor involved in wheat defense against stripe rust. Front. Plant Sci. 2021, 12, 783388. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic acid: Biosynthesis and signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef] [PubMed]
- Noutoshi, Y.; Okazaki, M.; Kida, T.; Nishina, Y.; Morishita, Y.; Ogawa, T.; Suzuki, H.; Shibata, D.; Jikumaru, Y.; Hanada, A. Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 2012, 24, 3795–3804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.-J.; Zhang, K. S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Lukan, T.; Coll, A. Intertwined roles of reactive oxygen species and salicylic acid signaling are crucial for the plant response to biotic stress. Int. J. Mol. Sci. 2022, 23, 5568. [Google Scholar] [CrossRef]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef]
- Cao, Z.; Jing, J.; Wang, M.; Shang, H.; Li, Z. Relation analysis of stripe rust resistance gene in wheat important cultivar suwon 11, suwon 92 and hybrid 46. Acta Bot. Boreali-Occident. Sin. 2003, 23, 64–68. [Google Scholar]
- Tronchet, M.; Ranty, B.; Marco, Y.; Roby, D. HSR203 antisense suppression in tobacco accelerates development of hypersensitive cell death. Plant J. 2001, 27, 115–127. [Google Scholar] [CrossRef]
- Takahashi, Y.; Berberich, T.; Yamashita, K.; Uehara, Y.; Miyazaki, A.; Kusano, T. Identification of tobacco HIN1 and two closely related genes as spermine-responsive genes and their differential expression during the Tobacco mosaic virus-induced hypersensitive response and during leaf- and flower-senescence. Plant Mol. Biol. 2004, 54, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.D.; Goodwin, P.H.; Hsiang, T. Induction of glutathione S-transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. J. Exp. Bot. 2005, 56, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bi, W.; Jing, G.; Yu, X.; Wang, H.; Liu, D. Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agric. 2018, 17, 2468–2477. [Google Scholar] [CrossRef]
- Zhang, J.; Du, X.; Wang, Q.; Chen, X.; Lv, D.; Xu, K.; Qu, S.; Zhang, Z. Expression of pathogenesis related genes in response to salicylic acid, methyl jasmonate and 1-aminocyclopropane-1-carboxylic acid in Malus hupehensis (Pamp.) Rehd. BMC Res. Notes 2010, 3, 208. [Google Scholar] [CrossRef]
- Froidure, S.; Canonne, J.; Daniel, X.; Jauneau, A.; Briere, C.; Roby, D.; Rivas, S. AtsPLA(2)-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc. Natl. Acad. Sci. USA 2018, 115, E3326–E3328. [Google Scholar]
- Vailleau, F.; Daniel, X.; Tronchet, M.; Montillet, J.L.; Triantaphylides, C.; Roby, D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 2002, 99, 10179–10184. [Google Scholar] [CrossRef]
- Zhou, J.M.; Loh, Y.T.; Bressan, R.A.; Martin, G.B. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 1995, 83, 925–935. [Google Scholar] [CrossRef]
- Shigenaga, A.M.; Berens, M.L.; Tsuda, K.; Argueso, C.T. Towards engineering of hormonal crosstalk in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.-M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 2003, 15, 2551–2565. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, K.; Chern, M.; Liu, Y.; Zhu, Z.; Liu, J.; Zhu, X.; Yin, J.; Ran, L.; Xiong, J.; et al. Sclerenchyma cell thickening through enhanced lignification induced by OsMYB30 prevents fungal penetration of rice leaves. New Phytol. 2020, 226, 1850–1863. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, J.; Luo, Q.; Yang, C.; Yang, H.; Cheng, Y. CsMYB96 enhances citrus fruit resistance against fungal pathogen by activating salicylic acid biosynthesis and facilitating defense metabolite accumulation. J. Plant Physiol. 2021, 264, 153472. [Google Scholar] [CrossRef] [PubMed]
- Hayta, S.; Smedley, M.A.; Demir, S.U.; Blundell, R.; Hinchliffe, A.; Atkinson, N.; Harwood, W.A. An efficient and reproducible Agrobacterium-mediated transformation method for hexaploid wheat (Triticum aestivum L.). Plant Methods 2019, 15, 152. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M. Challenges and solutions for stripe rust control in the United States. Aust. J. Agric. Res. 2007, 58, 648–655. [Google Scholar] [CrossRef]
- Wang, M.; Yuan, J.; Qin, L.; Shi, W.; Xia, G.; Liu, S. TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnol. J. 2020, 18, 791–804. [Google Scholar] [CrossRef]
- Wang, M.; Wang, M.; Zhao, M.; Wang, M.; Liu, S.; Tian, Y.; Moon, B.; Liang, C.; Li, C.; Shi, W.; et al. TaSRO1 plays a dual role in suppressing TaSIP1 to fine tune mitochondrial retrograde signalling and enhance salinity stress tolerance. New Phytol. 2022, 236, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Q.; Peng, L.; Tu, H.; Lee, L.Y.; Gelvin, S.B.; Pan, S.Q. Agrobacterium-delivered VirE2 interacts with host nucleoporin CG1 to facilitate the nuclear import of VirE2-coated T complex. Proc. Natl. Acad. Sci. USA 2020, 117, 26389–26397. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Guo, L.; Lang, X.; Li, M.; Wu, G.; Wu, R.; Wang, L.; Zhao, M.; Qing, L. Geminivirus C4 proteins inhibit GA signaling via prevention of NbGAI degradation, to promote viral infection and symptom development in N. benthamiana. PLoS Pathog. 2022, 18, e1010217. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Zhou, W.; Liu, J.; Tan, N.; Zhou, J.M.; Huang, L. A receptor-like protein from Nicotiana benthamiana mediates VmE02 PAMP-triggered immunity. New Phytol. 2021, 229, 2260–2272. [Google Scholar] [CrossRef]
- Liang, Z.; Zong, Y.; Gao, C. An efficient targeted mutagenesis system using CRISPR/Cas in Monocotyledons. Curr. Protoc. Plant Biol. 2016, 1, 329–344. [Google Scholar] [CrossRef]
- Bos, J.I.; Kanneganti, T.D.; Young, C.; Cakir, C.; Huitema, E.; Win, J.; Armstrong, M.R.; Birch, P.R.; Kamoun, S. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 2006, 48, 165–176. [Google Scholar] [CrossRef]
- Yu, X.; Tang, J.; Wang, Q.; Ye, W.; Tao, K.; Duan, S.; Lu, C.; Yang, X.; Dong, S.; Zheng, X. The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 2012, 196, 247–260. [Google Scholar] [CrossRef]
- Du, Q.; Wang, K.; Zou, C.; Xu, C.; Li, W.-X. The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol. 2018, 177, 1743–1753. [Google Scholar] [CrossRef]
- Hao, Q.; Wang, W.; Han, X.; Wu, J.; Lyu, B.; Chen, F.; Caplan, A.; Li, C.; Wu, J.; Wang, W.; et al. Isochorismate-based salicylic acid biosynthesis confers basal resistance to Fusarium graminearum in barley. Mol. Plant Pathol. 2018, 19, 1995–2010. [Google Scholar] [CrossRef]
- Wang, C.F.; Huang, L.L.; Buchenauer, H.; Han, Q.M.; Zhang, H.C.; Kang, Z.S. Histochemical studies on the accumulation of reactive oxygen species (O2− and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 2007, 71, 230–239. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hawku, M.D.; He, F.; Bai, X.; Islam, M.A.; Huang, X.; Kang, Z.; Guo, J. A R2R3 MYB Transcription Factor, TaMYB391, Is Positively Involved in Wheat Resistance to Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2022, 23, 14070. https://doi.org/10.3390/ijms232214070
Hawku MD, He F, Bai X, Islam MA, Huang X, Kang Z, Guo J. A R2R3 MYB Transcription Factor, TaMYB391, Is Positively Involved in Wheat Resistance to Puccinia striiformis f. sp. tritici. International Journal of Molecular Sciences. 2022; 23(22):14070. https://doi.org/10.3390/ijms232214070
Chicago/Turabian StyleHawku, Mehari Desta, Fuxin He, Xingxuan Bai, Md Ashraful Islam, Xueling Huang, Zhensheng Kang, and Jun Guo. 2022. "A R2R3 MYB Transcription Factor, TaMYB391, Is Positively Involved in Wheat Resistance to Puccinia striiformis f. sp. tritici" International Journal of Molecular Sciences 23, no. 22: 14070. https://doi.org/10.3390/ijms232214070
APA StyleHawku, M. D., He, F., Bai, X., Islam, M. A., Huang, X., Kang, Z., & Guo, J. (2022). A R2R3 MYB Transcription Factor, TaMYB391, Is Positively Involved in Wheat Resistance to Puccinia striiformis f. sp. tritici. International Journal of Molecular Sciences, 23(22), 14070. https://doi.org/10.3390/ijms232214070








