The SARS-CoV-2 Delta-Omicron Recombinant Lineage (XD) Exhibits Immune-Escape Properties Similar to the Omicron (BA.1) Variant
Abstract
1. Introduction
2. Results
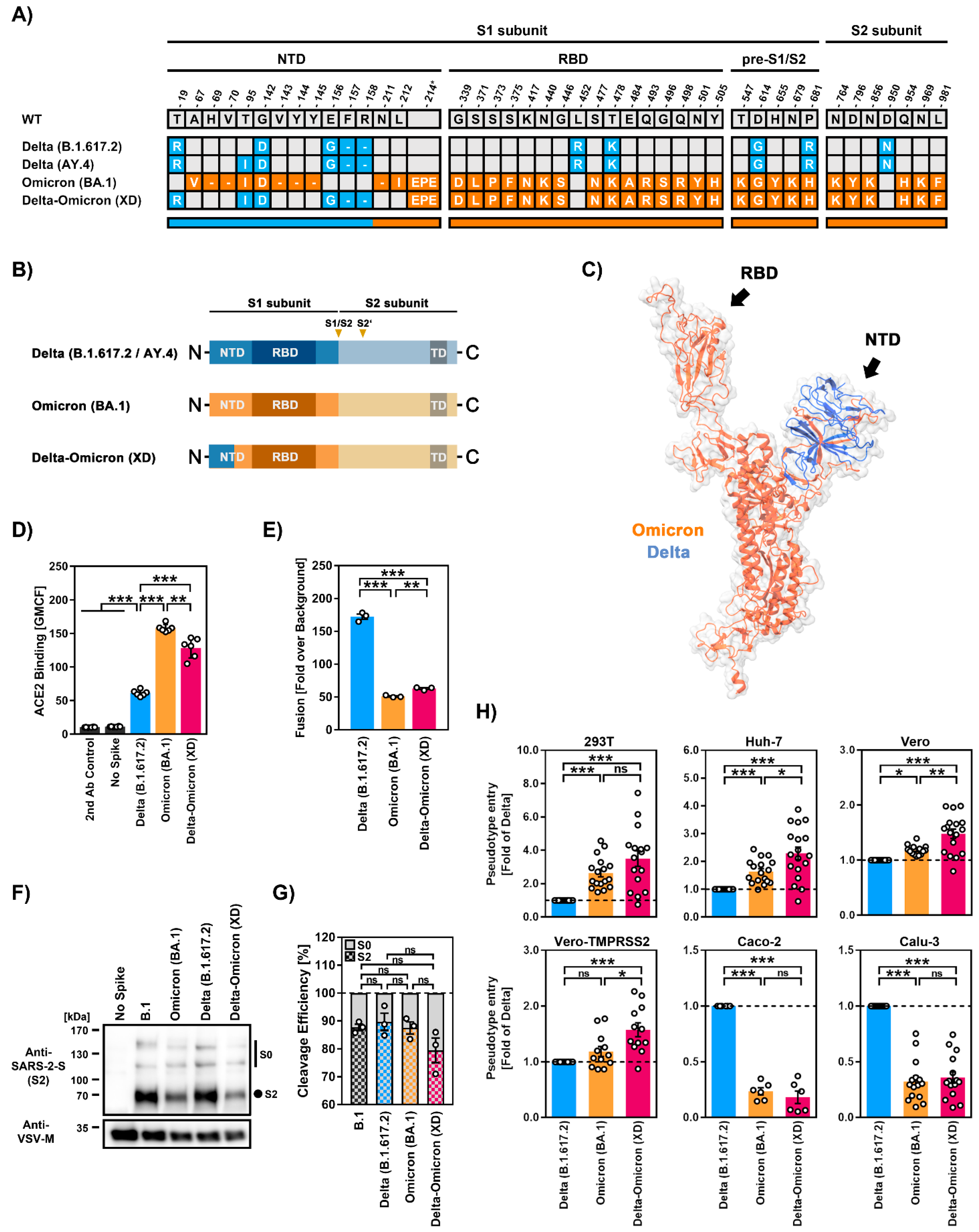
2.1. The S Protein of the Recombinant SARS-CoV-2 Lineage XD Efficiently Binds ACE2 but Has Low Capacity to Drive Cell–Cell Fusion
2.2. Similar Cell Line Tropism by SARS-CoV-2 XD and BA.1 (Omicron Variant) Lineages
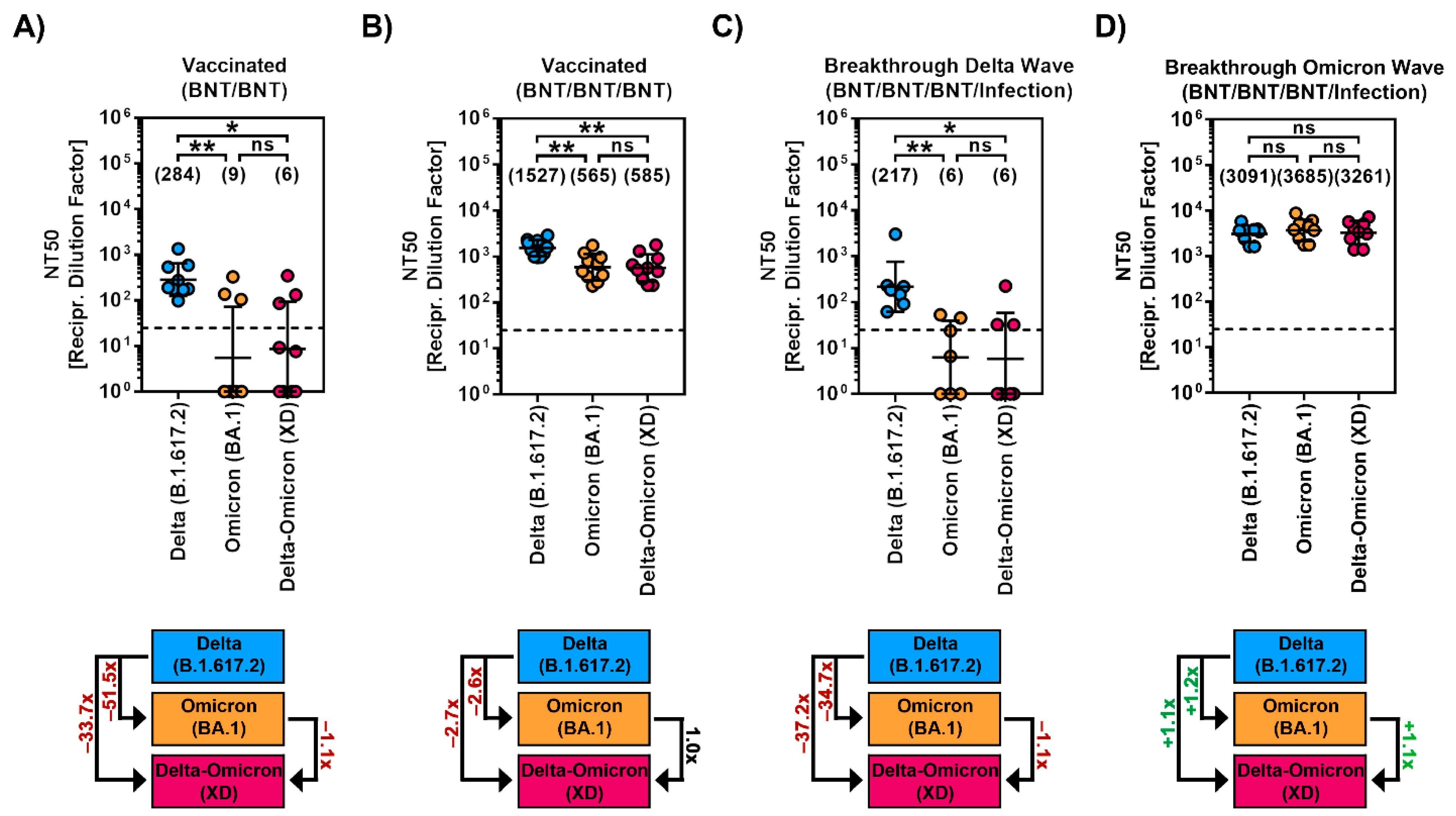
2.3. Diminished Sensitivity to Antibody-Mediated Neutralization by the Recombinant SARS-CoV-2 XD Lineage
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Expression Plasmids
4.3. Production of Soluble ACE2
4.4. ACE2 Binding
4.5. Production of VSV Pseudotypes
4.6. Immunoblot
4.7. Transduction of Target Cells
4.8. Neutralization Assay
4.9. Cell–Cell Fusion Assay
4.10. Protein Structures
4.11. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, F.; Tong, B.; Sun, L.; Shi, S.; Zheng, B.; Wang, Z.; Dong, X.; Zheng, P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. ELife 2021, 10, e69091. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Arora, P.; Gross, R.; Seidel, A.; Hornich, B.F.; Hahn, A.S.; Kruger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e12. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Consortium, C.-G.U.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022, 39, 110829. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Arora, P.; Zhang, L.; Rocha, C.; Sidarovich, A.; Kempf, A.; Schulz, S.; Cossmann, A.; Manger, B.; Baier, E.; Tampe, B.; et al. Comparable neutralisation evasion of SARS-CoV-2 omicron subvariants BA.1, BA.2, and BA.3. Lancet Infect. Dis. 2022, 22, 766–767. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; St Denis, K.J.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kruger, N.; Schulz, S.; Cossmann, A.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.S.; Winkler, M.S.; et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 2022, 185, 447–456.e11. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradnik, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef]
- Bager, P.; Wohlfahrt, J.; Bhatt, S.; Stegger, M.; Legarth, R.; Moller, C.H.; Skov, R.L.; Valentiner-Branth, P.; Voldstedlund, M.; Fischer, T.K.; et al. Risk of hospitalisation associated with infection with SARS-CoV-2 omicron variant versus delta variant in Denmark: An observational cohort study. Lancet Infect. Dis. 2022, 22, 967–976. [Google Scholar] [CrossRef]
- Menni, C.; Valdes, A.M.; Polidori, L.; Antonelli, M.; Penamakuri, S.; Nogal, A.; Louca, P.; May, A.; Figueiredo, J.C.; Hu, C.; et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: A prospective observational study from the ZOE COVID Study. Lancet 2022, 399, 1618–1624. [Google Scholar] [CrossRef]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef]
- Colson, P.; Fournier, P.E.; Delerce, J.; Million, M.; Bedotto, M.; Houhamdi, L.; Yahi, N.; Bayette, J.; Levasseur, A.; Fantini, J.; et al. Culture and identification of a “Deltamicron” SARS-CoV-2 in a three cases cluster in southern France. J. Med. Virol. 2022, 94, 3739–3749. [Google Scholar] [CrossRef] [PubMed]
- Moisan, A.; Mastrovito, B.; De Oliveira, F.; Martel, M.; Hedin, H.; Leoz, M.; Nesi, N.; Schaeffer, J.; Ar Gouilh, M.; Plantier, J.C. Evidence of transmission and circulation of Deltacron XD recombinant SARS-CoV-2 in Northwest France. Clin. Infect. Dis. 2022. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, H.Y.; Li, J.Y.; Cheng, Y.X.; Zhang, S.; Aliyari, S.; Wu, A.; Cheng, G. Potential intervariant and intravariant recombination of Delta and Omicron variants. J. Med. Virol. 2022, 94, 4830–4838. [Google Scholar] [CrossRef]
- Evans, J.P.; Qu, P.; Zeng, C.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Lozanski, G.; Mallampalli, R.K.; Saif, L.J.; Oltz, E.M.; et al. Neutralization of the SARS-CoV-2 Deltacron and BA.3 Variants. N. Engl. J. Med. 2022, 386, 2340–2342. [Google Scholar] [CrossRef]
- Kurhade, C.; Zou, J.; Xia, H.; Liu, M.; Yang, Q.; Cutler, M.; Cooper, D.; Muik, A.; Sahin, U.; Jansen, K.U.; et al. Neutralization of Omicron sublineages and Deltacron SARS-CoV-2 by three doses of BNT162b2 vaccine or BA.1 infection. Emerg. Microbes. Infect. 2022, 11, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Weisblum, Y.; Muecksch, F.; Hoffmann, H.H.; Michailidis, E.; Lorenzi, J.C.C.; Mendoza, P.; Rutkowska, M.; Bednarski, E.; Gaebler, C.; et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020, 217, e20201181. [Google Scholar] [CrossRef] [PubMed]
- Bussani, R.; Schneider, E.; Zentilin, L.; Collesi, C.; Ali, H.; Braga, L.; Volpe, M.C.; Colliva, A.; Zanconati, F.; Berlot, G.; et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine 2020, 61, 103104. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Arora, P.; Zhang, L.; Kruger, N.; Rocha, C.; Sidarovich, A.; Schulz, S.; Kempf, A.; Graichen, L.; Moldenhauer, A.S.; Cossmann, A.; et al. SARS-CoV-2 Omicron sublineages show comparable cell entry but differential neutralization by therapeutic antibodies. Cell Host Microbe 2022, 30, 1103–1111.e6. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Dong, Y.; Dai, T.; Wei, Y.; Zhang, L.; Zheng, M.; Zhou, F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020, 5, 237. [Google Scholar] [CrossRef]
- Arora, P.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Winkler, M.S.; Lier, M.; Schulz, S.; Jack, H.M.; Cossmann, A.; et al. The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell. Mol. Immunol. 2021, 18, 2673–2675. [Google Scholar] [CrossRef]
- Arora, P.; Sidarovich, A.; Kruger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.S.; Winkler, M.S.; Schulz, S.; Jack, H.M.; et al. B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021, 37, 109825. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Winkler, M.S.; Lier, M.; Schulz, S.; Jack, H.M.; Cossmann, A.; Stankov, M.V.; et al. SARS-CoV-2 variants C.1.2 and B.1.621 (Mu) partially evade neutralization by antibodies elicited upon infection or vaccination. Cell Rep. 2022, 39, 110754. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Schulz, S.R.; Cossmann, A.; Stankov, M.V.; Jack, H.M.; Behrens, G.M.N.; Pohlmann, S.; Hoffmann, M. Augmented neutralisation resistance of emerging omicron subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect. Dis. 2022, 22, 1117–1118. [Google Scholar] [CrossRef]
- Scheaffer, S.M.; Lee, D.; Whitener, B.; Ying, B.; Wu, K.; Liang, C.Y.; Jani, H.; Martin, P.; Amato, N.J.; Avena, L.E.; et al. Bivalent SARS-CoV-2 mRNA vaccines increase breadth of neutralization and protect against the BA.5 Omicron variant in mice. Nat. Med. 2022. [Google Scholar] [CrossRef]
- Callaway, E. COVID ‘variant soup’ is making winter surges hard to predict. Nature 2022, 611, 213–214. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Brinkmann, C.; Hoffmann, M.; Lubke, A.; Nehlmeier, I.; Kramer-Kuhl, A.; Winkler, M.; Pohlmann, S. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS ONE 2017, 12, e0189073. [Google Scholar] [CrossRef]
- Hoffmann, M.; Zhang, L.; Kruger, N.; Graichen, L.; Kleine-Weber, H.; Hofmann-Winkler, H.; Kempf, A.; Nessler, S.; Riggert, J.; Winkler, M.S.; et al. SARS-CoV-2 mutations acquired in mink reduce antibody-mediated neutralization. Cell Rep. 2021, 35, 109017. [Google Scholar] [CrossRef]
- Kleine-Weber, H.; Elzayat, M.T.; Wang, L.; Graham, B.S.; Muller, M.A.; Drosten, C.; Pohlmann, S.; Hoffmann, M. Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization. J. Virol. 2019, 93, e01381-18. [Google Scholar] [CrossRef]
- Berger Rentsch, M.; Zimmer, G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE 2011, 6, e25858. [Google Scholar] [CrossRef]
- Arora, P.; Nehlmeier, I.; Kempf, A.; Cossmann, A.; Schulz, S.R.; Dopfer-Jablonka, A.; Baier, E.; Tampe, B.; Moerer, O.; Dickel, S.; et al. Lung cell entry, cell-cell fusion capacity, and neutralisation sensitivity of omicron sublineage BA.2.75. Lancet Infect. Dis. 2022, 22, 1537–1538. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arora, P.; Zhang, L.; Rocha, C.; Graichen, L.; Nehlmeier, I.; Kempf, A.; Cossmann, A.; Ramos, G.M.; Baier, E.; Tampe, B.; et al. The SARS-CoV-2 Delta-Omicron Recombinant Lineage (XD) Exhibits Immune-Escape Properties Similar to the Omicron (BA.1) Variant. Int. J. Mol. Sci. 2022, 23, 14057. https://doi.org/10.3390/ijms232214057
Arora P, Zhang L, Rocha C, Graichen L, Nehlmeier I, Kempf A, Cossmann A, Ramos GM, Baier E, Tampe B, et al. The SARS-CoV-2 Delta-Omicron Recombinant Lineage (XD) Exhibits Immune-Escape Properties Similar to the Omicron (BA.1) Variant. International Journal of Molecular Sciences. 2022; 23(22):14057. https://doi.org/10.3390/ijms232214057
Chicago/Turabian StyleArora, Prerna, Lu Zhang, Cheila Rocha, Luise Graichen, Inga Nehlmeier, Amy Kempf, Anne Cossmann, Gema Morillas Ramos, Eva Baier, Björn Tampe, and et al. 2022. "The SARS-CoV-2 Delta-Omicron Recombinant Lineage (XD) Exhibits Immune-Escape Properties Similar to the Omicron (BA.1) Variant" International Journal of Molecular Sciences 23, no. 22: 14057. https://doi.org/10.3390/ijms232214057
APA StyleArora, P., Zhang, L., Rocha, C., Graichen, L., Nehlmeier, I., Kempf, A., Cossmann, A., Ramos, G. M., Baier, E., Tampe, B., Moerer, O., Dickel, S., Winkler, M. S., Behrens, G. M. N., Pöhlmann, S., & Hoffmann, M. (2022). The SARS-CoV-2 Delta-Omicron Recombinant Lineage (XD) Exhibits Immune-Escape Properties Similar to the Omicron (BA.1) Variant. International Journal of Molecular Sciences, 23(22), 14057. https://doi.org/10.3390/ijms232214057





