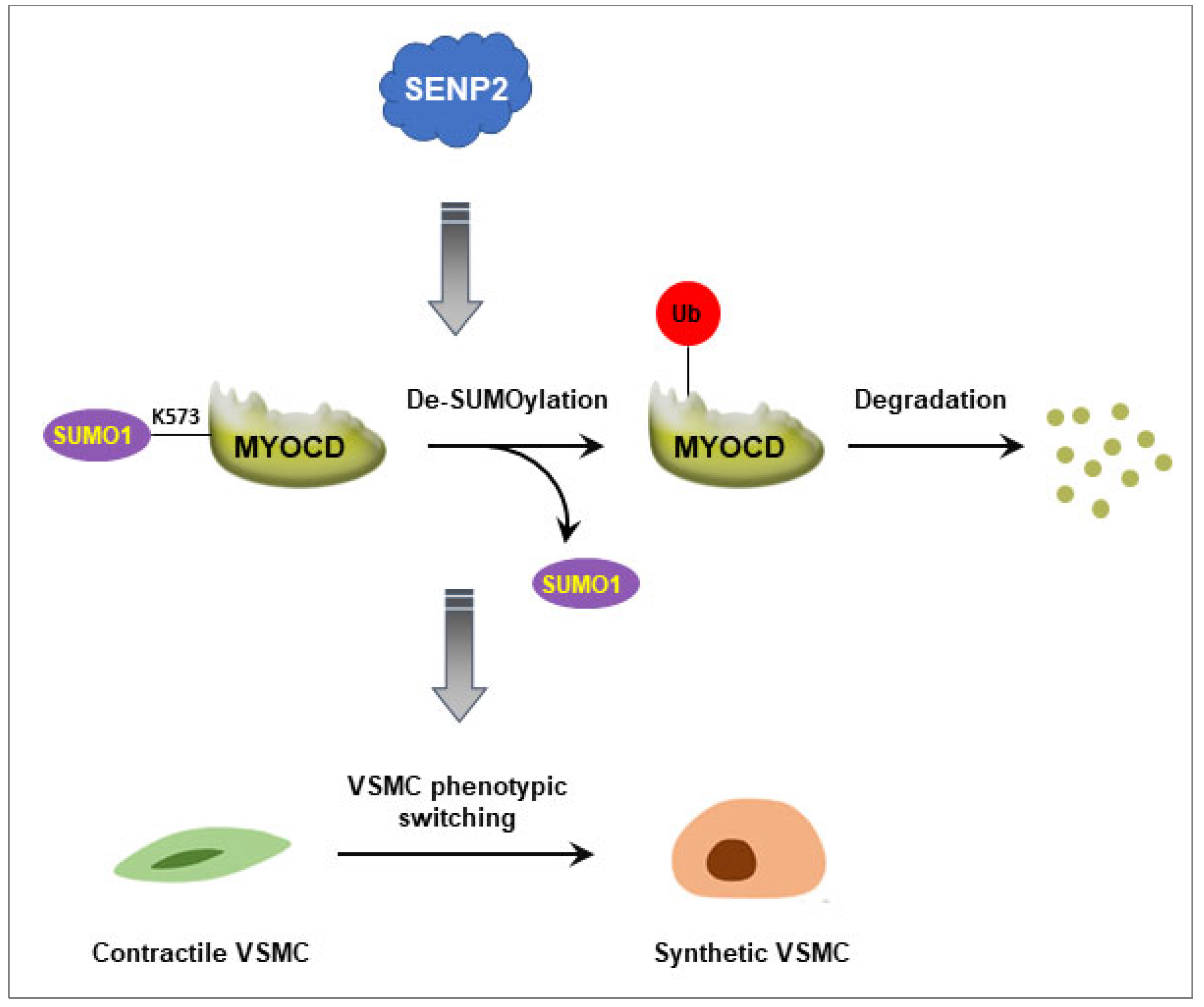
SENP2 Promotes VSMC Phenotypic Switching via Myocardin De-SUMOylation
Abstract
1. Introduction
2. Results
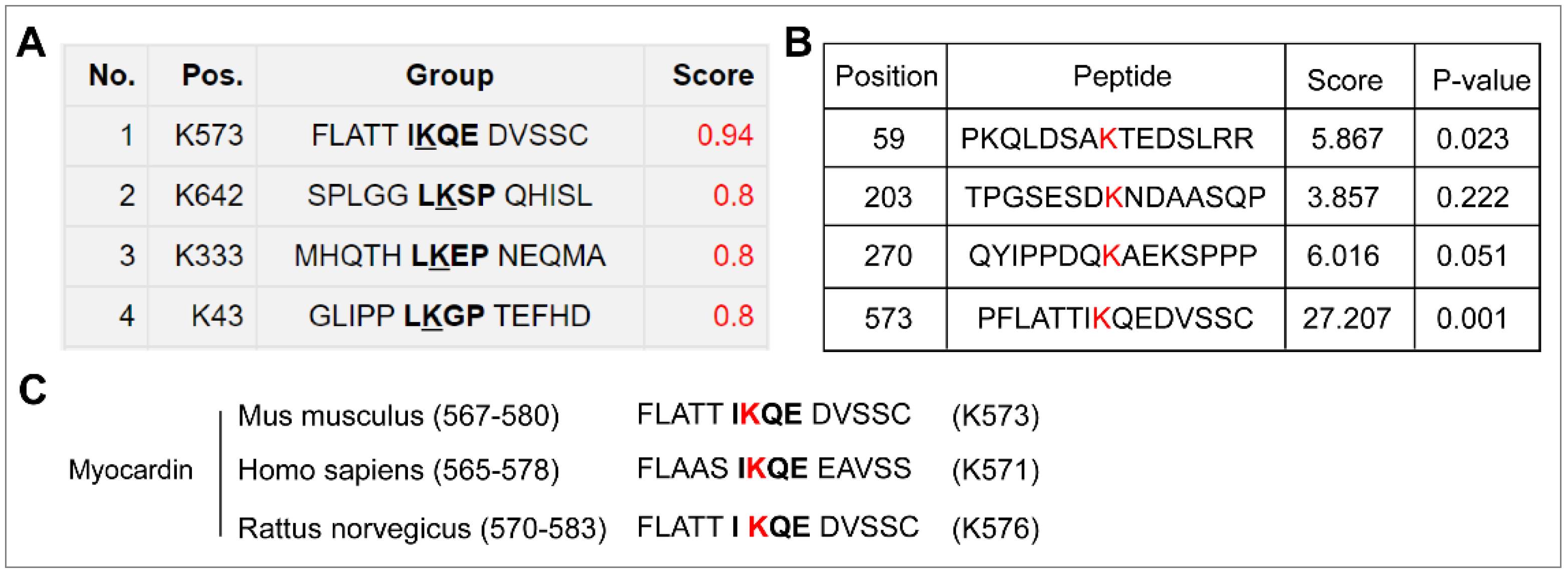
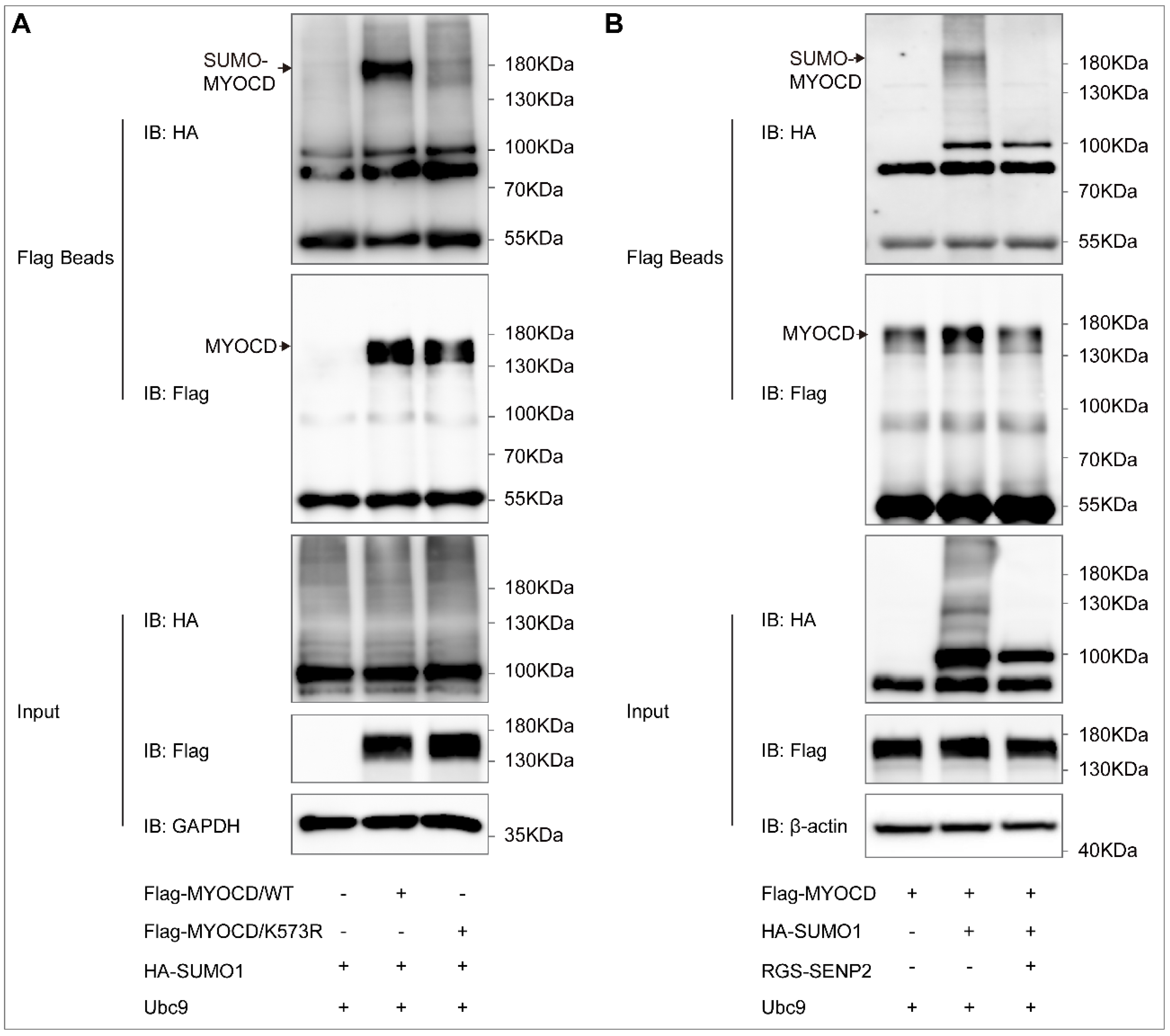
2.1. Myocardin Is SUMOylated at Lysine 573, Which Can Be De-SUMOylated by SENP2
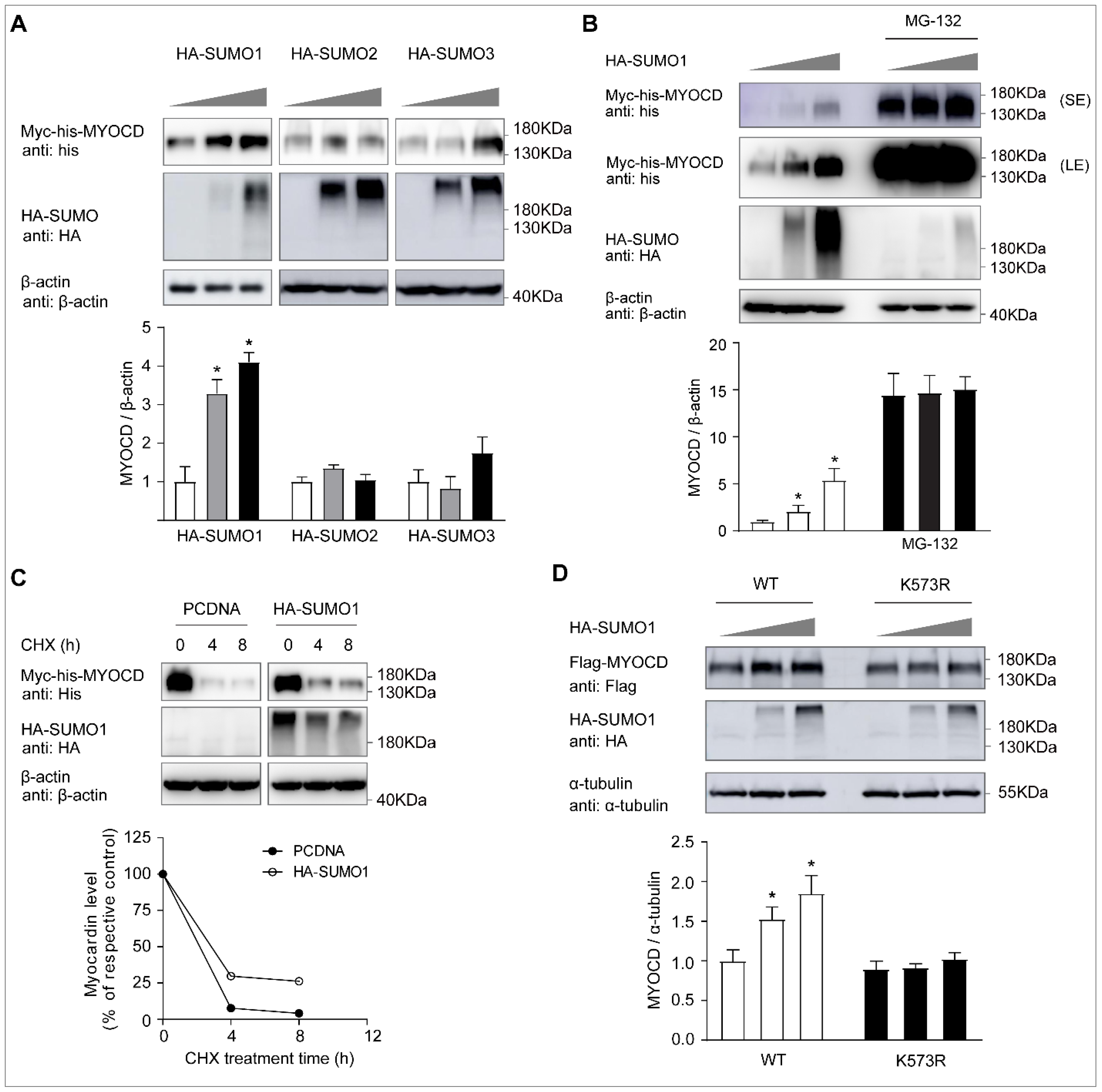
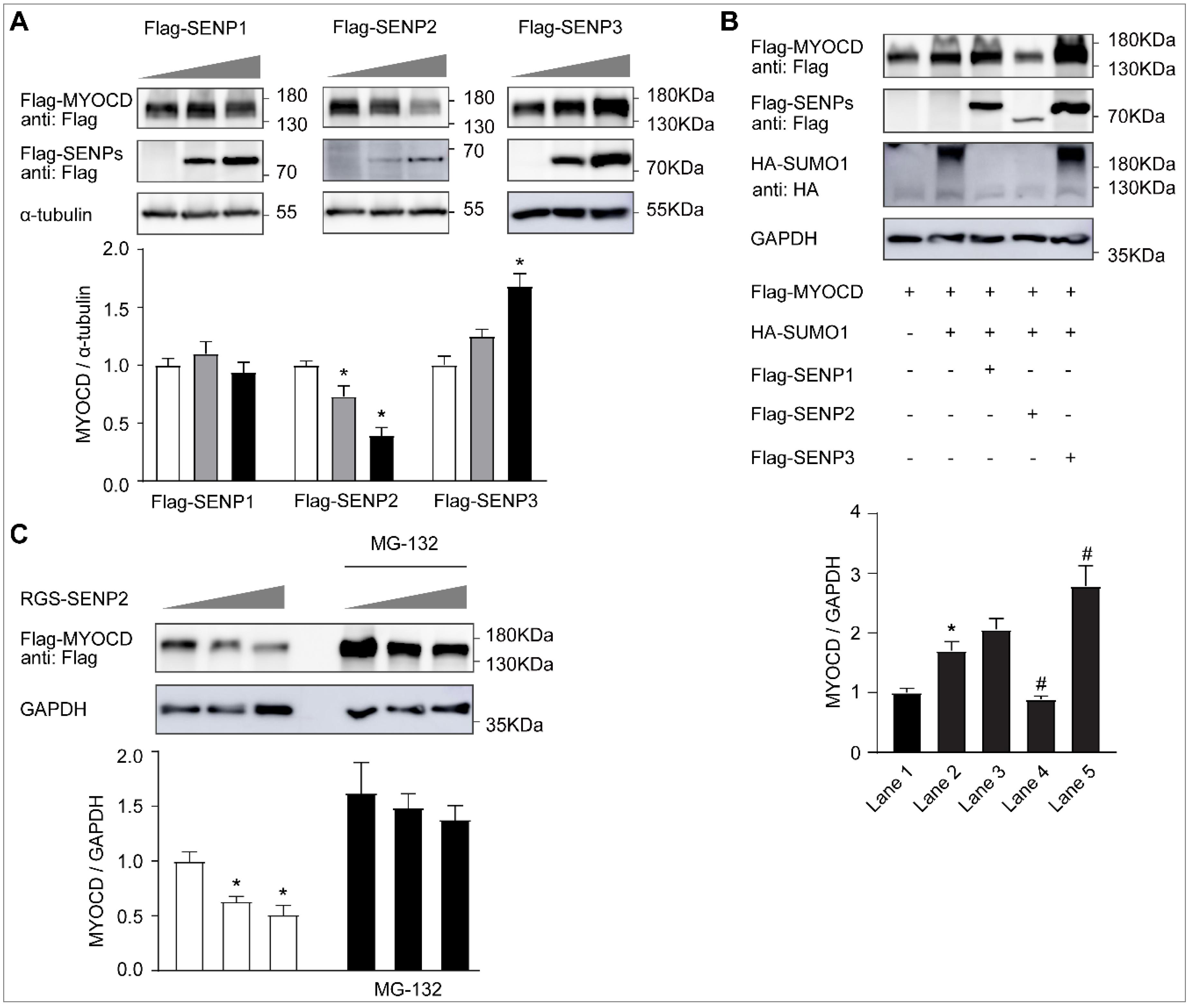
2.2. SUMO-1 Promotes Myocardin Stability, Whereas SENP2 Facilitates Its Proteasome-Dependent Degradation
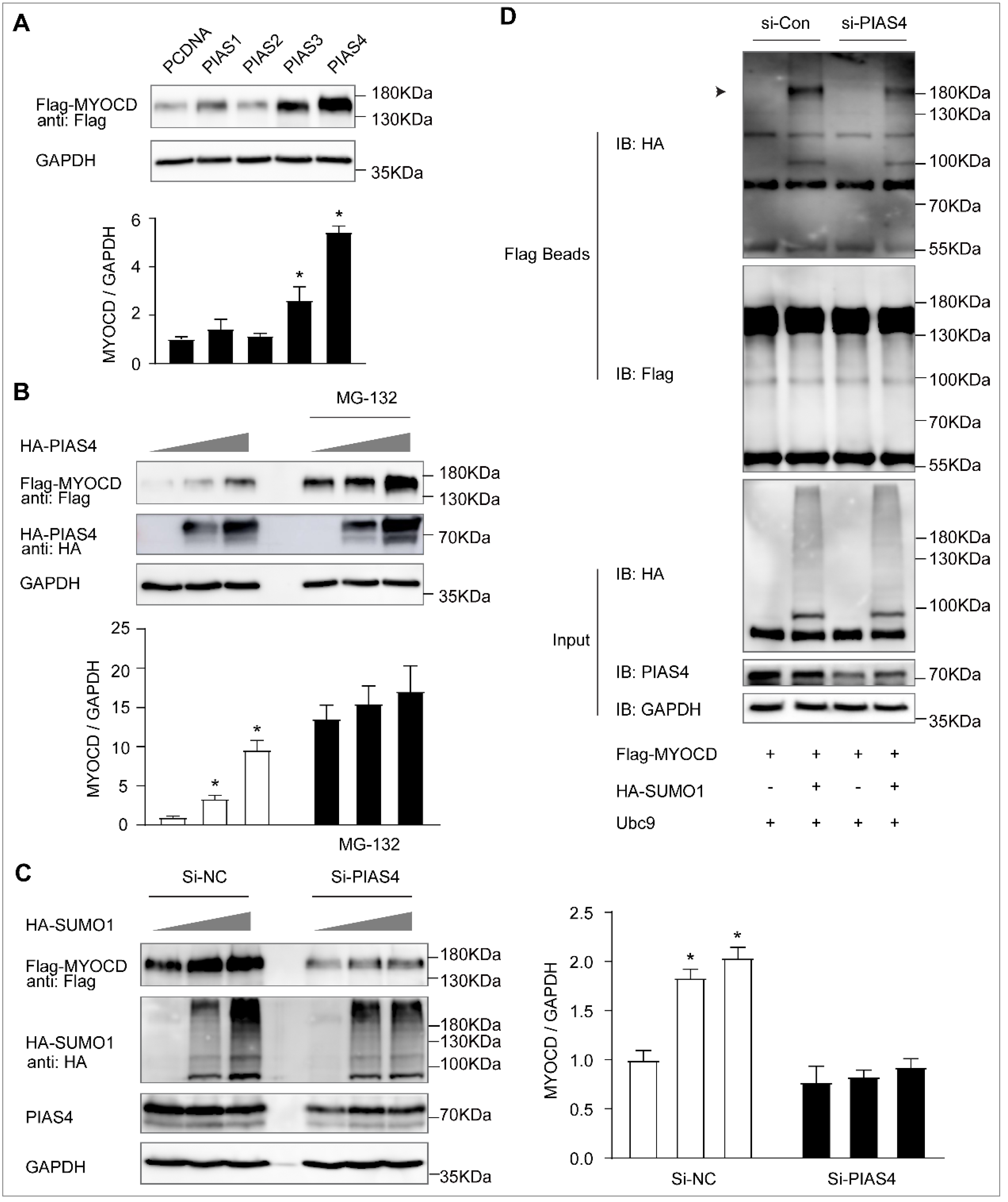
2.3. PIAS4 Is the SUMO E3 Ligase That Mediates Myocardin SUMOylation
2.4. SENP2 Promotes Phenotypic Switching of VSMC In Vitro
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmid Constructs and Transfection
4.3. Lentiviral Infection
4.4. Adenovirus Infection
4.5. Western Blotting
4.6. Flag Immunoprecipitation Assay
4.7. RNA Extraction and Real-Time Quantitative PCR
4.8. In Vitro Scratch-Wound Assay
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, D.Z.; Chang, P.S.; Wang, Z.; Sutherland, L.; Richardson, J.A.; Small, E.; Krieg, P.A.; Olson, E.N. Activation of Cardiac Gene Expression by Myocardin, a Transcriptional Cofactor for Serum Response Factor. Cell 2001, 105, 851–862. [Google Scholar] [CrossRef]
- Tang, R.H.; Zheng, X.L.; Callis, T.E.; Stansfield, W.E.; He, J.; Baldwin, A.S.; Wang, D.Z.; Selzman, C.H. Myocardin inhibits cellular pro-liferation by inhibiting NF-kappaB(p65)-dependent cell cycle progression. Proc. Natl. Acad. Sci. USA 2008, 105, 3362–3367. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yin, H.; Jiang, Y.; Radhakrishnan, S.K.; Huang, Z.P.; Li, J.; Shi, Z.; Kilsdonk, E.P.; Gui, Y.; Wang, D.Z.; et al. Induction of MicroRNA-1 by Myocardin in Smooth Muscle Cells Inhibits Cell Proliferation. Arter. Thromb. Vasc. Biol. 2011, 31, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Ackers-Johnson, M.; Talasila, A.; Sage, A.P.; Long, X.; Bot, I.; Morrell, N.W.; Bennett, M.R.; Miano, J.M.; Sinha, S. Myocardin Regulates Vascular Smooth Muscle Cell Inflammatory Activation and Disease. Arter. Thromb. Vasc. Biol. 2015, 35, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.D.; Zhou, Z.; Yu, X.H.; Zheng, X.L.; Tang, C.K. Myocardin: A novel player in atherosclerosis. Atherosclerosis 2017, 257, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Lee, J.J.; Stoll, S.; Ma, B.; Wiener, R.; Wang, C.; Costa, K.D.; Qiu, H. Inhibition of SRF/myocardin reduces aortic stiffness by targeting vascular smooth muscle cell stiffening in hypertension. Cardiovasc. Res. 2017, 113, 171–182. [Google Scholar] [CrossRef]
- Huang, J.; Wang, T.; Wright, A.C.; Yang, J.; Zhou, S.; Li, L.; Yang, J.; Small, A.; Parmacek, M.S. Myocardin is required for maintenance of vascular and visceral smooth muscle homeostasis during postnatal development. Proc. Natl. Acad. Sci. USA 2015, 112, 4447–4452. [Google Scholar] [CrossRef]
- Taurin, S.; Sandbo, N.; Yau, D.M.; Sethakorn, N.; Kach, J.; Dulin, N.O. Phosphorylation of myocardin by extracellular signal-regulated kinase. J. Biol. Chem. 2009, 284, 33789–33794. [Google Scholar] [CrossRef]
- Badorff, C.; Seeger, F.H.; Zeiher, A.M.; Dimmeler, S. Glycogen synthase kinase 3beta inhibits myocardin-dependent transcription and hypertrophy induction through site-specific phosphorylation. Circ. Res. 2005, 97, 645–654. [Google Scholar] [CrossRef]
- Cao, D.; Wang, C.; Tang, R.; Chen, H.; Zhang, Z.; Tatsuguchi, M.; Wang, D.Z. Acetylation of Myocardin Is Required for the Activation of Cardiac and Smooth Muscle Genes. J. Biol. Chem. 2012, 287, 38495–38504. [Google Scholar] [CrossRef]
- Wang, J.; Li, A.; Wang, Z.; Feng, X.; Olson, E.N.; Schwartz, R.J. Myocardin SUMOylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell. Biol. 2007, 27, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Geiss-Friedlander, R.; Melchior, F. Concepts in SUMOylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007, 8, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Wang, Z.; Yuan, R.; Cui, M.; Lao, Y.; Wang, Y.; Nie, P.; Shen, L.; Yi, J.; He, B. Redox-sensitive enzyme SENP3 mediates vascular remodeling via de-SUMOylation of beta-catenin and regulation of its stability. EBioMedicine 2021, 67, 103386. [Google Scholar] [CrossRef]
- Rabellino, A.; Andreani, C.; Scaglioni, P.P. The Role of PIAS SUMO E3-Ligases in Cancer. Cancer Res. 2017, 77, 1542–1547. [Google Scholar] [CrossRef]
- Kumar, A.; Zhang, K.Y. Advances in the development of SUMO specific protease (SENP) inhibitors. Comput. Struct. Biotechnol. J. 2015, 13, 204–211. [Google Scholar] [CrossRef]
- Huang, C.; Han, Y.; Wang, Y.; Sun, X.; Yan, S.; Yeh, E.T.H.; Chen, Y.; Cang, H.; Li, H.; Shi, G.; et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009, 28, 2748–2762. [Google Scholar] [CrossRef]
- Han, Y.; Huang, C.; Sun, X.; Xiang, B.; Wang, M.; Yeh, E.T.; Chen, Y.; Li, H.; Shi, G.; Cang, H.; et al. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J. Biol. Chem. 2010, 285, 12906–12915. [Google Scholar] [CrossRef]
- Ren, Y.H.; Liu, K.J.; Wang, M.; Yu, Y.N.; Yang, K.; Chen, Q.; Yu, B.; Wang, W.; Li, Q.W.; Wang, J.; et al. De-SUMOylation of FOXC2 by SENP3 promotes the epithelial-mesenchymal transition in gastric cancer cells. Oncotarget 2014, 5, 7093–7104. [Google Scholar] [CrossRef]
- Bawa-Khalfe, T.; Lu, L.S.; Zuo, Y.; Huang, C.; Dere, R.; Lin, F.M.; Yeh, E.T.H. Differential expression of SUMO-specific protease 7 variants regulates epithelial–mesenchymal transition. Proc. Natl. Acad. Sci. USA 2012, 109, 17466–17471. [Google Scholar] [CrossRef]
- Gu, J.; Fan, Y.; Liu, X.; Zhou, L.; Cheng, J.; Cai, R.; Xue, S. SENP1 protects against myocardial ischaemia/reperfusion injury via a HIF1α-dependent pathway. Cardiovasc. Res. 2014, 104, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Cashman, R.; Cohen, H.; Ben-Hamo, R.; Zilberberg, A.; Efroni, S. SENP5 mediates breast cancer invasion via a TGFbetaRI SUMOylation cascade. Oncotarget 2014, 5, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Wang, Y.; Zhao, H.; Qin, L.; Shi, Y.; Zhu, X.; Song, L.; Zhou, X.; Chen, J.; Zhou, H.; et al. The critical role of SENP1-mediated GATA2 deSUMOylation in promoting endothelial activation in graft arteriosclerosis. Nat. Commun. 2017, 8, 15426. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.S.; Chang, E.; Le, N.T.; Cushman, H.; Yeh, E.T.; Fujiwara, K.; Abe, J. De-SUMOylation enzyme of sentrin/SUMO-specific protease 2 regulates disturbed flow-induced SUMOylation of ERK5 and p53 that leads to endothelial dysfunction and ather-osclerosis. Circ Res. 2013, 112, 911–923. [Google Scholar] [CrossRef]
- Heo, K.S.; Le, N.T.; Cushman, H.J.; Giancursio, C.J.; Chang, E.; Woo, C.H.; Sullivan, M.A.; Taunton, J.; Yeh, E.T.; Fujiwara, K.; et al. Disturbed flow-activated p90RSK kinase accelerates atherosclerosis by inhibiting SENP2 function. J. Clin. Investig. 2015, 125, 1299–1310. [Google Scholar] [CrossRef]
- Zhu, X.; Ding, S.; Qiu, C.; Shi, Y.; Song, L.; Wang, Y.; Wang, Y.; Li, J.; Wang, Y.; Sun, Y.; et al. SUMOylation Negatively Regulates Angiogenesis by Targeting Endothelial NOTCH Signaling. Circ. Res. 2017, 121, 636–649. [Google Scholar] [CrossRef]
- Zhu, X.; Qiu, C.; Wang, Y.; Jiang, Y.; Chen, Y.; Fan, L.; Ren, R.; Wang, Y.; Chen, Y.; Feng, Y.; et al. FGFR1 SUMOylation coordinates endothelial angiogenic signaling in angiogenesis. Proc. Natl. Acad. Sci. USA 2022, 119, 2202631119. [Google Scholar] [CrossRef]
- Schimmel, J.; Larsen, K.M.; Matic, I.; van Hagen, M.; Cox, J.; Mann, M.; Andersen, J.S.; Vertegaal, A.C. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell. Proteom. MCP 2008, 7, 2107–2122. [Google Scholar] [CrossRef]
- Sriramachandran, A.M.; Dohmen, R.J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 2014, 1843, 75–85. [Google Scholar] [CrossRef]
- Geoffroy, M.C.; Hay, R.T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009, 10, 564–568. [Google Scholar] [CrossRef]
- Wang, M.; Sang, J.; Ren, Y.; Liu, K.; Liu, X.; Zhang, J.; Wang, H.; Wang, J.; Orian, A.; Yang, J.; et al. SENP3 regulates the global protein turnover and the Sp1 level via antagonizing SUMO2/3-targeted ubiquitination and degradation. Protein Cell 2016, 7, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, J.; Hu, G.; Aman, S.; Li, B.; Li, Y.; Xia, K.; Yang, Y.; Ahmad, B.; Wang, M.; et al. SUMOylation of MCL1 protein enhances its stability by regulating the ubiquitin-proteasome pathway. Cell. Signal. 2020, 73, 109686. [Google Scholar] [CrossRef] [PubMed]
- Kotaja, N.; Karvonen, U.; Jänne, O.A.; Palvimo, J.J. PIAS Proteins Modulate Transcription Factors by Functioning as SUMO-1 Ligases. Mol. Cell. Biol. 2002, 22, 5222–5234. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; Muller, S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 2002, 99, 2872–2877. [Google Scholar] [CrossRef]
- Cui, M.; Cai, Z.; Chu, S.; Sun, Z.; Wang, X.; Hu, L.; Yi, J.; Shen, L.; He, B. Orphan Nuclear Receptor Nur77 Inhibits Angiotensin II-Induced Vascular Remodeling via Downregulation of beta-Catenin. Hypertension 2016, 67, 153–162. [Google Scholar] [CrossRef]
- Yu, Y.; Cai, Z.; Cui, M.; Nie, P.; Sun, Z.; Sun, S.; Chu, S.; Wang, X.; Hu, L.; Yi, J.; et al. The orphan nuclear receptor Nur77 inhibits low shear stress-induced carotid artery remodeling in mice. Int. J. Mol. Med. 2015, 36, 1547–1555. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, M.; Cai, Z.; Jiang, Y.; Huo, H.; Shen, L.; He, B. SENP2 Promotes VSMC Phenotypic Switching via Myocardin De-SUMOylation. Int. J. Mol. Sci. 2022, 23, 12637. https://doi.org/10.3390/ijms232012637
Liang M, Cai Z, Jiang Y, Huo H, Shen L, He B. SENP2 Promotes VSMC Phenotypic Switching via Myocardin De-SUMOylation. International Journal of Molecular Sciences. 2022; 23(20):12637. https://doi.org/10.3390/ijms232012637
Chicago/Turabian StyleLiang, Min, Zhaohua Cai, Yangjing Jiang, Huanhuan Huo, Linghong Shen, and Ben He. 2022. "SENP2 Promotes VSMC Phenotypic Switching via Myocardin De-SUMOylation" International Journal of Molecular Sciences 23, no. 20: 12637. https://doi.org/10.3390/ijms232012637
APA StyleLiang, M., Cai, Z., Jiang, Y., Huo, H., Shen, L., & He, B. (2022). SENP2 Promotes VSMC Phenotypic Switching via Myocardin De-SUMOylation. International Journal of Molecular Sciences, 23(20), 12637. https://doi.org/10.3390/ijms232012637









