Physiologically Based Pharmacokinetic Modeling of Nanoparticle Biodistribution: A Review of Existing Models, Simulation Software, and Data Analysis Tools
Abstract
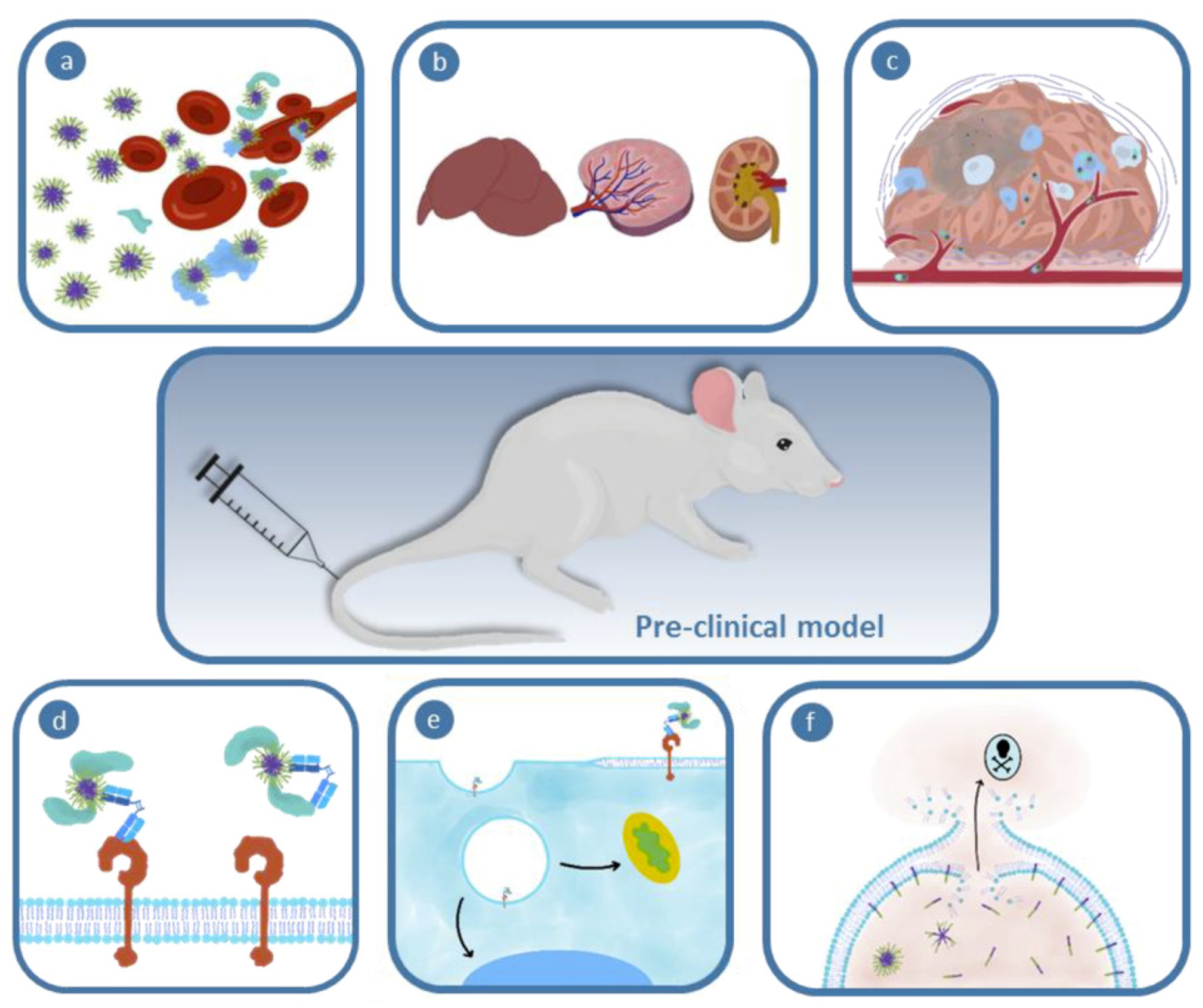
1. Introduction
2. Importance of Mathematical Modeling in Nanomedicine
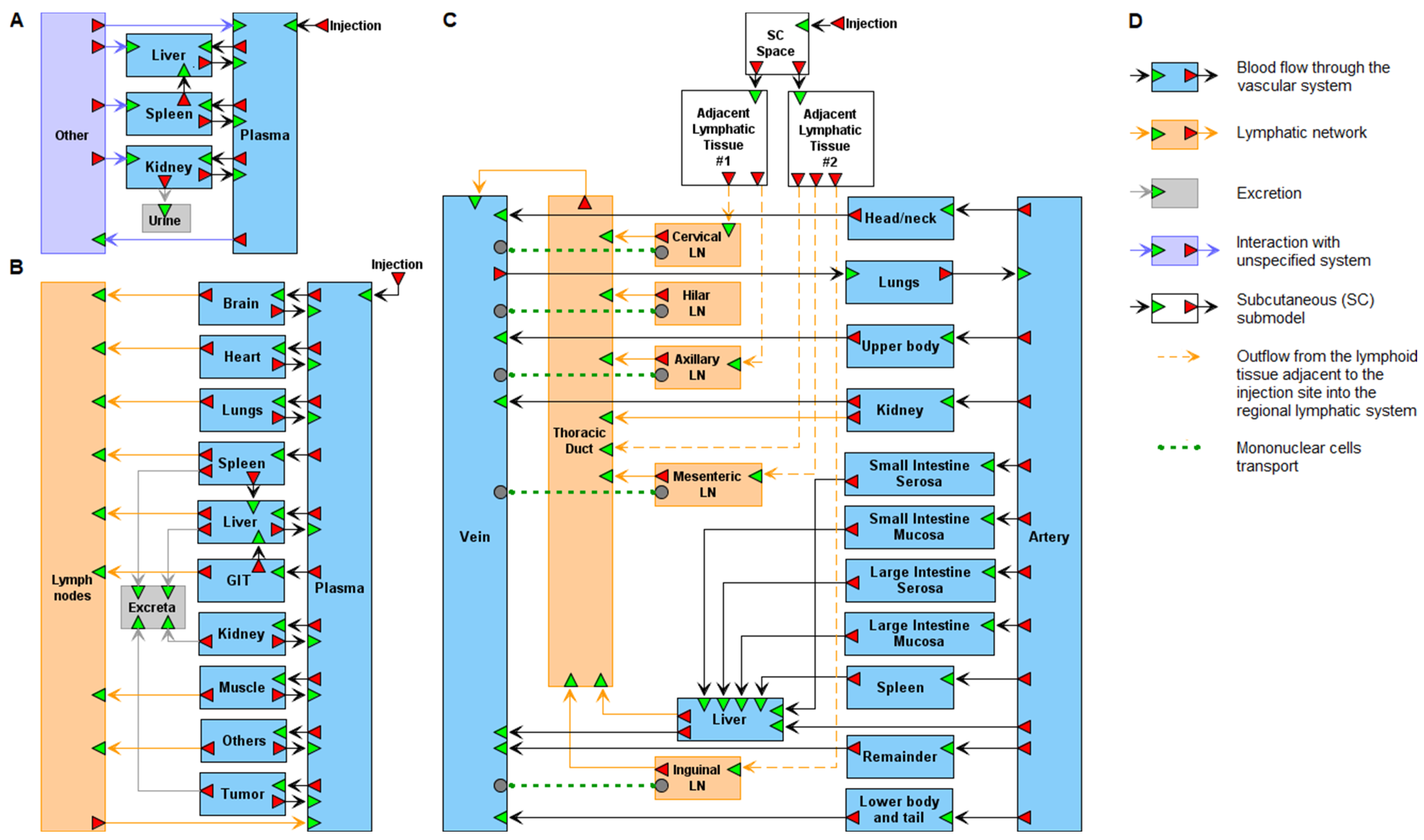
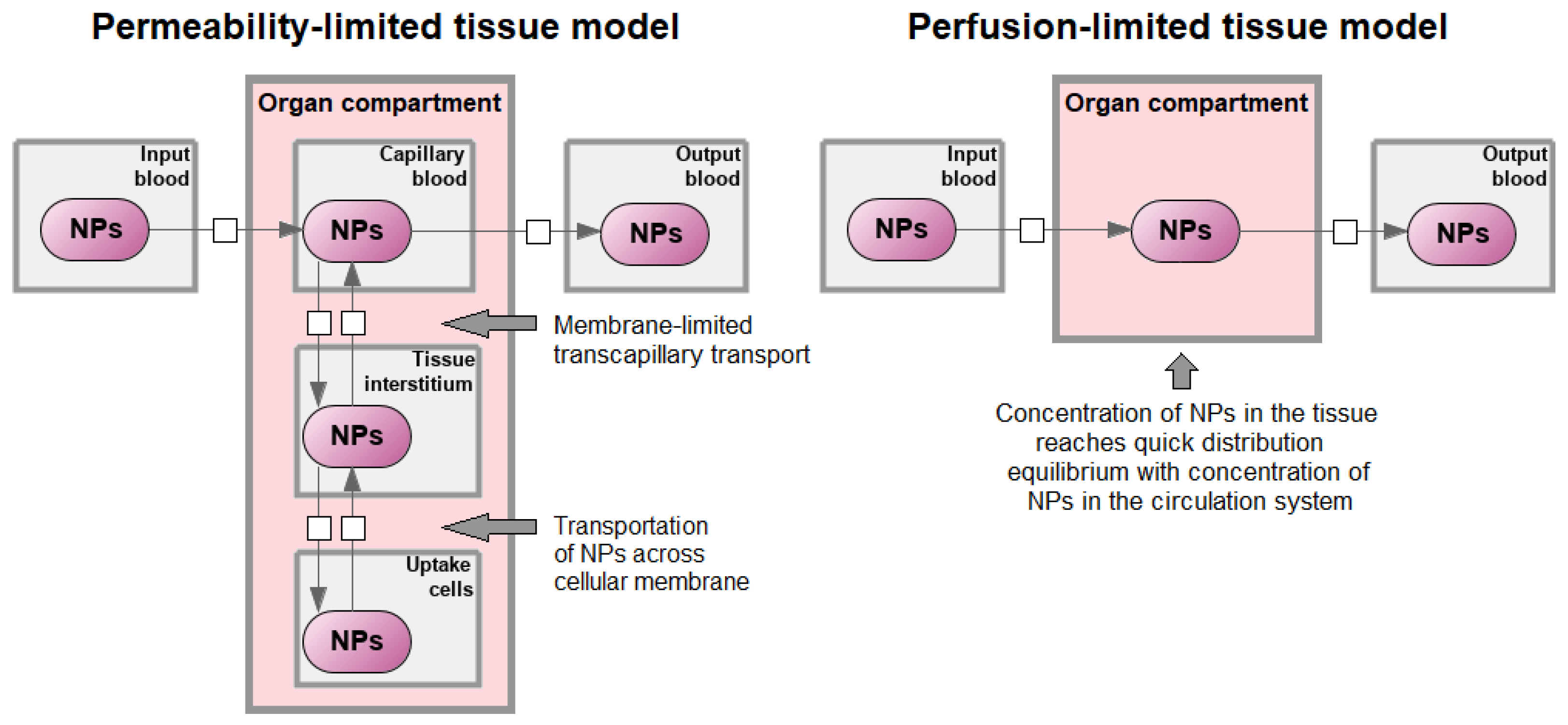
3. Principles of PBPK Modeling of Nanoparticles
4. Main PBPK Modeling Software
5. Auxiliary PBPK Modeling Software
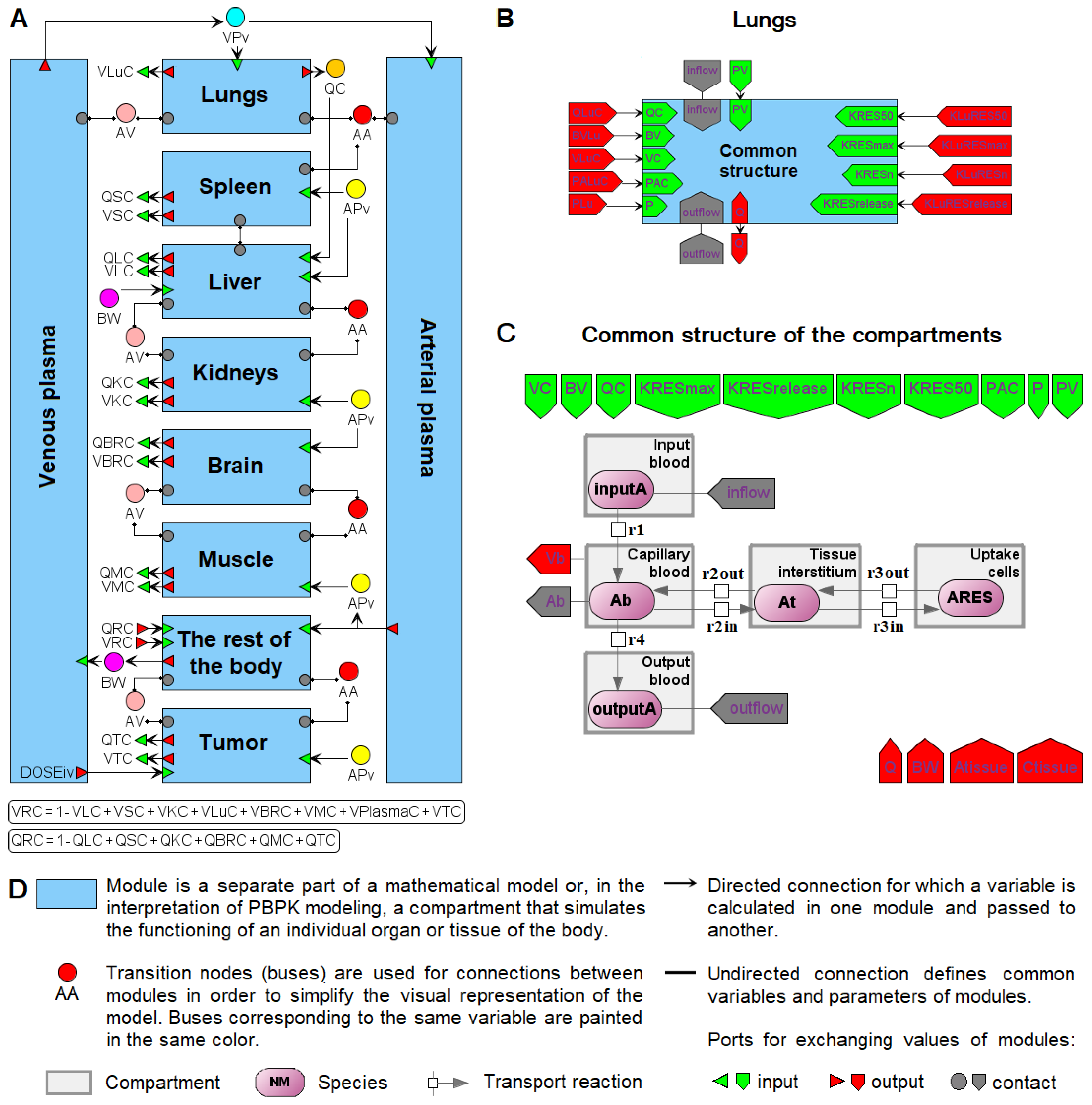
6. Modular Representation of PBPK Models in BioUML
7. Limitation of the Review
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, D.; Bria, E.; Costantini, A.; Di Maio, M.; Rosti, G.; Mancuso, A. Patients’ perception of chemotherapy side effects: Expectations, doctor–patient communication and impact on quality of life–An Italian survey. Eur. J. Cancer Care 2017, 26, e12618. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, V.; Moghe, P.V.; Roth, C.M. Targeting tumor metastases: Drug delivery mechanisms and technologies. J. Control. Release 2015, 219, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H. The 35th Anniversary of the Discovery of EPR Effect: A new wave of nanomedicines for tumor-targeted drug delivery—Personal remarks and future prospects. J. Pers. Med. 2021, 11, 229. [Google Scholar] [CrossRef]
- El-Readi, M.Z.; Althubiti, M.A. Cancer nanomedicine: A new era of successful targeted therapy. J. Nanomater. 2019, 2019, 4927312. [Google Scholar] [CrossRef]
- Parodi, A.; Kostyushev, D.; Brezgin, S.; Kostyusheva, A.; Borodina, T.; Akasov, R.; Frolova, A.; Chulanov, V.; Zamyatnin, A.A., Jr. Biomimetic approaches for targeting tumor inflammation. Semin. Cancer Biol. 2022, 86 Pt 2, 555–567. [Google Scholar] [CrossRef]
- Hejmady, S.; Pradhan, R.; Alexander, A.; Agrawal, M.; Singhvi, G.; Gorain, B.; Tiwari, S.; Kesharwani, P.; Dubey, S.K. Recent advances in targeted nanomedicine as promising antitumor therapeutics. Drug Discov. Today 2020, 25, 2227–2244. [Google Scholar] [CrossRef]
- Shi, Y.; Van der Meel, R.; Chen, X.; Lammers, T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics 2020, 10, 7921. [Google Scholar] [CrossRef]
- Yu, M.K.; Park, J.; Jon, S. Targeting strategies for multifunctional nanoparticles in cancer imaging and therapy. Theranostics 2012, 2, 3. [Google Scholar] [CrossRef]
- Kaddi, C.D.; Phan, J.H.; Wang, M.D. Computational nanomedicine: Modeling of nanoparticle-mediated hyperthermal cancer therapy. Nanomedicine 2013, 8, 1323–1333. [Google Scholar] [CrossRef]
- Li, W.; Peng, A.; Wu, H.; Quan, Y.; Li, Y.; Lu, L.; Cui, M. Anti-cancer nanomedicines: A revolution of tumor immunotherapy. Front. Immunol. 2020, 11, 601497. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Burgess, D.J. Nanomedicine-based drug delivery towards tumor biological and immunological microenvironment. Acta Pharm. Sin. B 2020, 10, 2110–2124. [Google Scholar] [CrossRef] [PubMed]
- Samaridou, E.; Heyes, J.; Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020, 154, 37–63. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.-j.; Xu, S.; Wang, H.-m.; Ling, Y.; Dong, J.; Xia, R.-d.; Sun, X.-h. Nanoparticles: Oral delivery for protein and peptide drugs. Aaps Pharmscitech 2019, 20, 1–11. [Google Scholar] [CrossRef]
- Boateng, F.; Ngwa, W. Delivery of nanoparticle-based radiosensitizers for radiotherapy applications. Int. J. Mol. Sci. 2019, 21, 273. [Google Scholar] [CrossRef]
- Núñez, C.; Estévez, S.V.; del Pilar Chantada, M. Inorganic nanoparticles in diagnosis and treatment of breast cancer. J. Biol. Inorg. Chem. 2018, 23, 331–345. [Google Scholar] [CrossRef]
- Rafique, R.; Kailasa, S.K.; Park, T.J. Recent advances of upconversion nanoparticles in theranostics and bioimaging applications. Trends Anal. Chem. 2019, 120, 115646. [Google Scholar] [CrossRef]
- Vervald, A.M.; Burikov, S.A.; Scherbakov, A.M.; Kudryavtsev, O.S.; Kalyagina, N.A.; Vlasov, I.I.; Ekimov, E.A.; Dolenko, T.A. Boron-doped nanodiamonds as anticancer agents: En route to hyperthermia/thermoablation therapy. ACS Biomater. Sci. Eng. 2020, 6, 4446–4453. [Google Scholar] [CrossRef]
- Sivasubramanian, M.; Chuang, Y.C.; Lo, L.-W. Evolution of nanoparticle-mediated photodynamic therapy: From superficial to deep-seated cancers. Molecules 2019, 24, 520. [Google Scholar] [CrossRef]
- Parodi, A.; Buzaeva, P.; Nigovora, D.; Baldin, A.; Kostyushev, D.; Chulanov, V.; Savvateeva, L.V.; Zamyatnin, A.A. Nanomedicine for increasing the oral bioavailability of cancer treatments. J. Nanobiotechnol. 2021, 19, 354. [Google Scholar] [CrossRef]
- Borodina, T.; Kostyushev, D.; Zamyatnin, A.A., Jr.; Parodi, A. Nanomedicine for treating diabetic retinopathy vascular degeneration. Int. J. Transl. Med. 2021, 1, 306–322. [Google Scholar] [CrossRef]
- Parodi, A.; Evangelopoulos, M.; Arrighetti, N.; Cevenini, A.; Livingston, M.; Khaled, S.Z.; Brown, B.S.; Yazdi, I.K.; Paradiso, F.; Campa-Carranza, J.N. Endosomal escape of polymer-coated silica nanoparticles in endothelial cells. Small 2020, 16, 1907693. [Google Scholar] [CrossRef] [PubMed]
- Khalid, K.; Tan, X.; Mohd Zaid, H.F.; Tao, Y.; Lye Chew, C.; Chu, D.-T.; Lam, M.K.; Ho, Y.-C.; Lim, J.W.; Chin Wei, L. Advanced in developmental organic and inorganic nanomaterial: A review. Bioengineered 2020, 11, 328–355. [Google Scholar] [CrossRef]
- Li, M.; Al-Jamal, K.T.; Kostarelos, K.; Reineke, J. Physiologically based pharmacokinetic modeling of nanoparticles. ACS Nano 2010, 4, 6303–6317. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; He, H.; Wu, Y.; Fan, J.; Cao, Y. Physiologically based pharmacokinetic modeling of nanoparticles. J. Pharm. Sci. 2019, 108, 58–72. [Google Scholar] [CrossRef]
- Li, M.; Zou, P.; Tyner, K.; Lee, S. Physiologically based pharmacokinetic (PBPK) modeling of pharmaceutical nanoparticles. AAPS J. 2017, 19, 26–42. [Google Scholar] [CrossRef]
- Wang, W.; Ouyang, D. Opportunities and challenges of physiologically based pharmacokinetic modeling in drug delivery. Drug Discov. Today 2022, 27, 2100–2120. [Google Scholar] [CrossRef]
- Singh, A.V.; Ansari, M.H.D.; Rosenkranz, D.; Maharjan, R.S.; Kriegel, F.L.; Gandhi, K.; Kanase, A.; Singh, R.; Laux, P.; Luch, A. Artificial intelligence and machine learning in computational nanotoxicology: Unlocking and empowering nanomedicine. Adv. Healthc. Mater. 2020, 9, e1901862. [Google Scholar] [CrossRef]
- Lin, Z.; Chou, W.C.; Cheng, Y.H.; He, C.; Monteiro-Riviere, N.A.; Riviere, J.E. Predicting nanoparticle delivery to tumors using machine learning and artificial intelligence approaches. Int. J. Nanomed. 2022, 17, 1365–1379. [Google Scholar] [CrossRef]
- Singh, A.V.; Chandrasekar, V.; Janapareddy, P.; Mathews, D.E.; Laux, P.; Luch, A.; Yang, Y.; Garcia-Canibano, B.; Balakrishnan, S.; Abinahed, J.; et al. Emerging application of nanorobotics and artificial intelligence to cross the BBB: Advances in design, controlled maneuvering, and targeting of the barriers. ACS Chem. Neurosci. 2021, 12, 1835–1853. [Google Scholar] [CrossRef]
- Ji, Z.; Guo, W.; Wood, E.L.; Liu, J.; Sakkiah, S.; Xu, X.; Patterson, T.A.; Hong, H. Machine learning models for predicting cytotoxicity of nanomaterials. Chem. Res. Toxicol. 2022, 35, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Hucka, M.; Bergmann, F.T.; Chaouiya, C.; Dräger, A.; Hoops, S.; Keating, S.M.; König, M.; Le Novère, N.; Myers, C.J.; Olivier, B.G. The systems biology markup language (SBML): Language specification for level 3 version 2 core release 2. J. Integr. Bioinform. 2019, 16, 20190021. [Google Scholar] [CrossRef] [PubMed]
- Novère, N.L.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.; Wegner, K.; Aladjem, M.I.; Wimalaratne, S.M. The systems biology graphical notation. Nat. Biotechnol. 2009, 27, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Utembe, W.; Clewell, H.; Sanabria, N.; Doganis, P.; Gulumian, M. Current approaches and techniques in physiologically based pharmacokinetic (PBPK) modelling of nanomaterials. Nanomaterials 2020, 10, 1267. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Monteiro-Riviere, N.A.; Riviere, J.E. Pharmacokinetics of metallic nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 189–217. [Google Scholar] [CrossRef]
- Wakaskar, R.R. Promising effects of nanomedicine in cancer drug delivery. J. Drug Target. 2018, 26, 319–324. [Google Scholar] [CrossRef]
- Abdifetah, O.; Na-Bangchang, K. Pharmacokinetic studies of nanoparticles as a delivery system for conventional drugs and herb-derived compounds for cancer therapy: A systematic review. Int. J. Nanomed. 2019, 14, 5659. [Google Scholar] [CrossRef]
- Salvioni, L.; Rizzuto, M.A.; Bertolini, J.A.; Pandolfi, L.; Colombo, M.; Prosperi, D. Thirty years of cancer nanomedicine: Success, frustration, and hope. Cancers 2019, 11, 1855. [Google Scholar] [CrossRef]
- Alexis, F.; Pridgen, E.; Molnar, L.K.; Farokhzad, O.C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 2008, 5, 505–515. [Google Scholar] [CrossRef]
- Haute, D.V.; Berlin, J.M. Challenges in realizing selectivity for nanoparticle biodistribution and clearance: Lessons from gold nanoparticles. Ther. Deliv. 2017, 8, 763–774. [Google Scholar] [CrossRef]
- Dogra, P.; Butner, J.D.; Chuang, Y.-l.; Caserta, S.; Goel, S.; Brinker, C.J.; Cristini, V.; Wang, Z. Mathematical modeling in cancer nanomedicine: A review. Biomed. Microdevices 2019, 21, 40. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Simpson, J.D.; Smith, S.A.; Thurecht, K.J.; Such, G. Engineered polymeric materials for biological applications: Overcoming challenges of the bio–nano interface. Polymers 2019, 11, 1441. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wang, J.; Mu, Q.; Su, G. In vivo protein corona formation: Characterizations, effects on engineered nanoparticles’ biobehaviors, and applications. Front. Bioeng. Biotechnol. 2021, 9, 646708. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Wang, Z.; Yue, T.; Su, G.; Teng, C.; Yan, B. Crossing biological barriers by engineered nanoparticles. Chem. Res. Toxicol. 2020, 33, 1055–1060. [Google Scholar] [CrossRef]
- Cheng, Z.; Li, M.; Dey, R.; Chen, Y. Nanomaterials for cancer therapy: Current progress and perspectives. J. Hematol. Oncol. 2021, 14, 85. [Google Scholar] [CrossRef]
- Frieboes, H.B.; Raghavan, S.; Godin, B. Modeling of nanotherapy response as a function of the tumor microenvironment: Focus on liver metastasis. Front. Bioeng. Biotechnol. 2020, 8, 1011. [Google Scholar] [CrossRef]
- Qu, Z.; Garfinkel, A.; Weiss, J.N.; Nivala, M. Multi-scale modeling in biology: How to bridge the gaps between scales? Prog. Biophys. Mol. Biol. 2011, 107, 21–31. [Google Scholar] [CrossRef]
- Liu, Y.; Shah, S.; Tan, J. Computational modeling of nanoparticle targeted drug delivery. Rev. Nanosci. Nanotechnol. 2012, 1, 66–83. [Google Scholar] [CrossRef]
- Cheng, Y.-H.; He, C.; Riviere, J.E.; Monteiro-Riviere, N.A.; Lin, Z. Meta-analysis of nanoparticle delivery to tumors using a physiologically based pharmacokinetic modeling and simulation approach. ACS Nano 2020, 14, 3075–3095. [Google Scholar] [CrossRef]
- Levit, S.L.; Tang, C. Polymeric nanoparticle delivery of combination therapy with synergistic effects in ovarian cancer. Nanomaterials 2021, 11, 1048. [Google Scholar] [CrossRef] [PubMed]
- Gerlowski, L.E.; Jain, R.K. Physiologically based pharmacokinetic modeling: Principles and applications. J. Pharm. Sci. 1983, 72, 1103–1127. [Google Scholar] [CrossRef] [PubMed]
- Gilkey, M.; Krishnan, V.; Scheetz, L.; Jia, X.; Rajasekaran, A.; Dhurjati, P. Physiologically based pharmacokinetic modeling of fluorescently labeled block copolymer nanoparticles for controlled drug delivery in leukemia therapy. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Dogra, P.; Butner, J.D.; Ramírez, J.R.; Chuang, Y.-l.; Noureddine, A.; Brinker, C.J.; Cristini, V.; Wang, Z. A mathematical model to predict nanomedicine pharmacokinetics and tumor delivery. Comput. Struct. Biotechnol. J. 2020, 18, 518–531. [Google Scholar] [CrossRef] [PubMed]
- Howell, B.A.; Chauhan, A. A physiologically based pharmacokinetic (PBPK) model for predicting the efficacy of drug overdose treatment with liposomes in man. J. Pharm. Sci. 2010, 99, 3601–3619. [Google Scholar] [CrossRef]
- Perazzolo, S.; Shen, D.D.; Ho, R.J. Physiologically based pharmacokinetic modeling of 3 HIV drugs in combination and the role of lymphatic system after subcutaneous dosing. Part 2: Model for the drug-combination nanoparticles. J. Pharm. Sci. 2022, 111, 825–837. [Google Scholar] [CrossRef]
- Rajoli, R.K.; Back, D.J.; Rannard, S.; Freel Meyers, C.L.; Flexner, C.; Owen, A.; Siccardi, M. Physiologically based pharmacokinetic modelling to inform development of intramuscular long-acting nanoformulations for HIV. Clin. Pharmacokinet. 2015, 54, 639–650. [Google Scholar] [CrossRef]
- Pery, A.R.; Brochot, C.; Hoet, P.H.; Nemmar, A.; Bois, F.Y. Development of a physiologically based kinetic model for 99 m-technetium-labelled carbon nanoparticles inhaled by humans. Inhal. Toxicol. 2009, 21, 1099–1107. [Google Scholar] [CrossRef]
- Jones, H.; Rowland-Yeo, K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, 1–12. [Google Scholar] [CrossRef]
- Lin, Z.; Monteiro-Riviere, N.A.; Riviere, J.E. A physiologically based pharmacokinetic model for polyethylene glycol-coated gold nanoparticles of different sizes in adult mice. Nanotoxicology 2016, 10, 162–172. [Google Scholar] [CrossRef]
- Li, M.; Panagi, Z.; Avgoustakis, K.; Reineke, J. Physiologically based pharmacokinetic modeling of PLGA nanoparticles with varied mPEG content. Int. J. Nanomed. 2012, 7, 1345. [Google Scholar]
- Chou, W.C.; Cheng, Y.H.; Riviere, J.E.; Monteiro-Riviere, N.A.; Kreyling, W.G.; Lin, Z. Development of a multi-route physiologically based pharmacokinetic (PBPK) model for nanomaterials: A comparison between a traditional versus a new route-specific approach using gold nanoparticles in rats. Part. Fibre Toxicol. 2022, 19, 47. [Google Scholar] [CrossRef]
- Moore, J.A.; Chow, J.C. Recent progress and applications of gold nanotechnology in medical biophysics using artificial intelligence and mathematical modeling. Nano Express 2021, 2, 022001. [Google Scholar] [CrossRef]
- Brown, L.V.; Coles, M.C.; McConnell, M.; Ratushny, A.V.; Gaffney, E.A. Analysis of cellular kinetic models suggest that physiologically based model parameters may be inherently, practically unidentifiable. J. Pharmacokinet. Pharmacodyn. 2022, 49, 539–556. [Google Scholar] [CrossRef]
- Tiwari, K.; Kananathan, S.; Roberts, M.G.; Meyer, J.P.; Sharif Shohan, M.U.; Xavier, A.; Maire, M.; Zyoud, A.; Men, J.; Ng, S. Reproducibility in systems biology modelling. Mol. Syst. Biol. 2021, 17, e9982. [Google Scholar] [CrossRef]
- Porubsky, V.L.; Goldberg, A.P.; Rampadarath, A.K.; Nickerson, D.P.; Karr, J.R.; Sauro, H.M. Best practices for making reproducible biochemical models. Cell Syst. 2020, 11, 109–120. [Google Scholar] [CrossRef]
- The MathWorks, Inc., USA. MATLAB. Available online: http://www.mathworks.com (accessed on 11 July 2022).
- Schmidt, H.; Jirstrand, M. Systems Biology Toolbox for MATLAB: A computational platform for research in systems biology. Bioinformatics 2006, 22, 514–515. [Google Scholar] [CrossRef]
- Lin, Z.; Jaberi-Douraki, M.; He, C.; Jin, S.; Yang, R.S.; Fisher, J.W.; Riviere, J.E. Performance assessment and translation of physiologically based pharmacokinetic models from acslX to Berkeley Madonna, MATLAB, and R Language: Oxytetracycline and gold nanoparticles as case examples. Toxicol. Sci. 2017, 158, 23–35. [Google Scholar] [CrossRef]
- Aborig, M.; Malik, P.R.; Nambiar, S.; Chelle, P.; Darko, J.; Mutsaers, A.; Edginton, A.N.; Fleck, A.; Osei, E.; Wettig, S. Biodistribution and physiologically-based pharmacokinetic modeling of gold nanoparticles in mice with interspecies extrapolation. Pharmaceutics 2019, 11, 179. [Google Scholar] [CrossRef]
- Dong, D.; Wang, X.; Wang, H.; Zhang, X.; Wang, Y.; Wu, B. Elucidating the in vivo fate of nanocrystals using a physiologically based pharmacokinetic model: A case study with the anticancer agent SNX-2112. Int. J. Nanomed. 2015, 10, 2521. [Google Scholar]
- Glass, E.; Kulkarni, S.; Eng, C.; Feng, S.; Malavia, A.; Radhakrishnan, R. Physiologically based multiphysics pharmacokinetic model for determining the temporal biodistribution of targeted nanoparticles. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kagan, L.; Gershkovich, P.; Wasan, K.M.; Mager, D.E. Dual physiologically based pharmacokinetic model of liposomal and nonliposomal amphotericin B disposition. Pharm. Res. 2014, 31, 35–45. [Google Scholar] [CrossRef]
- Lu, X.-F.; Bi, K.; Chen, X. Physiologically based pharmacokinetic model of docetaxel and interspecies scaling: Comparison of simple injection with folate receptor-targeting amphiphilic copolymer-modified liposomes. Xenobiotica 2016, 46, 1093–1104. [Google Scholar] [CrossRef]
- MacCalman, L.; CL, T.; Kuempel, E. Development of a bio-mathematical model in rats to describe clearance, retention and translocation of inhaled nano particles throughout the body. J. Phys. Conf. Ser. 2009, 151, 012028. [Google Scholar] [CrossRef]
- Opitz, A.W.; Wickstrom, E.; Thakur, M.L.; Wagner, N.J. Physiologically based pharmacokinetics of molecular imaging nanoparticles for mRNA detection determined in tumor-bearing mice. Oligonucleotides 2010, 20, 117–125. [Google Scholar] [CrossRef]
- Kasyanova, V.; Bazhukova, I. Modeling of cerium oxide nanoparticles pharmacokinetics. AIP Conf. Proc. 2020, 2313, 080015. [Google Scholar]
- Silva, A.H.; Lima, E., Jr.; Mansilla, M.V.; Zysler, R.D.; Pisciotti, M.L.M.; Locatelli, C.; Rajoli, R.K.R.; Owen, A.; Creczynski-Pasa, T.B.; Siccardi, M. A physiologically based pharmacokinetic model to predict the superparamagnetic iron oxide nanoparticles (SPIONs) accumulation in vivo. Eur. J. Nanomed. 2017, 9, 79–90. [Google Scholar]
- Chen, J.; Yuan, M.; Madison, C.A.; Eitan, S.; Wang, Y. Blood-brain barrier crossing using magnetic stimulated nanoparticles. J. Control. Release 2022, 345, 557–571. [Google Scholar] [CrossRef]
- Dubaj, T.; Kozics, K.; Sramkova, M.; Manova, A.; Bastús, N.G.; Moriones, O.H.; Kohl, Y.; Dusinska, M.; Runden-Pran, E.; Puntes, V.; et al. Pharmacokinetics of PEGylated gold nanoparticles: In vitro-in vivo correlation. Nanomaterials 2022, 12, 511. [Google Scholar] [CrossRef]
- Maiwald, T.; Timmer, J. Dynamical modeling and multi-experiment fitting with PottersWheel. Bioinformatics 2008, 24, 2037–2043. [Google Scholar] [CrossRef]
- Mager, D.E.; Mody, V.; Xu, C.; Forrest, A.; Lesniak, W.G.; Nigavekar, S.S.; Kariapper, M.T.; Minc, L.; Khan, M.K.; Balogh, L.P. Physiologically based pharmacokinetic model for composite nanodevices: Effect of charge and size on in vivo disposition. Pharm. Res. 2012, 29, 2534–2542. [Google Scholar] [CrossRef]
- Dogra, P.; Adolphi, N.L.; Wang, Z.; Lin, Y.-S.; Butler, K.S.; Durfee, P.N.; Croissant, J.G.; Noureddine, A.; Coker, E.N.; Bearer, E.L. Establishing the effects of mesoporous silica nanoparticle properties on in vivo disposition using imaging-based pharmacokinetics. Nat. Commun. 2018, 9, 4551. [Google Scholar] [CrossRef]
- Toy, R.; Hayden, E.; Shoup, C.; Baskaran, H.; Karathanasis, E. The effects of particle size, density and shape on margination of nanoparticles in microcirculation. Nanotechnology 2011, 22, 115101. [Google Scholar] [CrossRef]
- Klapproth, A.P.; Shevtsov, M.; Stangl, S.; Li, W.B.; Multhoff, G. A new pharmacokinetic model describing the biodistribution of intravenously and intratumorally administered superparamagnetic iron oxide nanoparticles (SPIONs) in a GL261 xenograft glioblastoma model. Int. J. Nanomed. 2020, 15, 4677. [Google Scholar] [CrossRef]
- Verma, J.; Lal, S.; Van Noorden, C.J. Nanoparticles for hyperthermic therapy: Synthesis strategies and applications in glioblastoma. Int. J. Nanomed. 2014, 9, 2863. [Google Scholar]
- University of California at Berkeley, USA. Berkeley Madonna. Available online: https://berkeley-madonna.myshopify.com/ (accessed on 12 July 2022).
- Marcoline, F.V.; Furth, J.; Nayak, S.; Grabe, M.; Macey, R.I. Berkeley Madonna Version 10–A simulation package for solving mathematical models. CPT Pharmacomet. Syst. Pharmacol. 2022, 11, 290–301. [Google Scholar] [CrossRef]
- Kutumova, E.; Akberdin, I.; Kiselev, I.; Sharipov, R.; Kolpakov, F. Modular representation of physiologically based pharmacokinetic models: Nanoparticle delivery to solid tumors in mice as an example. Mathematics 2022, 10, 1176. [Google Scholar] [CrossRef]
- Carlander, U.; Li, D.; Jolliet, O.; Emond, C.; Johanson, G. Toward a general physiologically-based pharmacokinetic model for intravenously injected nanoparticles. Int. J. Nanomed. 2016, 11, 625. [Google Scholar] [CrossRef]
- Li, D.; Johanson, G.; Emond, C.; Carlander, U.; Philbert, M.; Jolliet, O. Physiologically based pharmacokinetic modeling of polyethylene glycol-coated polyacrylamide nanoparticles in rats. Nanotoxicology 2014, 8, 128–137. [Google Scholar] [CrossRef]
- Li, D.; Morishita, M.; Wagner, J.G.; Fatouraie, M.; Wooldridge, M.; Eagle, W.E.; Barres, J.; Carlander, U.; Emond, C.; Jolliet, O. In vivo biodistribution and physiologically based pharmacokinetic modeling of inhaled fresh and aged cerium oxide nanoparticles in rats. Part. Fibre Toxicol. 2015, 13, 45. [Google Scholar]
- Li, L.; He, H.; Jiang, S.; Qi, J.; Lu, Y.; Ding, N.; Lin, H.-S.; Wu, W.; Xiang, X. Simulation of the in vivo fate of polymeric nanoparticles traced by environment-responsive near-infrared dye: A physiologically based pharmacokinetic modelling approach. Molecules 2021, 26, 1271. [Google Scholar] [CrossRef]
- Liang, X.; Wang, H.; Grice, J.E.; Li, L.; Liu, X.; Xu, Z.P.; Roberts, M.S. Physiologically based pharmacokinetic model for long-circulating inorganic nanoparticles. Nano Lett. 2016, 16, 939–945. [Google Scholar] [CrossRef]
- Lin, P.; Chen, J.-W.; Chang, L.W.; Wu, J.-P.; Redding, L.; Chang, H.; Yeh, T.-K.; Yang, C.S.; Tsai, M.-H.; Wang, H.-J. Computational and ultrastructural toxicology of a nanoparticle, quantum dot 705, in mice. Environ. Sci. Technol. 2008, 42, 6264–6270. [Google Scholar] [CrossRef]
- Wenger, Y.; Schneider II, R.J.; Reddy, G.R.; Kopelman, R.; Jolliet, O.; Philbert, M.A. Tissue distribution and pharmacokinetics of stable polyacrylamide nanoparticles following intravenous injection in the rat. Toxicol. Appl. Pharmacol. 2011, 251, 181–190. [Google Scholar] [CrossRef]
- Zhang, L.; Su, H.; Wang, H.; Li, Q.; Li, X.; Zhou, C.; Xu, J.; Chai, Y.; Liang, X.; Xiong, L. Tumor chemo-radiotherapy with rod-shaped and spherical gold nano probes: Shape and active targeting both matter. Theranostics 2019, 9, 1893. [Google Scholar] [CrossRef]
- Chen, W.-Y.; Cheng, Y.-H.; Hsieh, N.-H.; Wu, B.-C.; Chou, W.-C.; Ho, C.-C.; Chen, J.-K.; Liao, C.-M.; Lin, P. Physiologically based pharmacokinetic modeling of zinc oxide nanoparticles and zinc nitrate in mice. Int. J. Nanomed. 2015, 10, 6277. [Google Scholar]
- R Core Team. R language. Available online: https://www.r-project.org/ (accessed on 12 July 2022).
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- Mitchell, E.E.L.; Gauthier, J.S. Advanced Continuous Simulation Language (ACSL). Simulation 1976, 25, 72–78. [Google Scholar] [CrossRef]
- Carlander, U.; Moto, T.P.; Desalegn, A.A.; Yokel, R.A.; Johanson, G. Physiologically based pharmacokinetic modeling of nanoceria systemic distribution in rats suggests dose-and route-dependent biokinetics. Int. J. Nanomed. 2018, 13, 2631. [Google Scholar] [CrossRef]
- Lin, Z.; Monteiro-Riviere, N.A.; Kannan, R.; Riviere, J.E. A computational framework for interspecies pharmacokinetics, exposure and toxicity assessment of gold nanoparticles. Nanomedicine 2016, 11, 107–119. [Google Scholar] [CrossRef]
- Lankveld, D.P.; Oomen, A.G.; Krystek, P.; Neigh, A.; Troost–de Jong, A.; Noorlander, C.; Van Eijkeren, J.; Geertsma, R.; De Jong, W. The kinetics of the tissue distribution of silver nanoparticles of different sizes. Biomaterials 2010, 31, 8350–8361. [Google Scholar] [CrossRef]
- Sweeney, L.M.; MacCalman, L.; Haber, L.T.; Kuempel, E.D.; Tran, C.L. Bayesian evaluation of a physiologically-based pharmacokinetic (PBPK) model of long-term kinetics of metal nanoparticles in rats. Regul. Toxicol. Pharmacol. 2015, 73, 151–163. [Google Scholar] [CrossRef]
- Biosoft.ru, Ltd., Russia. BioUML. Available online: https://www.biouml.org (accessed on 14 July 2022).
- Kolpakov, F.; Akberdin, I.; Kiselev, I.; Kolmykov, S.; Kondrakhin, Y.; Kulyashov, M.; Kutumova, E.; Pintus, S.; Ryabova, A.; Sharipov, R. BioUML—Towards a universal research platform. Nucleic Acids Res. 2022, 50, W124–W131. [Google Scholar] [CrossRef]
- Kutumova, E.; Kiselev, I.; Sharipov, R.; Lifshits, G.; Kolpakov, F. Thoroughly calibrated modular agent-based model of the human cardiovascular and renal systems for blood pressure regulation in health and disease. Front. Physiol. 2021, 12, 746300. [Google Scholar] [CrossRef]
- Akberdin, I.R.; Kiselev, I.N.; Pintus, S.S.; Sharipov, R.N.; Vertyshev, A.Y.; Vinogradova, O.L.; Popov, D.V.; Kolpakov, F.A. A modular mathematical model of exercise-induced changes in metabolism, signaling, and gene expression in human skeletal muscle. Int. J. Mol. Sci. 2021, 22, 10353. [Google Scholar] [CrossRef]
- Certara, LP, USA. Simcyp. Available online: https://www.certara.com/software/simcyp-pbpk/ (accessed on 22 September 2022).
- Jamei, M.; Marciniak, S.; Feng, K.; Barnett, A.; Tucker, G.; Rostami-Hodjegan, A. The Simcyp population-based ADME simulator. Expert Opin. Drug Metab. Toxicol. 2009, 5, 211–223. [Google Scholar] [CrossRef]
- Jamei, M.; Marciniak, S.; Edwards, D.; Wragg, K.; Feng, K.; Barnett, A.; Rostami-Hodjegan, A. The simcyp population based simulator: Architecture, implementation, and quality assurance. Silico Pharmacol. 2013, 1, 9. [Google Scholar] [CrossRef]
- Ezuruike, U.; Zhang, M.; Pansari, A.; De Sousa Mendes, M.; Pan, X.; Neuhoff, S.; Gardner, I. Guide to development of compound files for PBPK modeling in the Simcyp population-based simulator. CPT: Pharmacomet. Syst. Pharmacol. 2022, 11, 805–821. [Google Scholar] [CrossRef]
- Kostewicz, E.S.; Aarons, L.; Bergstrand, M.; Bolger, M.B.; Galetin, A.; Hatley, O.; Jamei, M.; Lloyd, R.; Pepin, X.; Rostami-Hodjegan, A.; et al. PBPK models for the prediction of in vivo performance of oral dosage forms. Eur. J. Pharm. Sci. 2014, 57, 300–321. [Google Scholar] [CrossRef]
- Lin, Z.; Cheng, Y.H.; Chou, W.C.; Li, M. Chapter 10—Physiologically based pharmacokinetic model calibration, evaluation, and performance assessment. In Physiologically Based Pharmacokinetic (PBPK) Modeling: Methods and Applications in Toxicology and Risk Assessment; Academic Press: Cambridge, MA, USA, 2020; pp. 243–279. [Google Scholar]
- El-Khateeb, E.; Burkhill, S.; Murby, S.; Amirat, H.; Rostami-Hodjegan, A.; Ahmad, A. Physiological-based pharmacokinetic modeling trends in pharmaceutical drug development over the last 20-years; in-depth analysis of applications, organizations, and platforms. Biopharm. Drug Dispos. 2021, 42, 107–117. [Google Scholar] [CrossRef]
- Litou, C.; Patel, N.; Turner, D.B.; Kostewicz, E.; Kuentz, M.; Box, K.J.; Dressman, J. Combining biorelevant in vitro and in silico tools to simulate and better understand the in vivo performance of a nano-sized formulation of aprepitant in the fasted and fed states. Eur. J. Pharm. Sci. 2019, 138, 105031. [Google Scholar] [CrossRef]
- Simulations Plus, Inc., USA. GastroPlus. Available online: https://www.simulations-plus.com/software/gastroplus/ (accessed on 23 September 2022).
- Romero, R.M.; Bolger, M.B.; Morningstar-Kywi, N.; Haworth, I.S. Teaching of biopharmaceutics in a drug design course: Use of GastroPlus as educational software. J. Chem. Educ. 2020, 97, 2212–2220. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, S.K. In silico-in vitro-in vivo studies of experimentally designed carvedilol loaded silk fibroin-casein nanoparticles using physiological based pharmacokinetic model. Int. J. Biol. Macromol. 2017, 96, 403–420. [Google Scholar] [CrossRef]
- Mahdi, W.A.; Hussain, A.; Ramzan, M. 5-Fluorouracil loaded biogenic and albumin capped gold nanoparticles using bacterial enzyme –- in vitro-in silico Gastroplus® simulation and prediction. Processes 2020, 8, 1579. [Google Scholar] [CrossRef]
- Stewart, A.M.; Grass, M.E. Practical approach to modeling the impact of amorphous drug nanoparticles on the oral absorption of poorly soluble drugs. Mol. Pharm. 2020, 17, 180–189. [Google Scholar] [CrossRef]
- Ali, H.; Verma, P.R.P.; Dubey, S.K.; Venkatesan, J.; Seo, Y.; Kim, S.K.; Singh, S.K. In vitro–in vivo and pharmacokinetic evaluation of solid lipid nanoparticles of furosemide using Gastroplus™. RSC Adv. 2017, 7, 33314–33326. [Google Scholar] [CrossRef]
- Bayer Technology Services, GmbH, Germany. PK-sim and MoBi. Available online: https://www.open-systems-pharmacology.org/ (accessed on 23 September 2022).
- Eissing, T.; Kuepfer, L.; Becker, C.; Block, M.; Coboeken, K.; Gaub, T.; Goerlitz, L.; Jaeger, J.; Loosen, R.; Ludewig, B.; et al. A computational systems biology software platform for multiscale modeling and simulation: Integrating whole-body physiology, disease biology, and molecular reaction networks. Front. Physiol. 2011, 2, 4. [Google Scholar] [CrossRef]
- Willmann, S.; Lippert, J.; Sevestre, M.; Solodenko, J.; Fois, F.; Schmitt, W. PK-Sim®: A physiologically based pharmacokinetic ‘whole-body’ model. BIOSILICO 2003, 1, 121–124. [Google Scholar] [CrossRef]
- Kullenberg, F.; Degerstedt, O.; Calitz, C.; Pavlović, N.; Balgoma, D.; Gråsjö, J.; Sjögren, E.; Hedeland, M.; Heindryckx, F.; Lennernäs, H. In vitro cell toxicity and intracellular uptake of doxorubicin exposed as a solution or liposomes: Implications for treatment of hepatocellular carcinoma. Cells 2021, 10, 1717. [Google Scholar] [CrossRef]
- Julia. Available online: https://julialang.org (accessed on 14 July 2022).
- Beal, S.L., Sheiner, L.B. (University of California, USA), Bauer, R.J. (ICON Clinical Research, LLC, Ireland). NONMEM. Available online: https://www.iconplc.com/innovation/nonmem (accessed on 19 July 2022).
- Bauer, R.J. NONMEM tutorial part I: Description of commands and options, with simple examples of population analysis. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 525–537. [Google Scholar]
- Bauer, R.J. NONMEM tutorial part II: Estimation methods and advanced examples. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 538–556. [Google Scholar] [CrossRef] [PubMed]
- GNU Project, Free Software Foundation, Inc., USA. GNU MCSim. Available online: https://www.gnu.org/software/mcsim (accessed on 19 July 2022).
- Bois, F.Y. GNU MCSim: Bayesian statistical inference for SBML-coded systems biology models. Bioinformatics 2009, 25, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Elgrabli, D.; Beaudouin, R.; Jbilou, N.; Floriani, M.; Pery, A.; Rogerieux, F.; Lacroix, G. Biodistribution and clearance of TiO2 nanoparticles in rats after intravenous injection. PLoS ONE 2015, 10, e0124490. [Google Scholar] [CrossRef] [PubMed]
- Certara USA. Phoenix WiNonlin. Available online: https://www.certara.com/software/phoenix-winnonlin/ (accessed on 19 July 2022).
- Zazo, H.; Colino, C.I.; Gutiérrez-Millán, C.; Cordero, A.A.; Bartneck, M.; Lanao, J.M. Physiologically Based Pharmacokinetic (PBPK) Model of Gold Nanoparticle-Based Drug Delivery System for Stavudine Biodistribution. Pharmaceutics 2022, 14, 406. [Google Scholar] [CrossRef]
- GraphPad Software, Inc., USA. GraphPad Prism. Available online: https://www.graphpad.com/ (accessed on 19 July 2022).
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Decisioneering, Inc., USA (until March 2007); Hyperion, USA (until July 2007); Oracle, USA. Crystal Ball. Available online: https://www.oracle.com/applications/crystalball/ (accessed on 19 July 2022).
- AISN Software, Inc. (until 1990); Jandel Scientific Software, USA (until 1995); SPSS, Inc., USA (until 2004); Systat Software, Inc., USA. TableCurve 2D. Available online: https://systatsoftware.com/tablecurve2d/ (accessed on 14 July 2022).
- Moore, K. TableCurve 3.0. J. Chem. Inf. Comput. Sci. 1992, 32, 392. [Google Scholar] [CrossRef]
- Biomedical Simulations Resource (BMSR), USA. ADAPT. Available online: https://bmsr.usc.edu/software/adapt/ (accessed on 19 July 2022).
- Consortium of 16 European partners (from 12 EU countries: Cyprus, Denmark, Estonia, Finland, Germany, Greece, Ireland, Netherlands, Norway, Poland, Sweden and the UK) and 8 international partners from USA, Australia, South Africa, Japan and South Korea. NanoSolveIT. Available online: https://nanosolveit.eu/ (accessed on 19 July 2022).
- Afantitis, A.; Melagraki, G.; Isigonis, P.; Tsoumanis, A.; Varsou, D.D.; Valsami-Jones, E.; Papadiamantis, A.; Ellis, L.-J.A.; Sarimveis, H.; Doganis, P. NanoSolveIT Project: Driving nanoinformatics research to develop innovative and integrated tools for in silico nanosafety assessment. Comput. Struct. Biotechnol. J. 2020, 18, 583–602. [Google Scholar] [CrossRef]
- Tsiros, P.; Cheimarios, N.; Tsoumanis, A.; Jensen, A.Ø.; Melagraki, G.; Lynch, I.; Sarimveis, H.; Afantitis, A. Towards an in silico integrated approach for testing and assessment of nanomaterials: From predicted indoor air concentrations to lung dose and biodistribution. Environ. Sci. Nano 2022, 9, 1282–1297. [Google Scholar] [CrossRef]
- SPSS, Inc., USA (until 2009); IBM, USA. SPSS. Available online: https://www.ibm.com/products/spss-statistics (accessed on 19 July 2022).
- Microsoft, USA. Microsoft Excel. Available online: https://www.microsoft.com/en-us/microsoft-365/excel (accessed on 19 July 2022).
- Marino, D.J. Physiologically based pharmacokinetic modeling using microsoft excel and visual basic for applications. Toxicol. Mech. Methods 2005, 15, 137–154. [Google Scholar] [CrossRef]
- Bartels, M.; Rick, D.; Lowe, E.; Loizou, G.; Price, P.; Spendiff, M.; Arnold, S.; Cocker, J.; Ball, N. Development of PK-and PBPK-based modeling tools for derivation of biomonitoring guidance values. Comput. Methods Programs Biomed. 2012, 108, 773–788. [Google Scholar] [CrossRef]
- StatPac, Inc., USA. Statistics Calculator. Available online: https://statistics-calculator.software.informer.com/ (accessed on 29 September 2022).
- Bachler, G.; Losert, S.; Umehara, Y.; von Goetz, N.; Rodriguez-Lorenzo, L.; Petri-Fink, A.; Rothen-Rutishauser, B.; Hungerbuehler, K. Translocation of gold nanoparticles across the lung epithelial tissue barrier: Combining in vitro and in silico methods to substitute in vivo experiments. Part. Fibre Toxicol. 2015, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Minitab, LLC, USA. Minitab. Available online: https://www.minitab.com/en-us/ (accessed on 29 September 2022).
- COMSOL Inc., USA and other countries. COMSOL Multiphysics. Available online: https://www.comsol.com/ (accessed on 30 September 2022).
- OriginLab Corporation, USA. OriginPro. Available online: https://www.originlab.com/origin (accessed on 30 September 2022).
- Stevenson, K.J. Review of OriginPro 8.5. J. Am. Chem. Soc. 2011, 133, 5621. [Google Scholar] [CrossRef]
- Rohatgi, A. WebPlotDigitizer. Available online: https://automeris.io/WebPlotDigitizer/ (accessed on 21 July 2022).
- Huwaldt, J.A. PlotDigitizer. Available online: https://plot-digitizer.software.informer.com/ (accessed on 21 July 2022).
- Silk Scientific, Inc., USA. UN-SCAN-IT. Available online: https://www.silkscientific.com/graph-digitizer.htm (accessed on 21 July 2022).
- May, R.A.; Stevenson, K.J. Software review of UN-SCAN-IT: Graph digitizing software. J. Am. Chem. Soc. 2008, 130, 7516. [Google Scholar] [CrossRef]
- Lee, H.A.; Leavens, T.L.; Mason, S.E.; Monteiro-Riviere, N.A.; Riviere, J.E. Comparison of quantum dot biodistribution with a blood-flow-limited physiologically based pharmacokinetic model. Nano Lett. 2009, 9, 794–799. [Google Scholar] [CrossRef]
- Blinov, M.L.; Ruebenacker, O.; Moraru, I.I. Complexity and modularity of intracellular networks: A systematic approach for modelling and simulation. IET Syst. Biol. 2008, 2, 363–368. [Google Scholar] [CrossRef]
- Neal, M.L.; Cooling, M.T.; Smith, L.P.; Thompson, C.T.; Sauro, H.M.; Carlson, B.E.; Cook, D.L.; Gennari, J.H. A reappraisal of how to build modular, reusable models of biological systems. PLoS Comput. Biol. 2014, 10, e1003849. [Google Scholar] [CrossRef]
- Pan, M.; Gawthrop, P.J.; Cursons, J.; Crampin, E.J. Modular assembly of dynamic models in systems biology. PLoS Comput. Biol. 2021, 17, e1009513. [Google Scholar] [CrossRef]
- Oxford Instruments, PLC, USA. IMARIS. Available online: https://imaris.oxinst.com/), (accessed on 22 July 2022).
- NIH, USA. ImageJ. Available online: https://imagej.nih.gov/ij/ (accessed on 13 July 2022).
- Hu, T.; Xu, S.; Wei, L.; Zhang, X.; Wang, X. CellTracker: An automated toolbox for single-cell segmentation and tracking of time-lapse microscopy images. Bioinformatics 2021, 37, 285–287. [Google Scholar] [CrossRef]
- Creed, J.H.; Wilson, C.M.; Soupir, A.C.; Colin-Leitzinger, C.M.; Kimmel, G.J.; Ospina, O.E.; Chakiryan, N.H.; Markowitz, J.; Peres, L.C.; Coghill, A. spatialTIME and iTIME: R package and Shiny application for visualization and analysis of immunofluorescence data. Bioinformatics 2021, 37, 4584–4586. [Google Scholar] [CrossRef]
- Gómez-de-Mariscal, E.; García-López-de-Haro, C.; Ouyang, W.; Donati, L.; Lundberg, E.; Unser, M.; Muñoz-Barrutia, A.; Sage, D. DeepImageJ: A user-friendly environment to run deep learning models in ImageJ. Nat. Methods 2021, 18, 1192–1195. [Google Scholar]
- Schapiro, D.; Sokolov, A.; Yapp, C.; Chen, Y.-A.; Muhlich, J.L.; Hess, J.; Creason, A.L.; Nirmal, A.J.; Baker, G.J.; Nariya, M.K. MCMICRO: A scalable, modular image-processing pipeline for multiplexed tissue imaging. Nat. Methods 2022, 19, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Manz, T.; Gold, I.; Patterson, N.H.; McCallum, C.; Keller, M.S.; Herr, B.W.; Börner, K.; Spraggins, J.M.; Gehlenborg, N. Viv: Multiscale visualization of high-resolution multiplexed bioimaging data on the web. Nat. Methods 2022, 19, 515–516. [Google Scholar] [CrossRef] [PubMed]
- Dogra, P.; Ramírez, J.R.; Butner, J.D.; Peláez, M.J.; Chung, C.; Hooda-Nehra, A.; Pasqualini, R.; Arap, W.; Cristini, V.; Calin, G.A. Translational modeling identifies synergy between nanoparticle-delivered miRNA-22 and standard-of-care drugs in triple-negative breast cancer. Pharm. Res. 2022, 39, 511–528.136. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.; Carneiro, A.; Rosada, R.; Silva, C.; Santana, M. A mathematical model describing the kinetic of cationic liposome production from dried lipid films adsorbed in a multitubular system. Braz. J. Chem. Eng. 2007, 24, 477–486. [Google Scholar] [CrossRef][Green Version]
- Kiafar, F.; Shadbad, M.R.S.; Valizadeh, H. Filgrastim (G-CSF) loaded liposomes: Mathematical modeling and optimization of encapsulation efficiency and particle size. BioImpacts BI 2016, 6, 195. [Google Scholar] [CrossRef]
- Zhan, W.; Alamer, M.; Xu, X.Y. Computational modelling of drug delivery to solid tumour: Understanding the interplay between chemotherapeutics and biological system for optimised delivery systems. Adv. Drug Deliv. Rev. 2018, 132, 81–103. [Google Scholar] [CrossRef]
- Asemani, D.; Haemmerich, D. A unified mathematical model for nano-liposomal drug delivery to solid tumors. IEEE Trans. Nanobiosci. 2017, 17, 3–11. [Google Scholar] [CrossRef]
- Zhan, W.; Xu, X.Y. A mathematical model for thermosensitive liposomal delivery of doxorubicin to solid tumour. J. Drug Deliv. 2013, 2013, 172529. [Google Scholar] [CrossRef]
- Rezaeian, M.; Sedaghatkish, A.; Soltani, M. Numerical modeling of high-intensity focused ultrasound-mediated intraperitoneal delivery of thermosensitive liposomal doxorubicin for cancer chemotherapy. Drug Deliv. 2019, 26, 898–917. [Google Scholar] [CrossRef]
- Huang, Y.; Gu, B.; Liu, C.; Stebbing, J.; Gedroyc, W.; Thanou, M.; Xu, X.Y. Thermosensitive liposome-mediated drug delivery in chemotherapy: Mathematical modelling for spatio–temporal drug distribution and model-based optimisation. Pharmaceutics 2019, 11, 637. [Google Scholar] [CrossRef]
- Sedaghatkish, A.; Rezaeian, M.; Heydari, H.; Ranjbar, A.M.; Soltani, M. Acoustic streaming and thermosensitive liposomes for drug delivery into hepatocellular carcinoma tumor adjacent to major hepatic veins; an acoustics–thermal–fluid-mass transport coupling model. Int. J. Therm. Sci. 2020, 158, 106540. [Google Scholar] [CrossRef]
- Haghiralsadat, F.; Amoabediny, G.; Helder, M.N.; Naderinezhad, S.; Sheikhha, M.H.; Forouzanfar, T.; Zandieh-Doulabi, B. A comprehensive mathematical model of drug release kinetics from nano-liposomes, derived from optimization studies of cationic PEGylated liposomal doxorubicin formulations for drug-gene delivery. Artif. Cells Nanomed. Biotechnol. 2018, 46, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Korsmeyer, R.W.; Gurny, R.; Doelker, E.; Buri, P.; Peppas, N.A. Mechanisms of solute release from porous hydrophilic polymers. Int. J. Pharm. 1983, 15, 25–35. [Google Scholar]
- Arifin, D.Y.; Lee, L.Y.; Wang, C.-H. Mathematical modeling and simulation of drug release from microspheres: Implications to drug delivery systems. Adv. Drug Deliv. Rev. 2006, 58, 1274–1325. [Google Scholar] [CrossRef]
- Al Sawaftah, N.; Paul, V.; Awad, N.; Husseini, G.A. Modeling of anti-cancer drug release kinetics from liposomes and micelles: A review. IEEE Trans. Nanobiosci. 2021, 20, 565–576. [Google Scholar]
- Wadi, A.; Abdel-Hafez, M.; Husseini, G.A.; Paul, V. Multi-model investigation and adaptive estimation of the acoustic release of a model drug from liposomes. IEEE Trans. Nanobiosc. 2019, 19, 68–77. [Google Scholar] [CrossRef]
- Lu, T.; Ten Hagen, T.L. A novel kinetic model to describe the ultra-fast triggered release of thermosensitive liposomal drug delivery systems. J. Control. Release 2020, 324, 669–678. [Google Scholar] [CrossRef]
- Enden, G.; Schroeder, A. A mathematical model of drug release from liposomes by low frequency ultrasound. Ann. Biomed. Eng. 2009, 37, 2640–2645. [Google Scholar]
- Maojo, V.; Fritts, M.; de la Iglesia, D.; Cachau, R.E.; Garcia-Remesal, M.; Mitchell, J.A.; Kulikowski, C. Nanoinformatics: A new area of research in nanomedicine. Int. J. Nanomed. 2012, 7, 3867. [Google Scholar] [CrossRef]
- Barnard, A.; Motevalli, B.; Parker, A.; Fischer, J.; Feigl, C.; Opletal, G. Nanoinformatics, and the big challenges for the science of small things. Nanoscale 2019, 11, 19190–19201. [Google Scholar]
- Demina, P.A.; Sholina, N.V.; Akasov, R.A.; Khochenkov, D.A.; Arkharova, N.A.; Nechaev, A.V.; Khaydukov, E.V.; Generalova, A.N. A versatile platform for bioimaging based on colominic acid-decorated upconversion nanoparticles. Biomater. Sci. 2020, 8, 4570–4580. [Google Scholar] [CrossRef] [PubMed]
- Rocheva, V.; Savelyev, A.; Nechaev, A.; Generalova, A.; Semchishen, V.; Zvyagin, A.; Khaydukov, E. Three-dimensional luminescence tomographic visualization of biological tissues. Opt. Spectrosc. 2019, 126, 92–94. [Google Scholar] [CrossRef]
- Rudzińska, M.; Parodi, A.; Maslova, V.D.; Efremov, Y.M.; Gorokhovets, N.V.; Makarov, V.A.; Popkov, V.A.; Golovin, A.V.; Zernii, E.Y.; Zamyatnin, A.A. Cysteine cathepsins inhibition affects their expression and human renal cancer cell phenotype. Cancers 2020, 12, 1310. [Google Scholar] [CrossRef] [PubMed]
- Sevencan, C.; McCoy, R.S.A.; Ravisankar, P.; Liu, M.; Govindarajan, S.; Zhu, J.; Bay, B.H.; Leong, D.T. Cell membrane nanotherapeutics: From synthesis to applications emerging tools for personalized cancer therapy. Adv. Ther. 2020, 3, 1900201. [Google Scholar] [CrossRef]
- Vizirianakis, I.S.; Mystridis, G.A.; Avgoustakis, K.; Fatouros, D.G.; Spanakis, M. Enabling personalized cancer medicine decisions: The challenging pharmacological approach of PBPK models for nanomedicine and pharmacogenomics (Review). Oncol. Rep. 2016, 35, 1891–1904. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M. Frontiers in cancer nanomedicine: Directing mass transport through biological barriers. Trends Biotechnol. 2010, 28, 181–188. [Google Scholar] [CrossRef]
- Bachler, G.; von Goetz, N.; Hungerbühler, K. A physiologically based pharmacokinetic model for ionic silver and silver nanoparticles. Int. J. Nanomed. 2013, 8, 3365. [Google Scholar]
- Bachler, G.; von Goetz, N.; Hungerbuhler, K. Using physiologically based pharmacokinetic (PBPK) modeling for dietary risk assessment of titanium dioxide (TiO2) nanoparticles. Nanotoxicology 2015, 9, 373–380. [Google Scholar] [CrossRef]
- Deng, L.; Liu, H.; Ma, Y.; Miao, Y.; Fu, X.; Deng, Q. Endocytosis mechanism in physiologically-based pharmacokinetic modeling of nanoparticles. Toxicol. Appl. Pharmacol. 2019, 384, 114765. [Google Scholar]
- Rackauckas, C.; Nie, Q. Differentialequations. DifferentialEquations.jl—A performant and feature-rich ecosystem for solving differential equations in Julia. J. Open Res. Softw. 2017, 5, 15. [Google Scholar] [CrossRef]
- Giorgi, F.M.; Ceraolo, C.; Mercatelli, D. The R Language: An Engine for Bioinformatics and Data Science. Life 2022, 12, 648. [Google Scholar] [CrossRef] [PubMed]

| Criteria | MATLAB/ SimBiology | Berkeley Madonna | The R Language | acslX | BioUML | Simcyp Simulator | GastroPlus | PK-Sim/MoBi |
|---|---|---|---|---|---|---|---|---|
| Specialized PBPK software | – | – | – | – | – | + | + | + |
| General purpose software | + | + | + | + | + | – | – | + |
| Free | – | – | + | + | + | – | – | + |
| Open source | – | – | + | – | + | – | – | + |
| Currently supported | + | + | + | – | + | + | + | + |
| Stand-alone edition | + | + | + | + | + | + | + | + |
| Web-edition | – | – | – | – | + | + | + | – |
| Windows | + | + | + | + | + | + | + | + |
| Linux | + | – | + | – | + | – | – | – |
| MacOS | + | + | + | – | + | – | + 1 | – |
| Parallel computing | + | – | + | + | + | + | + | + |
| Requires programming skills | + | + | + | + | – | – | – | – |
| User-friendly interface | + | + | – | + | + | + | + | + |
| Interactive web-based interface of a model 2 | + | – | + | – | + | + | + | – |
| Visual modeling of the PBPK structure | + | – | – | – | + | – | + | + |
| Database of models | + | – | – | – | – | + | + | + |
| Model structural changes | + | + | + | + | + | – | – | + |
| Monte Carlo simulation | + | + | + | + | – | + | + | + |
| Parameter estimation | + | + | + | + | + | + | + | + |
| Sensitivity analysis | + | + | + | + | + | + | + | + |
| SBML support | + | – | + | + | + | – | – | + |
| Preferred for NPs 3 | + | + | + | + | + | – | – | + |
| Preferred for small molecules 3 | – | – | – | – | – | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kutumova, E.O.; Akberdin, I.R.; Kiselev, I.N.; Sharipov, R.N.; Egorova, V.S.; Syrocheva, A.O.; Parodi, A.; Zamyatnin, A.A., Jr.; Kolpakov, F.A. Physiologically Based Pharmacokinetic Modeling of Nanoparticle Biodistribution: A Review of Existing Models, Simulation Software, and Data Analysis Tools. Int. J. Mol. Sci. 2022, 23, 12560. https://doi.org/10.3390/ijms232012560
Kutumova EO, Akberdin IR, Kiselev IN, Sharipov RN, Egorova VS, Syrocheva AO, Parodi A, Zamyatnin AA Jr., Kolpakov FA. Physiologically Based Pharmacokinetic Modeling of Nanoparticle Biodistribution: A Review of Existing Models, Simulation Software, and Data Analysis Tools. International Journal of Molecular Sciences. 2022; 23(20):12560. https://doi.org/10.3390/ijms232012560
Chicago/Turabian StyleKutumova, Elena O., Ilya R. Akberdin, Ilya N. Kiselev, Ruslan N. Sharipov, Vera S. Egorova, Anastasiia O. Syrocheva, Alessandro Parodi, Andrey A. Zamyatnin, Jr., and Fedor A. Kolpakov. 2022. "Physiologically Based Pharmacokinetic Modeling of Nanoparticle Biodistribution: A Review of Existing Models, Simulation Software, and Data Analysis Tools" International Journal of Molecular Sciences 23, no. 20: 12560. https://doi.org/10.3390/ijms232012560
APA StyleKutumova, E. O., Akberdin, I. R., Kiselev, I. N., Sharipov, R. N., Egorova, V. S., Syrocheva, A. O., Parodi, A., Zamyatnin, A. A., Jr., & Kolpakov, F. A. (2022). Physiologically Based Pharmacokinetic Modeling of Nanoparticle Biodistribution: A Review of Existing Models, Simulation Software, and Data Analysis Tools. International Journal of Molecular Sciences, 23(20), 12560. https://doi.org/10.3390/ijms232012560









