Discovery of New Phenylacetone Monooxygenase Variants for the Development of Substituted Indigoids through Biocatalysis
Abstract
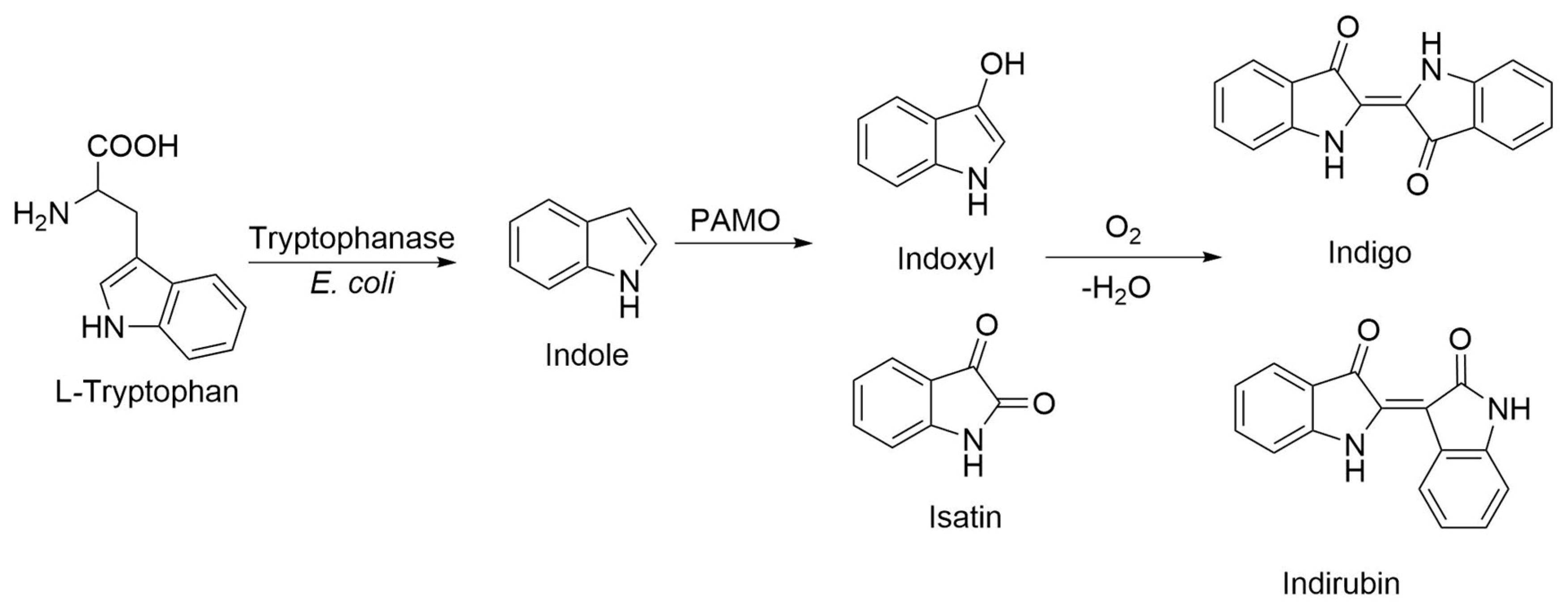
1. Introduction
2. Results
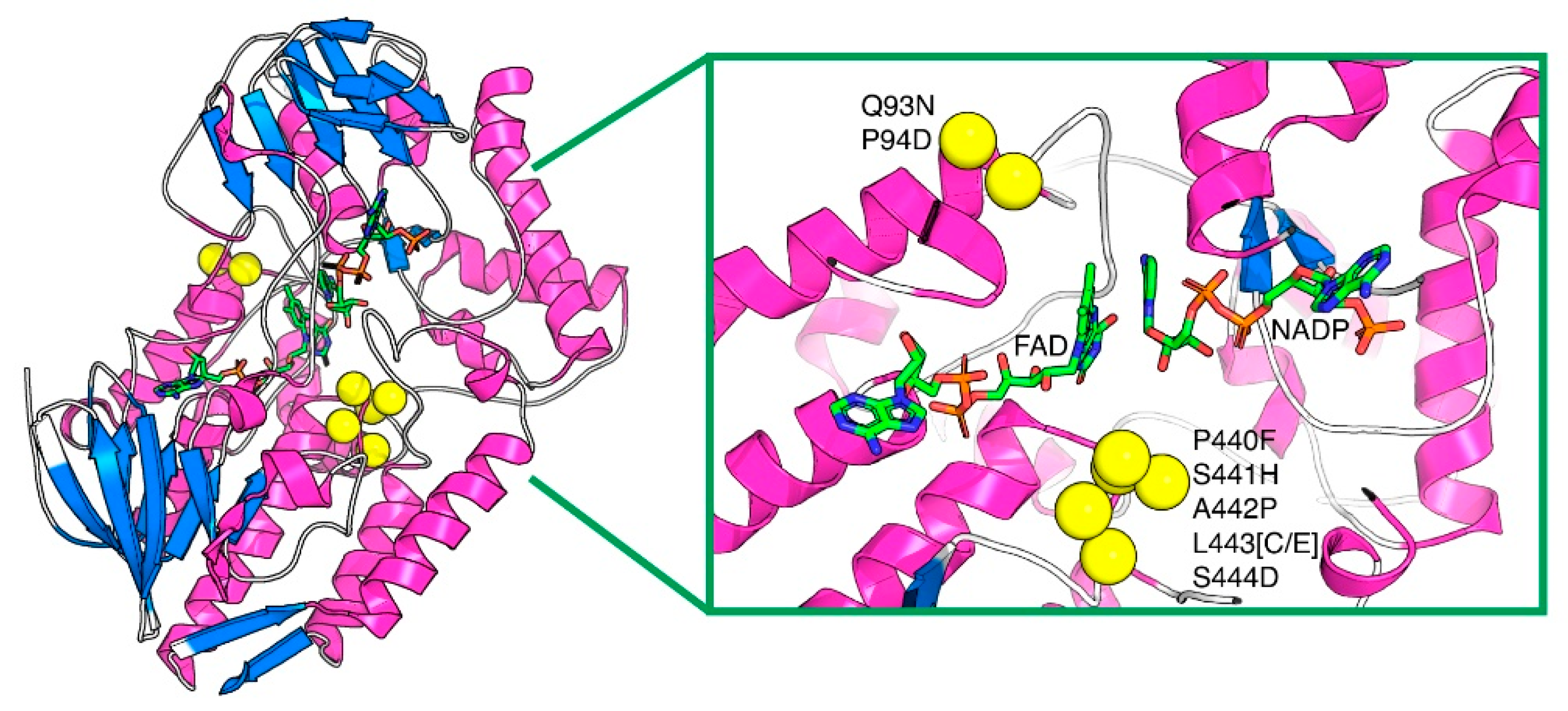
2.1. Thermostability of PAMO Variants
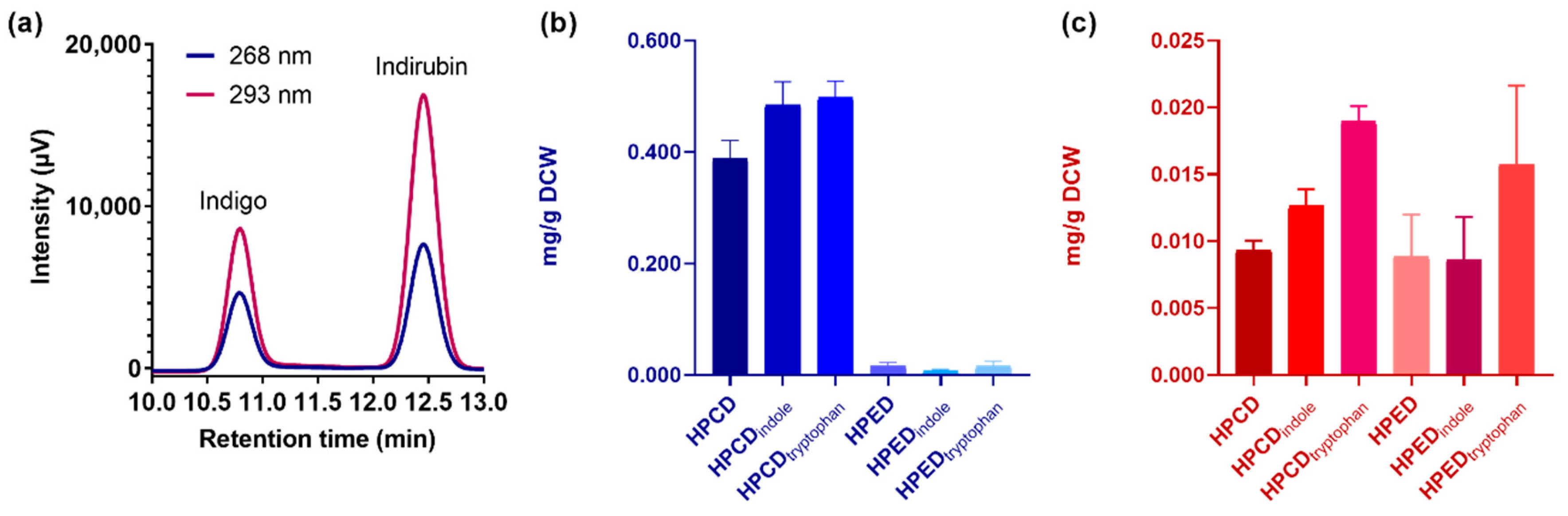
2.2. Structural and Quantitative Analysis of Whole-Cell Biosynthesis of Indigoids
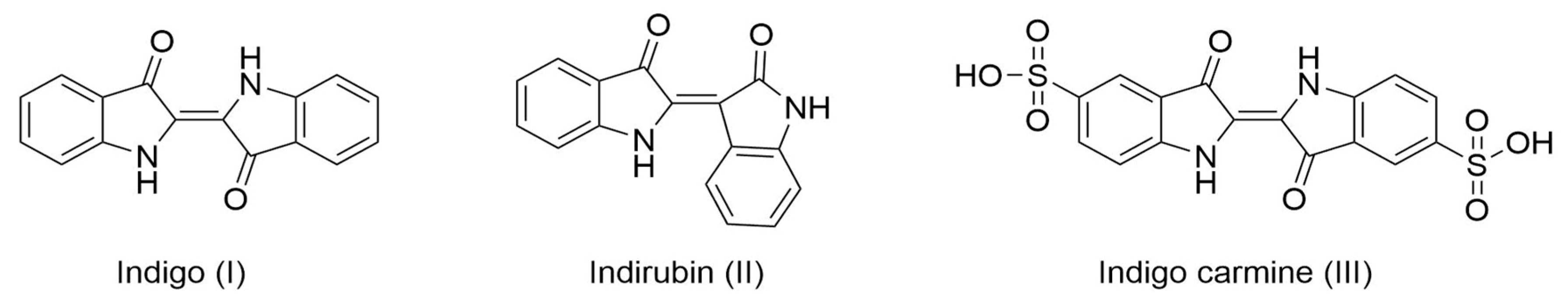
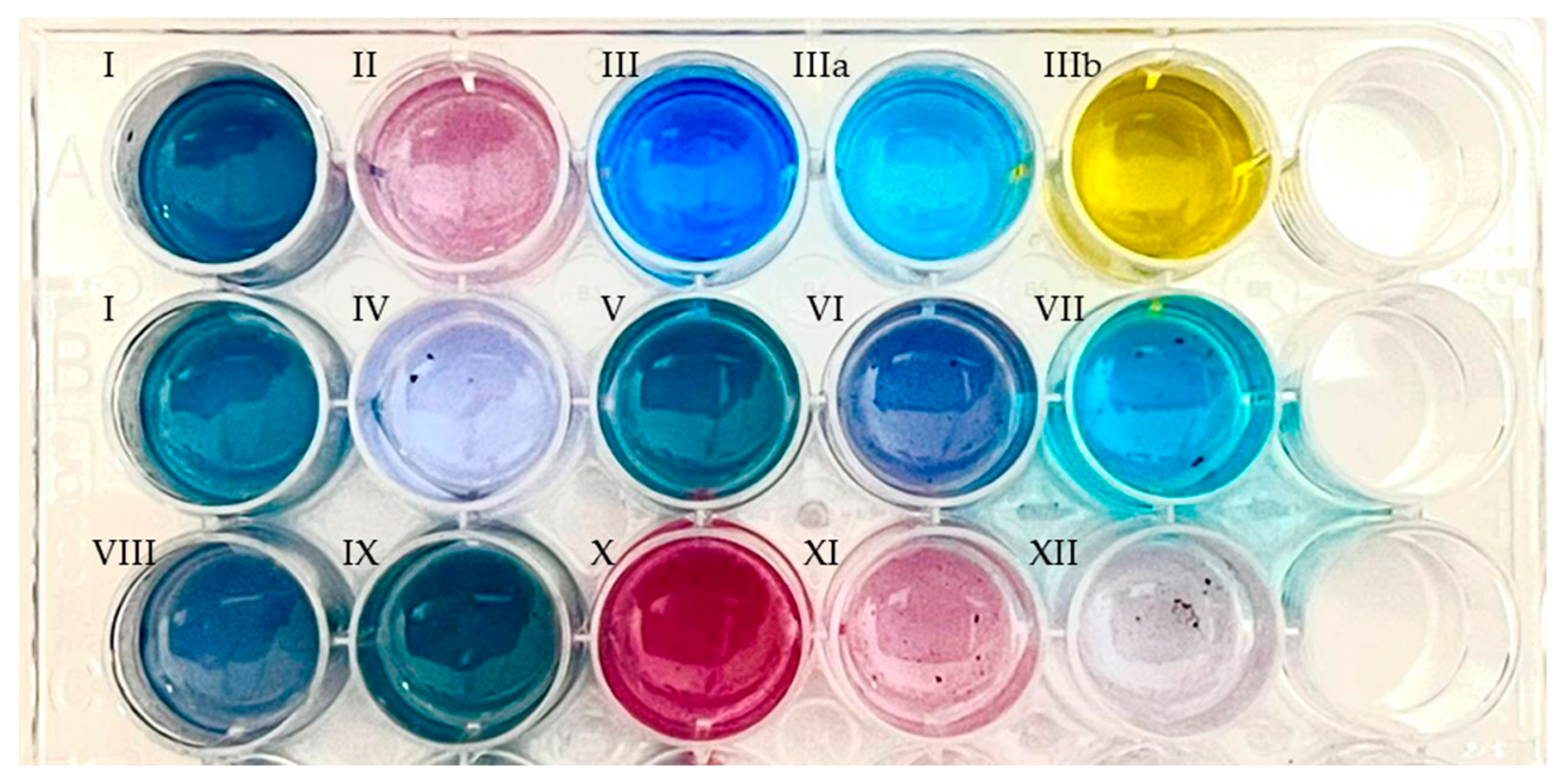
2.3. PAMOHPCD and PAMOHPED also Accept Substituted Indoles as Substrates
2.4. Steady-State Kinetic Parameters of the PAMO Variants
3. Discussion
4. Materials and Methods
4.1. Chemical and Reagents
4.2. Creation of PAMO Variants
4.3. Enzyme Characterization
4.4. Thermostability
4.5. Indigo Production Using E. coli Expressing PAMO Variants
4.6. Optimization of the Whole-Cell Reaction
4.7. Structural and Quantitative Analysis of Biosynthetically Produced Indigo
4.8. Steady-State Kinetics
4.9. Substituted Indoles Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balfour-Paul, J. Indigo: Egyptian Mummies to Blue Jeans; Firefly Books: Richmond Hill, ON, Canada, 2012; ISBN 1554079896. [Google Scholar]
- Yang, Q.-Y.; Zhang, T.; He, Y.-N.; Huang, S.-J.; Deng, X.; Han, L.; Xie, C.-G. From Natural Dye to Herbal Medicine: A Systematic Review of Chemical Constituents, Pharmacological Effects and Clinical Applications of Indigo Naturalis. Chin. Med. 2020, 15, 127. [Google Scholar] [CrossRef]
- Paul, J.B. Indigo and Blue: A Marriage Made in Heaven—ProQuest; University of Texas at Austin; University of Texas Press: Austin, TX, USA, 2020. [Google Scholar]
- Hsu, T.M.; Welner, D.H.; Russ, Z.N.; Cervantes, B.; Prathuri, R.L.; Adams, P.D.; Dueber, J.E. Employing a Biochemical Protecting Group for a Sustainable Indigo Dyeing Strategy. Nat. Chem. Biol. 2018, 14, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Głowacki, E.D.; Voss, G.; Sariciftci, N.S. 25th Anniversary Article: Progress in Chemistry and Applications of Functional Indigos for Organic Electronics. Adv. Mater. 2013, 25, 6783–6800. [Google Scholar] [CrossRef] [PubMed]
- Klimovich, I.V.; Zhilenkov, A.V.; Kuznetsova, L.I.; Frolova, L.A.; Yamilova, O.R.; Troyanov, S.I.; Lyssenko, K.A.; Troshin, P.A. Novel Functionalized Indigo Derivatives for Organic Electronics. Dye. Pigment. 2021, 186, 108966. [Google Scholar] [CrossRef]
- Gaitanis, G.; Magiatis, P.; Velegraki, A.; Bassukas, I.D. A Traditional Chinese Remedy Points to a Natural Skin Habitat: Indirubin (Indigo Naturalis) for Psoriasis and the Malassezia Metabolome. Br. J. Dermatol. 2018, 179, 800. [Google Scholar] [CrossRef]
- Min, G.-Y.; Kim, J.-H.; Kim, T.-I.; Cho, W.-K.; Yang, J.-H.; Ma, J.-Y. Indigo Pulverata Levis (Chung-Dae, Persicaria Tinctoria) Alleviates Atopic Dermatitis-like Inflammatory Responses In Vivo and In Vitro. Int. J. Mol. Sci. 2022, 23, 553. [Google Scholar] [CrossRef]
- Sugimoto, S.; Naganuma, M.; Kanai, T. Indole Compounds May Be Promising Medicines for Ulcerative Colitis. J. Gastroenterol. 2016, 51, 853–861. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, G.-X.; Niu, Y.-T.; Wang, Q.; Zheng, J.; Yang, J.-M.; Sun, T.; Niu, J.-G.; Yu, J.-Q. Anti-Inflammatory and Analgesic Activities of Indigo through Regulating the IKKβ/IκB/NF-ΚB Pathway in Mice. Food Funct. 2020, 11, 8537–8546. [Google Scholar] [CrossRef]
- Saling, P.; Kicherer, A.; Dittrich-Krämer, B.; Wittlinger, R.; Zombik, W.; Schmidt, I.; Schrott, W.; Schmidt, S. Eco-Efficiency Analysis by Basf: The Method. Int. J. Life Cycle Assess. 2002, 7, 203–218. [Google Scholar] [CrossRef]
- Wenda, S.; Illner, S.; Mell, A.; Kragl, U. Industrial Biotechnology—The Future of Green Chemistry? Green Chem. 2011, 13, 3007–3047. [Google Scholar] [CrossRef]
- Hartl, A.; Proaño Gaibor, A.N.; van Bommel, M.R.; Hofmann-de Keijzer, R. Searching for Blue: Experiments with Woad Fermentation Vats and an Explanation of the Colours through Dye Analysis. J. Archaeol. Sci. Rep. 2015, 2, 9–39. [Google Scholar] [CrossRef]
- Chen, T.; Wang, X.; Zhuang, L.; Shao, A.; Lu, Y.; Zhang, H. Development and Optimization of a Microbial Co-Culture System for Heterologous Indigo Biosynthesis. Microb. Cell Fact. 2021, 20, 154. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.; Lee, P.-G.; Kim, E.-J.; Kroutil, W.; Kim, B. Elucidating Cysteine-Assisted Synthesis of Indirubin by a Flavin-Containing Monooxygenase. ACS Catal. 2019, 9, 9539–9544. [Google Scholar] [CrossRef]
- Lin, Y.-K.; Leu, Y.-L.; Huang, T.-H.; Wu, Y.-H.; Chung, P.-J.; Su Pang, J.-H.; Hwang, T.-L. Anti-Inflammatory Effects of the Extract of Indigo Naturalis in Human Neutrophils. J. Ethnopharmacol. 2009, 125, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.; Madamwar, D. Biosynthesis of Indigo Dye by Newly Isolated Naphthalene-Degrading Strain Pseudomonas sp. HOB1 and its Application in Dyeing Cotton fabric. Appl. Biochem. Biotechnol. 2010, 160, 1616–1626. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Fan, J.; Zhang, Z.; Ma, Q.; Peng, X. Indigoids Biosynthesis from Indole by Two Phenol-Degrading Strains, Pseudomonas Sp. PI1 Acinetobacter Sp. PI2. Appl. Biochem. Biotechnol. 2015, 176, 1263–1276. [Google Scholar] [CrossRef]
- Liu, C.; Xu, J.; Gao, S.Q.; He, B.; Wei, C.W.; Wang, X.J.; Wang, Z.; Lin, Y.W. Green and Efficient Biosynthesis of Indigo from Indole by Engineered Myoglobins. RSC Adv. 2018, 8, 33325–33330. [Google Scholar] [CrossRef]
- Kim, H.J.; Jang, S.; Kim, J.; Yang, Y.H.; Kim, Y.G.; Kim, B.G.; Choi, K.Y. Biosynthesis of Indigo in Escherichia Coli Expressing Self-Sufficient CYP102A from Streptomyces Cattleya. Dye. Pigment. 2017, 140, 29–35. [Google Scholar] [CrossRef]
- Yin, H.; Chen, H.; Yan, M.; Li, Z.; Yang, R.; Li, Y.; Wang, Y.; Guan, J.; Mao, H.; Wang, Y.; et al. Efficient Bioproduction of Indigo and Indirubin by Optimizing a Novel Terpenoid Cyclase XiaI in Escherichia Coli. ACS Omega 2021, 6, 20569–20576. [Google Scholar] [CrossRef]
- Namgung, S.; Park, H.A.; Kim, J.; Lee, P.-G.; Kim, B.-G.; Yang, Y.-H.; Choi, K.-Y. Ecofriendly One-Pot Biosynthesis of Indigo Derivative Dyes Using CYP102G4 and PrnA Halogenase. Dye. Pigment. 2019, 162, 80–88. [Google Scholar] [CrossRef]
- Lončar, N.; van Beek, H.L.; Fraaije, M.W. Structure-Based Redesign of a Self-Sufficient Flavin-Containing Monooxygenase towards Indigo Production. Int. J. Mol. Sci. 2019, 20, 6148. [Google Scholar] [CrossRef] [PubMed]
- Mascotti, M.L.; Lapadula, W.J.; Juri Ayub, M. The Origin and Evolution of Baeyer—Villiger Monooxygenases (BVMOs): An Ancestral Family of Flavin Monooxygenases. PLoS ONE 2015, 10, e0132689. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, M.W.; Wu, J.; Heuts, D.P.H.M.; Van Hellemond, E.W.; Spelberg, J.H.L.; Janssen, D.B. Discovery of a Thermostable Baeyer-Villiger Monooxygenase by Genome Mining. Appl. Microbiol. Biotechnol. 2005, 66, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Torres Pazmiño, D.E.; Snajdrova, R.; Rial, D.V.; Mihovilovic, M.D.; Fraaije, M.W. Altering the Substrate Specificity and Enantioselectivity of Phenylacetone Monooxygenase by Structure-Inspired Enzyme Redesign. Adv. Synth. Catal. 2007, 349, 1361–1368. [Google Scholar] [CrossRef]
- Wu, S.; Acevedo, J.P.; Reetz, M.T. Induced Allostery in the Directed Evolution of an Enantioselective Baeyer–Villiger Monooxygenase. Proc. Natl. Acad. Sci. USA 2010, 107, 2775–2780. [Google Scholar] [CrossRef]
- Dudek, H.M.; Torres Pazmiño, D.E.; Rodríguez, C.; De Gonzalo, G.; Gotor, V.; Fraaije, M.W. Investigating the Coenzyme Specificity of Phenylacetone Monooxygenase from Thermobifida Fusca. Appl. Microbiol. Biotechnol. 2010, 88, 1135–1143. [Google Scholar] [CrossRef]
- Yang, G.; Cang, R.; Shen, L.-Q.; Xue, F.; Huang, H.; Zhang, Z.-G. Expanding the Substrate Scope of Phenylacetone Monooxygenase from Thermobifida Fusca towards Cyclohexanone by Protein Engineering. Catal. Commun. 2019, 119, 159–163. [Google Scholar] [CrossRef]
- Parra, L.P.; Acevedo, J.P.; Reetz, M.T. Directed Evolution of Phenylacetone Monooxygenase as an Active Catalyst for the Baeyer-Villiger Conversion of Cyclohexanone to Caprolactone. Biotechnol. Bioeng. 2015, 112, 1354–1364. [Google Scholar] [CrossRef]
- Johnson, C.M. Differential scanning calorimetry as a tool for protein folding and stability. Arch. Biochem. Biophys. 2013, 531, 100–109. [Google Scholar] [CrossRef]
- Li, G.; Young, K.D. Indole Production by the Tryptophanase TnaA in Escherichia Coli Is Determined by the Amount of Exogenous Tryptophan. Microbiology 2013, 159, 402–410. [Google Scholar] [CrossRef]
- Choi, H.S.; Kim, J.K.; Cho, E.H.; Kim, Y.C.; Kim, J.I.; Kim, S.W. A Novel Flavin-Containing Monooxygenase from Methylophaga Sp. Strain SK1 and Its Indigo Synthesis in Escherichia Coli. Biochem. Biophys. Res. Commun. 2003, 306, 930–936. [Google Scholar] [CrossRef]
- Ameria, S.P.L.; Jung, H.S.; Kim, H.S.; Han, S.S.; Kim, H.S.; Lee, J.H. Characterization of a Flavin-Containing Monooxygenase from Corynebacterium Glutamicum and Its Application to Production of Indigo and Indirubin. Biotechnol. Lett. 2015, 37, 1637–1644. [Google Scholar] [CrossRef] [PubMed]
- Han, G.H.; Gim, G.H.; Kim, W.; Seo, S.I.; Kim, S.W. Enhanced Indirubin Production in Recombinant Escherichia Coli Harboring a Flavin-Containing Monooxygenase Gene by Cysteine Supplementation. J. Biotechnol. 2013, 164, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Catucci, G.; Turella, S.; Cheropkina, H.; De Angelis, M.; Gilardi, G.; Sadeghi, S.J. Green Production of Indigo and Indirubin by an Engineered Baeyer–Villiger Monooxygenase. Biocatal. Agric. Biotechnol. 2022, 44, 102458. [Google Scholar] [CrossRef]
- Kim, H.; Kim, G.; Song, I.; Lee, J.; Abdullah, H.; Yang, C.; Oh, J.H. Ambipolar Organic Phototransistors Based on 6,6′-Dibromoindigo. RSC Adv. 2018, 8, 14747–14752. [Google Scholar] [CrossRef]
- McGovern, P.E.; Michel, R.H. Royal Purple Dye: The Chemical Reconstruction of the Ancient Mediterranean Industry. Acc. Chem. Res. 1990, 23, 152–158. [Google Scholar] [CrossRef]
- Guo, C.; Quinn, J.; Sun, B.; Li, Y. An Indigo-Based Polymer Bearing Thermocleavable Side Chains for n-Type Organic Thin Film Transistors. J. Mater. Chem. C 2015, 3, 5226–5232. [Google Scholar] [CrossRef]
- Niero, M.; Righetto, I.; Beneventi, E.; Polverino de Laureto, P.; Fraaije, M.W.; Filippini, F.; Bergantino, E. Unique Features of a New Baeyer–Villiger Monooxygenase from a Halophilic Archaeon. Catalysts 2020, 10, 128. [Google Scholar] [CrossRef]
- Torres Pazmiño, D.E.; Snajdrova, R.; Baas, B.-J.; Ghobrial, M.; Mihovilovic, M.D.; Fraaije, M.W. Self-Sufficient Baeyer-Villiger Monooxygenases: Effective Coenzyme Regeneration for Biooxygenation by Fusion Engineering. Angew. Chem. Int. Ed. Engl. 2008, 47, 2275–2278. [Google Scholar] [CrossRef]
- Lee, J.; Kim, J.; Song, J.E.; Song, W.-S.; Kim, E.-J.; Kim, Y.-G.; Jeong, H.-J.; Kim, H.R.; Choi, K.-Y.; Kim, B.-G. Production of Tyrian Purple Indigoid Dye from Tryptophan in Escherichia Coli. Nat. Chem. Biol. 2021, 17, 104–112. [Google Scholar] [CrossRef]
- Reetz, M.T.; Wu, S. Laboratory Evolution of Robust and Enantioselective Baeyer−Villiger Monooxygenases for Asymmetric Catalysis. J. Am. Chem. Soc. 2009, 131, 15424–15432. [Google Scholar] [CrossRef] [PubMed]
- Hogrefe, H.H.; Cline, J.; Youngblood, G.L.; Allen, R.M. Creating Randomized Amino Acid Libraries with the QuikChange ® Multi Site-Directed Mutagenesis Kit. Biotechniques 2002, 33, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Kille, S.; Acevedo-Rocha, C.G.; Parra, L.P.; Zhang, Z.-G.; Opperman, D.J.; Reetz, M.T.; Acevedo, J.P. Reducing Codon Redundancy and Screening Effort of Combinatorial Protein Libraries Created by Saturation Mutagenesis. ACS Synth. Biol. 2013, 2, 83–92. [Google Scholar] [CrossRef] [PubMed]
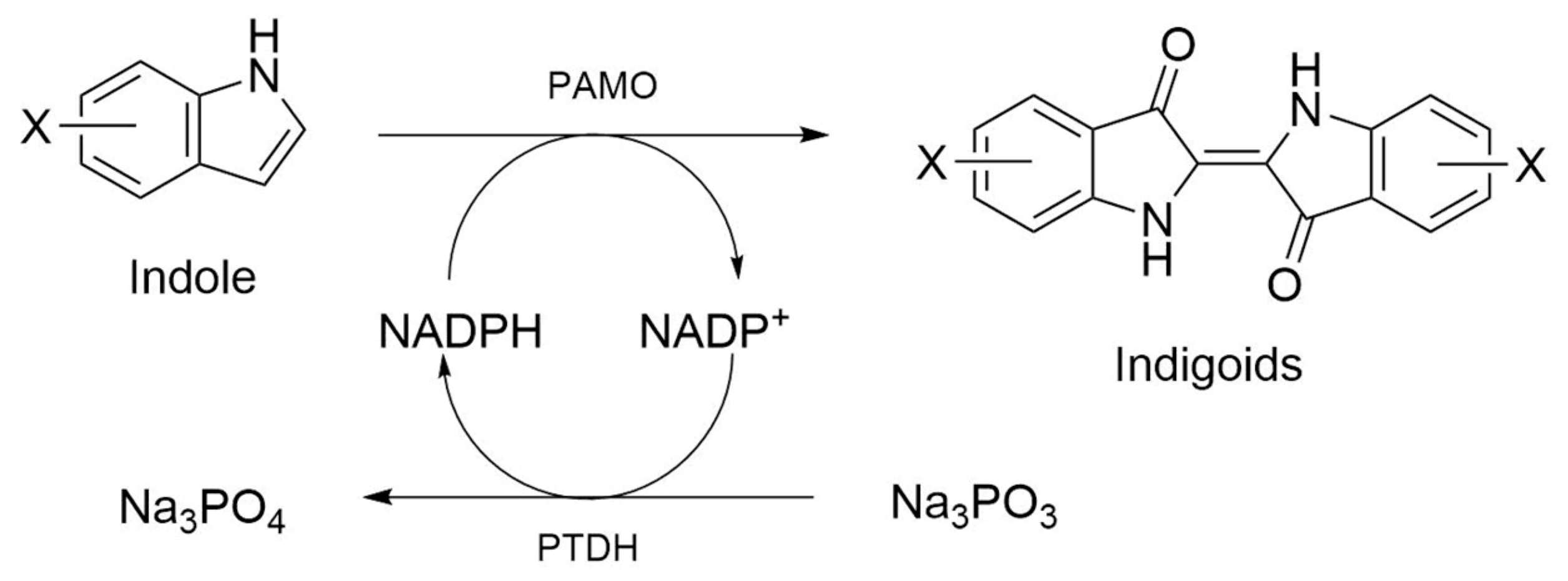
| HPCD | HPCDindole | HPCDtryptophan | HPED | HPEDindole | HPEDtryptophan | |
|---|---|---|---|---|---|---|
| Indigo (mg/g DCW) | 0.388 ± 0.009 | 0.484 ± 0.125 | 0.498 ± 0.024 | 0.015 ± 0.006 | 0.007 ± 0.002 | 0.016 ± 0.007 |
| Indirubin (mg/g DCW) | 0.009 ± 0.001 | 0.013 ± 0.001 | 0.019 ± 0.001 | 0.008 ± 0.002 | 0.009 ± 0.001 | 0.016 ± 0.001 |
| Substrate | HPCD | HPED | Product * | Exact Mass (g mol−1) | Product Nomenclature |
|---|---|---|---|---|---|
| Indole | + | + | Indigo | 262.0742 | I |
| 5-Cyanoindole | + | + | 5,5′-dicyanoindigo | 312.0647 | IV |
| 5-Fluroindole | + | + | 5,5′-difluoroindigo | 298.0554 | V |
| 5-Chloroindole | + | + | 5,5′-dichloroindigo | 329.9963 | VI |
| 5-Hidroxyindol | + | + | 5,5′-hydroxyindigo | 296.0641 | VII |
| 5-Methylindol | + | + | 5,5′-dimethylindigo | 290.1055 | VIII |
| 5-Methoxyindol | + | + | 5,5′-dimethoxyindigo | 322.0954 | IX |
| 6-Fluoroindole | + | + | 6,6′-difluoroindigo | 298.0554 | X |
| 6-Chloroindole | + | + | 6,6′-dichloroindigo | 329.9963 | XI |
| 6-Bromoindole | + | − | 6,6′-dibromoindigo | 417.8953 | XII |
| 5-Nitroindole | − | − | - | - | - |
| 5-Iodoindole | − | − | - | - | - |
| 5-(Benzyloxy)Indole | − | − | - | - | - |
| Indole-5-carboxaldehyde | − | − | - | - | - |
| NADPH | Indole | |||||
|---|---|---|---|---|---|---|
| Variant | kcat (s−1) | Km (µM) | kcat/Km (s−1M−1) | kcat (s−1) | Km (μM) | kcat/Km (s−1M−1) |
| PAMOWT | 4.8 | 0.9 | 5,200,000 | - | - | - |
| PAMOHPCD | 6.8 | 7.6 | 900,000 | 5.4 | 58.2 | 93,000 |
| PAMOHPED | 1.9 | 4.4 | 430,000 | 1.7 | 109.0 | 16,000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez-Navarro, N.; Salazar Muñoz, J.; Castillo, F.; Ramírez-Sarmiento, C.A.; Poblete-Castro, I.; Zacconi, F.C.; Parra, L.P. Discovery of New Phenylacetone Monooxygenase Variants for the Development of Substituted Indigoids through Biocatalysis. Int. J. Mol. Sci. 2022, 23, 12544. https://doi.org/10.3390/ijms232012544
Núñez-Navarro N, Salazar Muñoz J, Castillo F, Ramírez-Sarmiento CA, Poblete-Castro I, Zacconi FC, Parra LP. Discovery of New Phenylacetone Monooxygenase Variants for the Development of Substituted Indigoids through Biocatalysis. International Journal of Molecular Sciences. 2022; 23(20):12544. https://doi.org/10.3390/ijms232012544
Chicago/Turabian StyleNúñez-Navarro, Nicolás, Javier Salazar Muñoz, Francisco Castillo, César A. Ramírez-Sarmiento, Ignacio Poblete-Castro, Flavia C. Zacconi, and Loreto P. Parra. 2022. "Discovery of New Phenylacetone Monooxygenase Variants for the Development of Substituted Indigoids through Biocatalysis" International Journal of Molecular Sciences 23, no. 20: 12544. https://doi.org/10.3390/ijms232012544
APA StyleNúñez-Navarro, N., Salazar Muñoz, J., Castillo, F., Ramírez-Sarmiento, C. A., Poblete-Castro, I., Zacconi, F. C., & Parra, L. P. (2022). Discovery of New Phenylacetone Monooxygenase Variants for the Development of Substituted Indigoids through Biocatalysis. International Journal of Molecular Sciences, 23(20), 12544. https://doi.org/10.3390/ijms232012544







