Quenching of Protein Fluorescence by Fullerenol C60(OH)36 Nanoparticles
Abstract
1. Introduction
2. Results
2.1. Characterization of C60(OH)36 Aqueous Suspensions
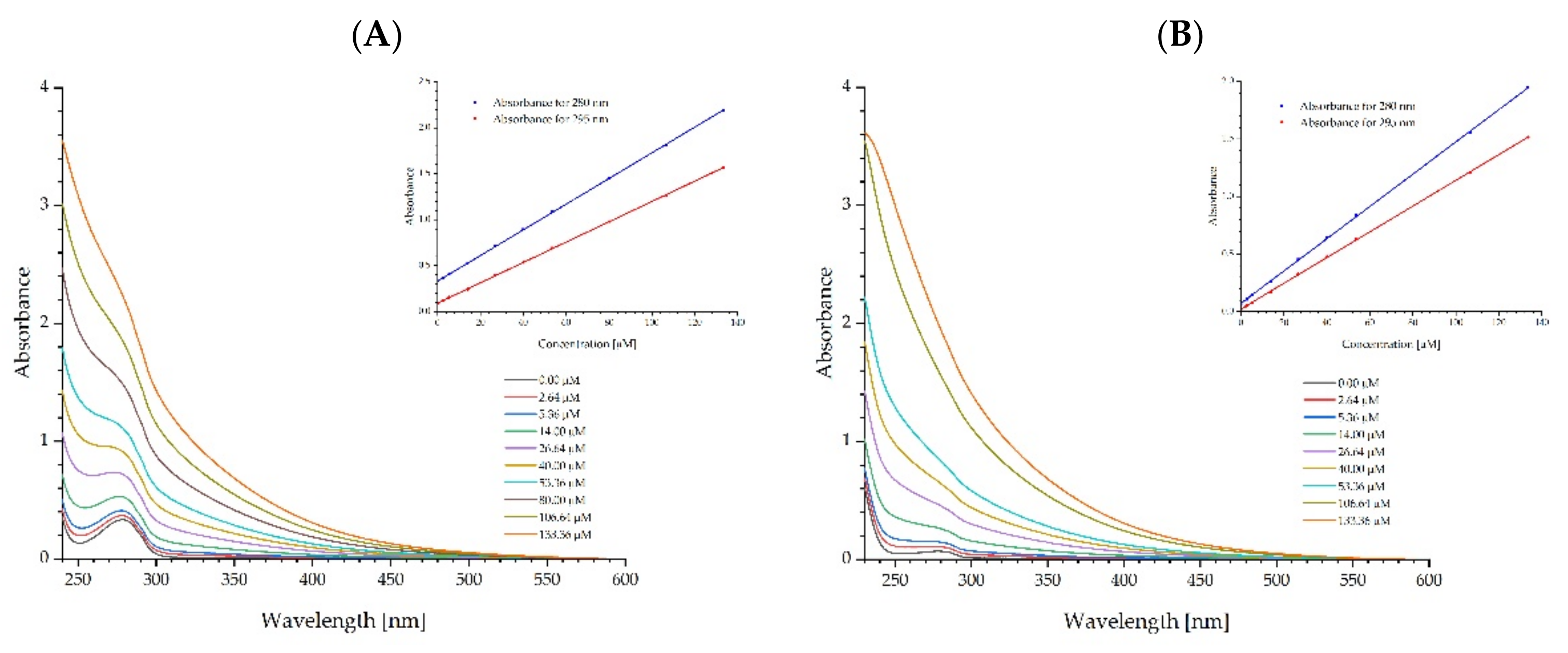
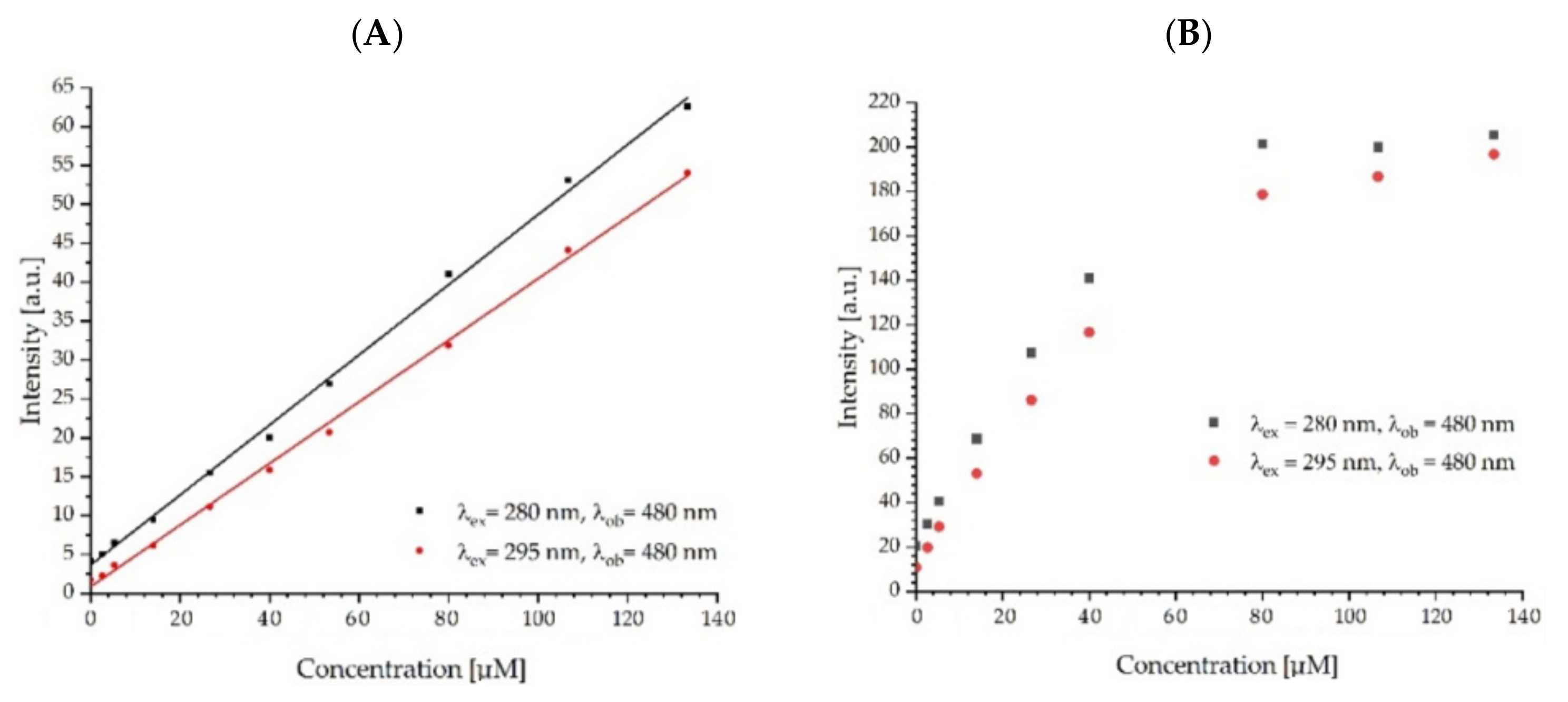
2.2. Spectral Characteristics of Fullerenol
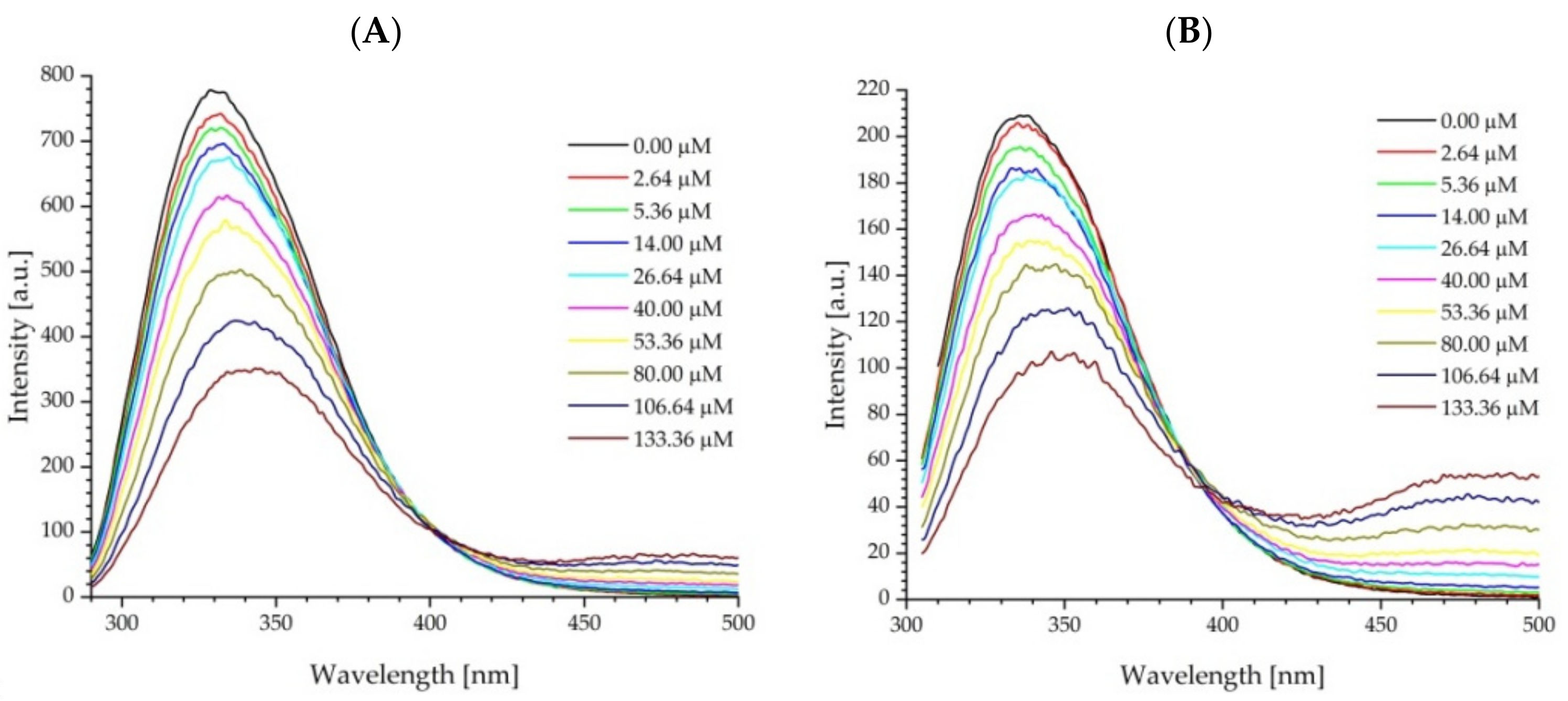
2.3. Protein Fluorescence Quenching by Fullerenol
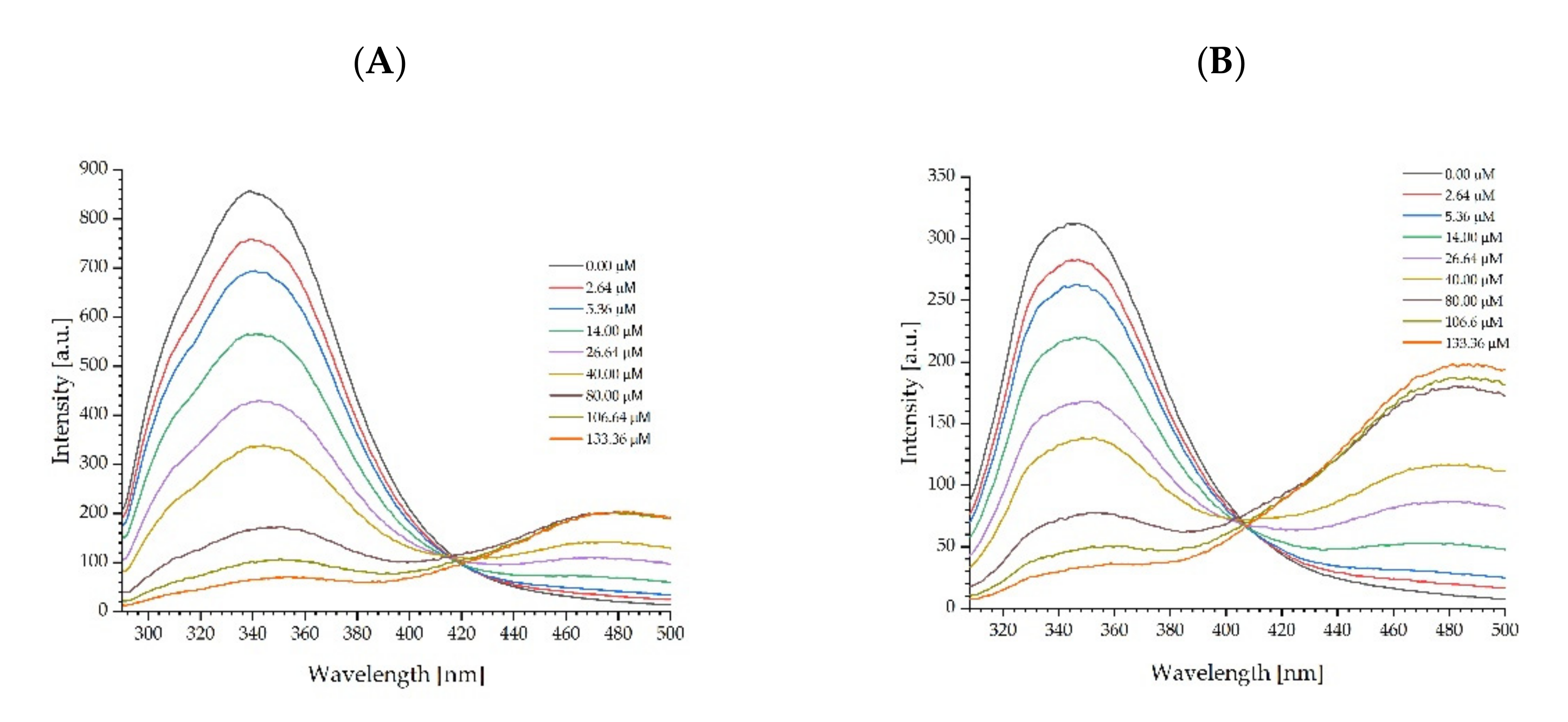
2.4. Mechanism of Fluorescence Quenching by Fullerenol
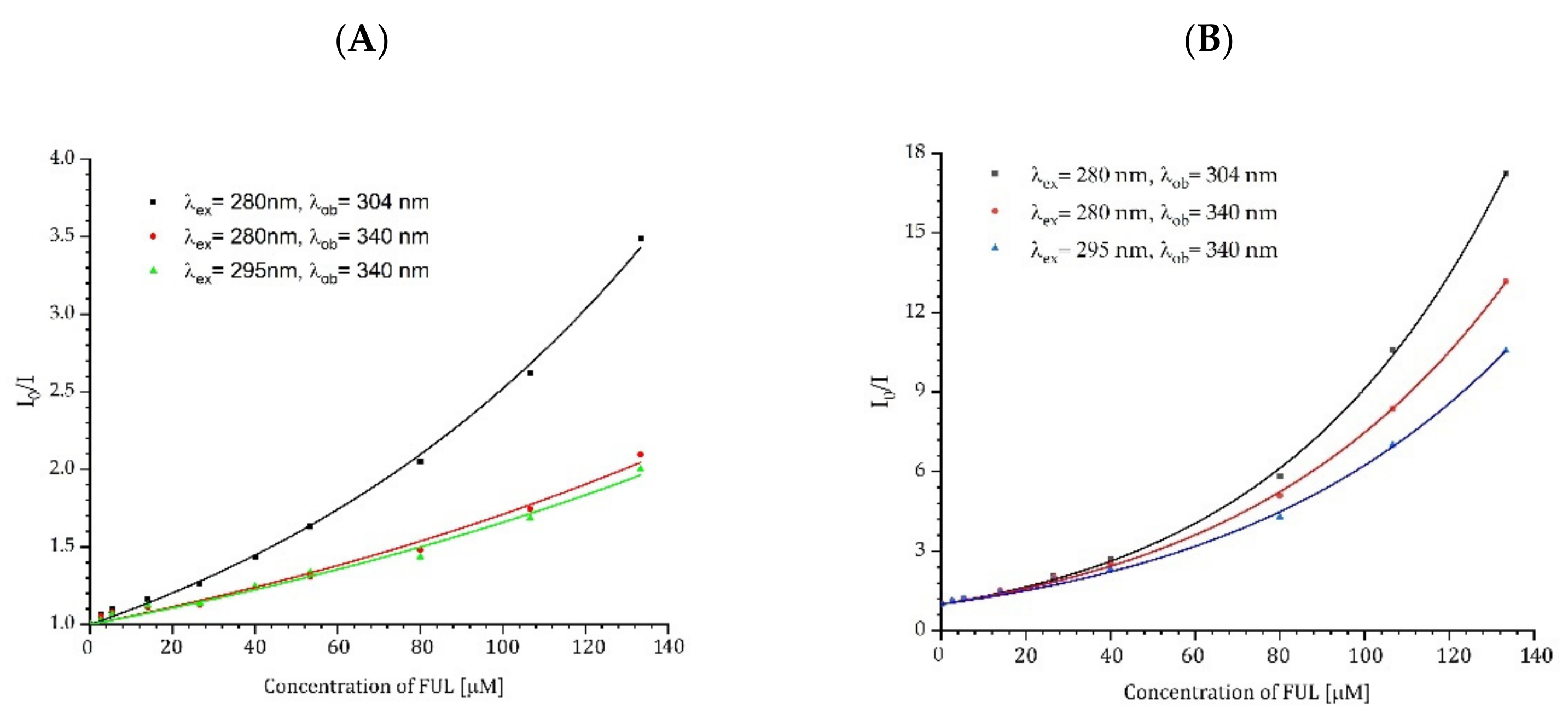
2.5. Determination of the Binding Parameters
2.6. Energy Transfer
3. Discussion
4. Materials and Methods
4.1. Reagents and Materials
4.2. Methods
4.2.1. Synthesis of Fullerenol Nanoparticles
4.2.2. Nanoparticle Tracking Analysis
4.2.3. ζ-Potential Measurements
4.2.4. Preparation of the Sample of Fullerenol with Proteins
4.2.5. Spectroscopic Measurements
4.2.6. Lifetime Measurements
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Russ, K.A.; Elvati, P.; Parsonage, T.L.; Dews, A.; Jarvis, J.A.; Ray, M.; Schneider, B.; Smith, P.J.S.; Williamson, P.T.F.; Violi, A.; et al. C60 fullerene localization and membrane interactions in RAW 264.7 immortalized mouse macrophages. Nanoscale 2016, 8, 4134–4144. [Google Scholar] [CrossRef] [PubMed]
- Torres, V.M.; Posa, M.; Srdjenovic, B.; Simplício, A.L. Solubilization of fullerene C60 in micellar solutions of different solubilizers. Colloids Surf. B BioInterfaces 2011, 82, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Bai, Y.; Mu, Y.; Pan, B.; Xing, B.; Lin, Y. Fluorescence quenching of fulvic acids by fullerene in water. Environ. Pollut. 2013, 172, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Ito, O.; D’Souza, F. Recent advances in photoinduced electron transfer processes of fullerene-based molecular assemblies and nanocomposites. Molecules 2012, 17, 5816–5835. [Google Scholar] [CrossRef]
- Brites, M.J.; Santos, C.; Nascimento, S.; Gigante, B.; Luftmann, H.; Fedorov, A.; Berberan-Santos, M.N. Synthesis and fluorescence properties of [60] and [70]fullerene-coumarin dyads: Efficient dipole-dipole resonance energy transfer from coumarin to fullerene. New J. Chem. 2006, 30, 1036–1045. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, Y.; Fang, Y. Quenching and enhancement of fluorescence of fullerene molecules on gold particle. Chem. Phys. 2006, 323, 169–172. [Google Scholar] [CrossRef]
- Djordjevic, A.; Šojić Merkulov, D.; Lazarević, M.; Borišev, I.; Medić, I.; Pavlović, V.; Miljević, B.; Abramović, B. Enhancement of nano titanium dioxide coatings by fullerene and polyhydroxy fullerene in the photocatalytic degradation of the herbicide mesotrione. Chemosphere 2018, 196, 145–152. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Arinishi, M.; Katayama, T.; Miyasaka, H.; Asahi, T. Femtosecond excited-state dynamics of fullerene-C60 nanoparticles in water. Phys. Chem. Chem. Phys. 2018, 20, 958–966. [Google Scholar] [CrossRef]
- Indeglia, P.A.; Georgieva, A.T.; Krishna, V.B.; Martyniuk, C.J.; Bonzongo, J.-C.C.J.C.J. Toxicity of functionalized fullerene and fullerene synthesis chemicals. Chemosphere 2018, 207, 1–9. [Google Scholar] [CrossRef]
- Kovel, E.S.; Sachkova, A.S.; Vnukova, N.G.; Churilov, G.N.; Knyazeva, E.M.; Kudryasheva, N.S. Antioxidant activity and toxicity of fullerenols via bioluminescence signaling: Role of oxygen substituents. Int. J. Mol. Sci. 2019, 20, 2324. [Google Scholar] [CrossRef]
- Rokitskaya, T.I.; Antonenko, Y.N. Fullerenol C60(OH)24 increases ion permeability of lipid membranes in a pH-dependent manner. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1165–1174. [Google Scholar] [CrossRef]
- Sharoyko, V.V.; Iamalova, N.R.; Ageev, S.V.; Meshcheriakov, A.A.; Iurev, G.O.; Petrov, A.V.; Nerukh, D.A.; Farafonov, V.S.; Vasina, L.V.; Penkova, A.V.; et al. In Vitro and In Silico Investigation of Water-Soluble Fullerenol C 60 (OH) 24: Bioactivity and Biocompatibility. J. Phys. Chem. B 2021, 125, 9197–9212. [Google Scholar] [CrossRef]
- Di Costanzo, L.; Geremia, S. Atomic Details of Carbon-Based Nanomolecules Interacting with Proteins. Molecules 2020, 25, 3555. [Google Scholar] [CrossRef]
- Ye, L.; Kollie, L.; Liu, X.; Guo, W.; Ying, X.; Zhu, J.; Yang, S.; Yu, M. Antitumor Activity and Potential Mechanism of Novel Fullerene Derivative Nanoparticles. Molecules 2021, 26, 3252. [Google Scholar] [CrossRef]
- Minami, K.; Song, J.; Shrestha, L.K.; Ariga, K. Nanoarchitectonics for fullerene biology. Appl. Mater. Today 2021, 23, 100989. [Google Scholar] [CrossRef]
- Feng, J.; Mack, S.; Shan, G.; Gee, S.; Hammock, B.D.; Kennedy, I.M. C 60 nanoparticle quenching used as a biological label. Genet. Eng. Opt. Probes Biomed. Appl. 2003, 4967, 156. [Google Scholar] [CrossRef]
- Andreoni, A.; Nardo, L.; Bondani, M.; Zhao, B.; Roberts, J.E. Time-resolved fluorescence studies of fullerene derivatives. J. Phys. Chem. B 2013, 117, 7203–7209. [Google Scholar] [CrossRef]
- Chun-Mao Lin; Tan-Yi Lu C60 Fullerene Derivatized Nanoparticles and their Application to Therapeutics. Recent Pat. Nanotechnol. 2012, 6, 105–113. [CrossRef]
- Nepomnyashchaya, E.; Savchenko, E.; Velichko, E.; Aksenov, E. Investigation of albumin-fullerenol interaction using laser correlation spectroscopy: The algorithm. J. Biomed. Photonics Eng. 2016, 2, 040309. [Google Scholar] [CrossRef]
- Khan, A.A.; Shrivastava, A.; Khurshid, M. Normal to cancer microbiome transformation and its implication in cancer diagnosis. Biochim. Biophys. Acta Rev. Cancer 2012, 1826, 331–337. [Google Scholar] [CrossRef]
- Sitar, M.E.; Aydin, S.; Çakatay, U. Human serum albumin and its relation with oxidative stress. Clin. Lab. 2013, 59, 945–952. [Google Scholar] [CrossRef]
- Vayá, I.; Bonancía, P.; Jiménez, M.C.; Markovitsi, D.; Gustavsson, T.; Miranda, M.A. Excited state interactions between flurbiprofen and tryptophan in drug-protein complexes and in model dyads. Fluorescence studies from the femtosecond to the nanosecond time domains. Phys. Chem. Chem. Phys. 2013, 15, 4727–4734. [Google Scholar] [CrossRef]
- Ryan, A.J.; Ghuman, J.; Zunszain, P.A.; Chung, C.W.; Curry, S. Structural basis of binding of fluorescent, site-specific dansylated amino acids to human serum albumin. J. Struct. Biol. 2011, 174, 84–91. [Google Scholar] [CrossRef]
- Khan, A.Y.; Hossain, M.; Suresh Kumar, G. Investigations on the interaction of the phototoxic alkaloid coralyne with serum albumins. Chemosphere 2012, 87, 775–781. [Google Scholar] [CrossRef]
- Lee, P.; Wu, X. Review: Modifications of Human Serum Albumin and their Binding Effect. Curr. Pharm. Des. 2015, 21, 1862–1865. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, X.; Huang, J.; Wang, H. Probing the Interaction between Human Serum Albumin and 9-Hydroxyphenanthrene: A Spectroscopic and Molecular Docking Study. ACS Omega 2020, 5, 16833–16840. [Google Scholar] [CrossRef]
- Zsila, F. Subdomain IB Is the Third Major Drug Binding Region of Human Serum Albumin: Toward the Three-Sites Model. Mol. Pharm. 2013, 10, 1668–1682. [Google Scholar] [CrossRef]
- Chaves, O.; Ferreira, R.; da Silva, L.; de Souza, B.; Cesarin-Sobrinho, D.; Netto-Ferreira, J.C.; Sant’Anna, C.; Ferreira, A. Multiple Spectroscopic and Theoretical Approaches to Study the Interaction between HSA and the Antiparasitic Drugs: Benznidazole, Metronidazole, Nifurtimox and Megazol. J. Braz. Chem. Soc. 2018, 29, 1551–1562. [Google Scholar] [CrossRef]
- Valojerdi, F.M.; Farasat, A.; Shariatifar, H.; Gheibi, N. Study of HSA interactions with arachidonic acid using spectroscopic methods revealing molecular dynamics of HSA-AA interactions. Biomed. Rep. 2019, 12, 125–133. [Google Scholar] [CrossRef]
- Sharoyko, V.V.; Ageev, S.V.; Meshcheriakov, A.A.; Akentiev, A.V.; Noskov, B.A.; Rakipov, I.T.; Charykov, N.A.; Kulenova, N.A.; Shaimardanova, B.K.; Podolsky, N.E.; et al. Physicochemical study of water-soluble C60(OH)24 fullerenol. J. Mol. Liq. 2020, 311, 113360. [Google Scholar] [CrossRef]
- Yang, L.-Y.; Hua, S.-Y.; Zhou, Z.-Q.; Wang, G.-C.; Jiang, F.-L.; Liu, Y. Characterization of fullerenol-protein interactions and an extended investigation on cytotoxicity. Colloids Surf. B Biointerfaces 2017, 157, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Raj, S.B.; Ramaswamy, S.; Plapp, B.V. Yeast Alcohol Dehydrogenase Structure and Catalysis. Biochemistry 2014, 53, 5791–5803. [Google Scholar] [CrossRef] [PubMed]
- de Smidt, O.; du Preez, J.C.; Albertyn, J. The alcohol dehydrogenases of Saccharomyces cerevisiae: A comprehensive review. FEMS Yeast Res. 2008, 8, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-F.; Xu, Z.-Q.; Ge, Y.-S.; Jiang, F.-L.; Liu, Y. Binding of fullerol to human serum albumin: Spectroscopic and electrochemical approach. J. Photochem. Photobiol. B Biol. 2012, 108, 34–43. [Google Scholar] [CrossRef]
- Leonis, G.; Avramopoulos, A.; Papavasileiou, K.D.; Reis, H.; Steinbrecher, T.; Papadopoulos, M.G. A Comprehensive Computational Study of the Interaction between Human Serum Albumin and Fullerenes. J. Phys. Chem. B 2015, 119, 14971–14985. [Google Scholar] [CrossRef]
- Gross, J.; Sayle, S.; Karow, A.R.; Bakowsky, U.; Garidel, P. Nanoparticle tracking analysis of particle size and concentration detection in suspensions of polymer and protein samples: Influence of experimental and data evaluation parameters. Eur. J. Pharm. Biopharm. 2016, 104, 30–41. [Google Scholar] [CrossRef]
- Groza, R.C.; Li, B.; Ryder, A.G. Anisotropy resolved multidimensional emission spectroscopy (ARMES): A new tool for protein analysis. Anal. Chim. Acta 2015, 886, 133–142. [Google Scholar] [CrossRef][Green Version]
- De Bertozo, L.C.; Tavares Neto, E.; de Oliveira, L.C.; Ximenes, V.F. Oxidative alteration of Trp-214 and Lys-199 in human serum albumin increases binding affinity with phenylbutazone: A combined experimental and computational investigation. Int. J. Mol. Sci. 2018, 19, 2868. [Google Scholar] [CrossRef]
- Plapp, B.V.; Savarimuthu, B.R.; Ramaswamy, S. Yeast alcohol dehydrogenase I, Saccharomyces Cerevisiae fermentative enzyme. 2014, 53, 5791–5803. PDB. [CrossRef]
- Sugio, S.; Mochizuki, S.; Noda, M.; Kashima, A. Crystal structure of human serum albumin. Nat. Struct. Mol. Biol. 1997, 5, 827–835. [Google Scholar] [CrossRef]
- Kasparek, A.; Smyk, B. A new approach to the old problem: Inner filter effect type I and II in fluorescence. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 198, 297–303. [Google Scholar] [CrossRef]
- Kimball, J.; Chavez, J.; Ceresa, L.; Kitchner, E.; Nurekeyev, Z.; Doan, H.; Szabelski, M.; Borejdo, J.; Gryczynski, I.; Gryczynski, Z. On the origin and correction for inner filter effects in fluorescence Part I: Primary inner filter effect-the proper approach for sample absorbance correction. Methods Appl. Fluoresc. 2020, 8, 033002. [Google Scholar] [CrossRef]
- Principles of Fluorescence Spectroscopy, 3rd ed. Lakowicz, J.R. (Ed.) Springer: Boston, MA, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Schlamadinger, D.E.; Kats, D.I.; Kim, J.E. Quenching of Tryptophan Fluorescence in Unfolded Cytochrome c: A Biophysics Experiment for Physical Chemistry Students. J. Chem. Educ. 2010, 87, 961–964. [Google Scholar] [CrossRef]
- Korzonek-Szlacheta, I.; Zubelewicz-Szkodzińska, B.; Gąsior, M. Płytki krwi—ogniwo łączące zakrzepicę ze stanem zapalnym. Folia Cardiol. 2018, 13, 303–308. [Google Scholar] [CrossRef][Green Version]
- Valeur, B.; Berberan-Santos, M.N. Molecular Fluorescence; Wiley: Hoboken, NJ, USA, 2012; p. 8376. ISBN 9783527328376. [Google Scholar]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981, 114, 199–227. [Google Scholar] [CrossRef]
- Sillen, A.; Engelborghs, Y. The Correct Use of “Average” Fluorescence Parameters. Photochem. Photobiol. 1998, 67, 475–486. [Google Scholar] [CrossRef]
- Kasparek, A.; Smyk, B. Spectroscopic demonstration of sinapic acid methyl ester complexes with serum albumins. RSC Adv. 2020, 10, 8810–8820. [Google Scholar] [CrossRef]
- Reviews in Fluorescence 2005. Geddes, C.D.; Lakowicz, J.R. (Eds.) Springer: Boston, MA, USA, 2005; Volume 2005, ISBN 978-0-387-23628-5. [Google Scholar]
- Palleria, C.; Di Paolo, A.; Giofrè, C.; Caglioti, C.; Leuzzi, G.; Siniscalchi, A.; De Sarro, G.; Gallelli, L. Pharmacokinetic drug-drug interaction and their implication in clinical management. J. Res. Med. Sci. 2013, 18, 601–610. [Google Scholar]
- An, X.; Zhao, J.; Cui, F.; Qu, G. The investigation of interaction between Thioguanine and human serum albumin by fluorescence and modeling. Arab. J. Chem. 2017, 10, S1781–S1787. [Google Scholar] [CrossRef]
- Krokosz, A.; Grebowski, J.; Rodacka, A.; Pasternak, B.; Puchala, M. The effect of fullerenol C60(OH)~30 on the alcohol dehydrogenase activity irradiated with X-rays. Radiat. Phys. Chem. 2014, 97, 102–106. [Google Scholar] [CrossRef]
- Ghisaidoobe, A.B.T.; Chung, S.J. Intrinsic Tryptophan Fluorescence in the Detection and Analysis of Proteins: A Focus on Förster Resonance Energy Transfer Techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Maliwal, B.P.; Gryczynski, Z.; Lakowicz, J.R. Long-Wavelength Long-Lifetime Luminophores. Anal. Chem. 2001, 73, 4277–4285. [Google Scholar] [CrossRef]
- Kang, J.S.; Piszczek, G.; Lakowicz, J.R. Enhanced Emission Induced by FRET from a Long-Lifetime, Low Quantum Yield Donor to a Long-Wavelength, High Quantum Yield Acceptor. J. Fluoresc. 2002, 12, 97–103. [Google Scholar] [CrossRef]
- Tang, L.; Li, J.; Pan, T.; Yin, Y.; Mei, Y.; Xiao, Q.; Wang, R.; Yan, Z.; Wang, W. Versatile carbon nanoplatforms for cancer treatment and diagnosis: Strategies, applications and future perspectives. Theranostics 2022, 12, 2290–2321. [Google Scholar] [CrossRef]
- Grebowski, J.; Litwinienko, G. Metallofullerenols in biomedical applications. Eur. J. Med. Chem. 2022, 238, 114481. [Google Scholar] [CrossRef]
- Lichota, A.; Piwoński, I.; Michlewska, S.; Krokosz, A. A Multiparametric Study of Internalization of Fullerenol C60(OH)36 Nanoparticles into Peripheral Blood Mononuclear Cells: Cytotoxicity in Oxidative Stress Induced by Ionizing Radiation. Int. J. Mol. Sci. 2020, 21, 2281. [Google Scholar] [CrossRef]






| Parameter | Water | 0.02 M Phosphate Buffer pH 7.4 |
|---|---|---|
| Mean (nm) ± SD | 186.5 ± 76.6 | 164.4 ± 65.6 |
| Mode (nm) | 132.8 | 117.3 |
| Concentration (particles/mL) | 2.65 e + 008 | 4.55 e + 008 |
| Zeta potential (mV) ± SD | −27.5 ± 1.0 | −37.4 ± 3.0 |
| Molar Extinction Coefficient ε280 [mol−1 l cm−1] | |
|---|---|
| ADH | 167,900 |
| HSA | 35,340 |
| FUL in 0.02 M phosphate buffer pH 7.4 | 43,336 |
| Concentration of FUL [µM] | τ1 [ns] (α1) | τ2 [ns] (α2) | τ3 [ns] (α3) | <τ> [ns] | ||
|---|---|---|---|---|---|---|
| ADH + FUL | 0 | 4.8040 | 1.8636 | 0.5017 | 2.6894 | 0.998 |
| (0.3909) | (0.3716) | (0.2376) | ||||
| 2.64 | 4.8590 | 2.0210 | 0.6145 | 2.7143 | 0.969 | |
| (0.3750) | (0.2611) | (0.2638) | ||||
| 5.36 | 4.7977 | 1.8792 | 0.5522 | 2.6606 | 0.987 | |
| (0.3793) | (0.3753) | (0.2454) | ||||
| 13.36 | 4.8332 | 1.9843 | 0.5832 | 2.6684 | 0.972 | |
| (0.3709) | (0.3631) | (0.2660) | ||||
| 26.64 | 4.8140 | 1.9055 | 0.5346 | 2.6182 | 0.986 | |
| (0.3669) | (0.3745) | (0.2586) | ||||
| 40.00 | 4.8168 | 1.9429 | 0.5514 | 2.6397 | 0.989 | |
| (0.3703) | (0.3658) | (0.2640) | ||||
| 53.36 | 4.8025 | 1.9342 | 0.5385 | 2.6075 | 1.018 | |
| (0.3654) | (0.3662) | (0.2684) | ||||
| 80.00 | 4.8393 | 2.0225 | 0.6241 | 2.6221 | 0.972 | |
| (0.3518) | (0.3683) | (0.2798) | ||||
| 106.64 | 4.8105 | 1.9846 | 0.5850 | 2.5839 | 0.994 | |
| (0.3507) | (0.3694) | (0.2799) | ||||
| 133.36 | 4.7757 | 1.9377 | 0.5571 | 2.5306 | 0.987 | |
| (0.3495) | (0.3617) | (0.2889 | ||||
| HSA + FUL | 0 | 7.2305 | 3.4098 | 0.8234 | 3.9624 | 1.010 |
| (0.3359) | (0.3815) | (0.2826) | ||||
| 2.64 | 7.2217 | 3.3001 | 0.7711 | 3.7433 | 1.007 | |
| (0.3175) | (0.3653) | (0.3171) | ||||
| 13.36 | 7.0044 | 2.8965 | 0.6909 | 3.4167 | 1.009 | |
| (0.3093) | (0.3504) | (0.3403) | ||||
| 40.00 | 6.9017 | 2.6751 | 0.6620 | 2.9743 | 0.982 | |
| (0.2574) | (0.3509) | (0.3918) | ||||
| 80.00 | 6.7892 | 2.5679 | 0.6879 | 2.6013 | 0.970 | |
| (0.2047) | (0.3534) | (0.4419) | ||||
| 133.36 | 6.5247 | 2.3054 | 0.6084 | 2.2367 | 0.963 | |
| (0.1720) | (0.3600) | (0.4681) |
| TIME-RESOLVED MEASUREMENTS | ||||
|---|---|---|---|---|
| Dynamic Quenching | ||||
| λex/λem [nm] | KSV [M−1] | R2 | ||
| ADH + FUL | 280/330 | 461 ± 43 | 0.9999 | |
| HSA + FUL | 280/340 | 10869 ± 989 | 0.9998 | |
| STEADY-STATE MEASUREMENTS | ||||
| Dynamic Quenching | Static Quenching | |||
|
λex/λem [nm] | KSV [M−1] | V [M−1] | R2 | |
| ADH + FUL | 280/304 | 461 | 8796 ± 68 | 0.9977 |
| 280/340 | 461 | 4917 ± 103 | 0.9887 | |
| 295/340 | 461 | 4610 ± 104 | 0.9872 | |
| HSA + FUL | 280/304 | 12353 ± 1919 | 14067 ± 753 | 0.9993 |
| 280/340 | 13097 ± 1528 | 11742 ± 583 | 0.9994 | |
| 295/340 | 10831 ± 1956 | 10966 ± 837 | 0.9988 | |
| λex/λem [nm] | Concentration of FUL [M] | Kb [M−1] | n | R2 | |
|---|---|---|---|---|---|
| ADH + FUL | 280/304 | 2.64 × 10−6–1.40 × 10−5 | 136.6 | 0.60 ± 0.03 | 0.9957 |
| 280/340 | 17.5 | 0.45 ± 0.04 | 0.9789 | ||
| 295/340 | 3616 | 0.91 ± 0.17 | 0.9663 | ||
| 280/304 | 2.66 × 10−5–5.34 × 10−5 | 1.25 × 105 | 1.24 ± 0.04 | 0.9992 | |
| 280/340 | 1.19 × 105 | 1.30 ± 0.15 | 0.9869 | ||
| 295/340 | 1.36 × 105 | 1.31 ± 0.10 | 0.9941 | ||
| 280/304 | 8.00 × 10−5–1.33 × 10−4 | 8.00 × 106 | 1.68 ± 0.11 | 0.9956 | |
| 280/340 | 1.82 × 106 | 1.61 ± 0.06 | 0.9985 | ||
| 295/340 | 1.95 × 106 | 1.62 ± 0.03 | 0.9997 | ||
| HSA + FUL | 280/304 | 2.64 × 10−6–1.40 × 10−5 | 8671 | 0.87 ± 0.02 | 0.9993 |
| 280/340 | 6246 | 0.84 ± 0.01 | 0.9999 | ||
| 295/340 | 3896 | 0.81 ± 0.03 | 0.9990 | ||
| 280/304 | 1.40 × 10−5–4.00 × 10−5 | 2.02 × 105 | 1.15 ± 0.02 | 0.9998 | |
| 280/340 | 6.23 × 104 | 1.05 ± 0.01 | 0.9999 | ||
| 295/340 | 6.34 × 104 | 1.06 ± 0.04 | 0.9987 | ||
| 280/304 | 8.00 × 10−5–1.33 × 10−4 | 2.72 × 1010 | 2.38 ± 0.02 | 0.9999 | |
| 280/340 | 2.02 × 109 | 2.12 ± 0.06 | 0.9992 | ||
| 295/340 | 1.20 × 109 | 2.09 ± 0.01 | 0.9999 |
| τ1 [ns](α3) | τ2 [ns](α1) | τ3 [ns](α2) | τ4 [ns](α3) | <τ>[ns] | ||
|---|---|---|---|---|---|---|
| FUL in 0.02 M Phosphate Buffer pH 7.4 | 6.4423 | 1.7801 | 0.3502 | 0.0531 | 0.545 | 1.040 |
| (0.0285) | (0.1304) | (0.2860) | (0.5552) | |||
| ADH + FUL | 6.3530 | 2.5694 | 0.9725 | 0.2078 | 1.533 | 0.960 |
| (0.0855) | (0.2614) | (0.2389) | (0.4142) | |||
| HSA + FUL | 8.667 | 3.061 | 1.1289 | 0.3005 | 1.637 | 0.959 |
| (0.0658) | (0.1869) | (0.3260) | (0.4212) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lichota, A.; Szabelski, M.; Krokosz, A. Quenching of Protein Fluorescence by Fullerenol C60(OH)36 Nanoparticles. Int. J. Mol. Sci. 2022, 23, 12382. https://doi.org/10.3390/ijms232012382
Lichota A, Szabelski M, Krokosz A. Quenching of Protein Fluorescence by Fullerenol C60(OH)36 Nanoparticles. International Journal of Molecular Sciences. 2022; 23(20):12382. https://doi.org/10.3390/ijms232012382
Chicago/Turabian StyleLichota, Anna, Mariusz Szabelski, and Anita Krokosz. 2022. "Quenching of Protein Fluorescence by Fullerenol C60(OH)36 Nanoparticles" International Journal of Molecular Sciences 23, no. 20: 12382. https://doi.org/10.3390/ijms232012382
APA StyleLichota, A., Szabelski, M., & Krokosz, A. (2022). Quenching of Protein Fluorescence by Fullerenol C60(OH)36 Nanoparticles. International Journal of Molecular Sciences, 23(20), 12382. https://doi.org/10.3390/ijms232012382






