Dietary Selenomethionine Reduce Mercury Tissue Levels and Modulate Methylmercury Induced Proteomic and Transcriptomic Alterations in Hippocampi of Adolescent BALB/c Mice
Abstract
1. Introduction
2. Results
2.1. Feed Intake and Body Weight
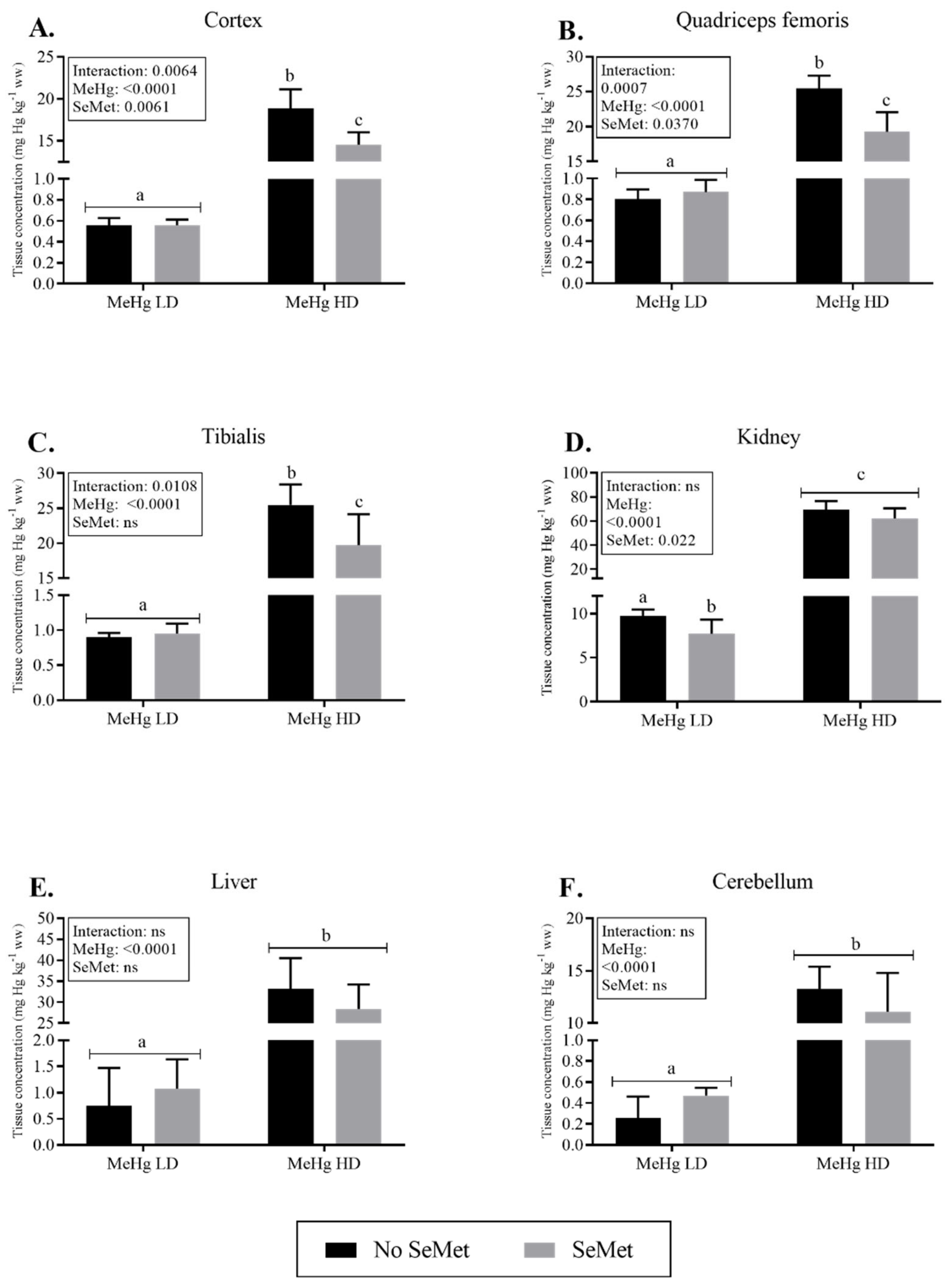
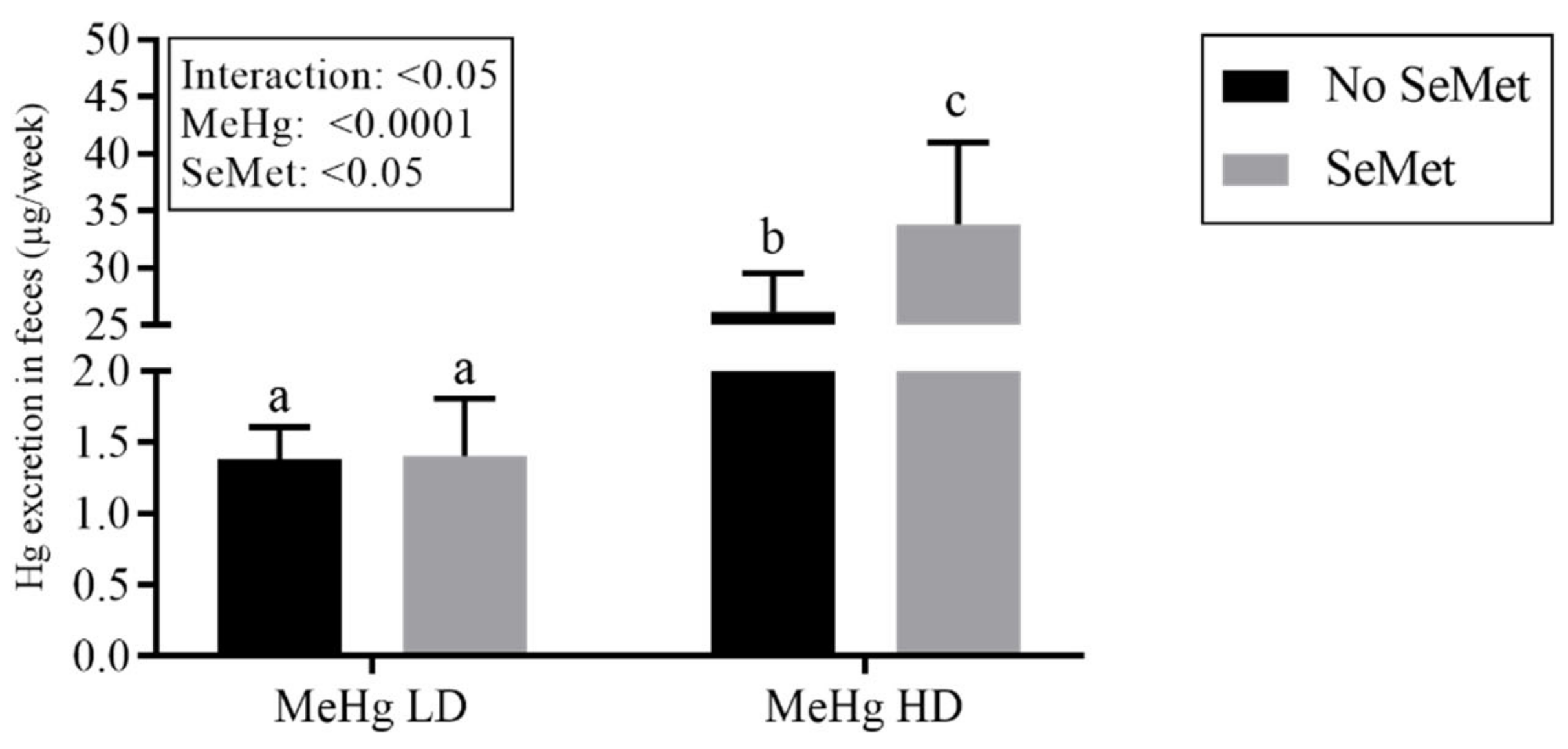
2.2. Mercury Tissue Levels and Excretion
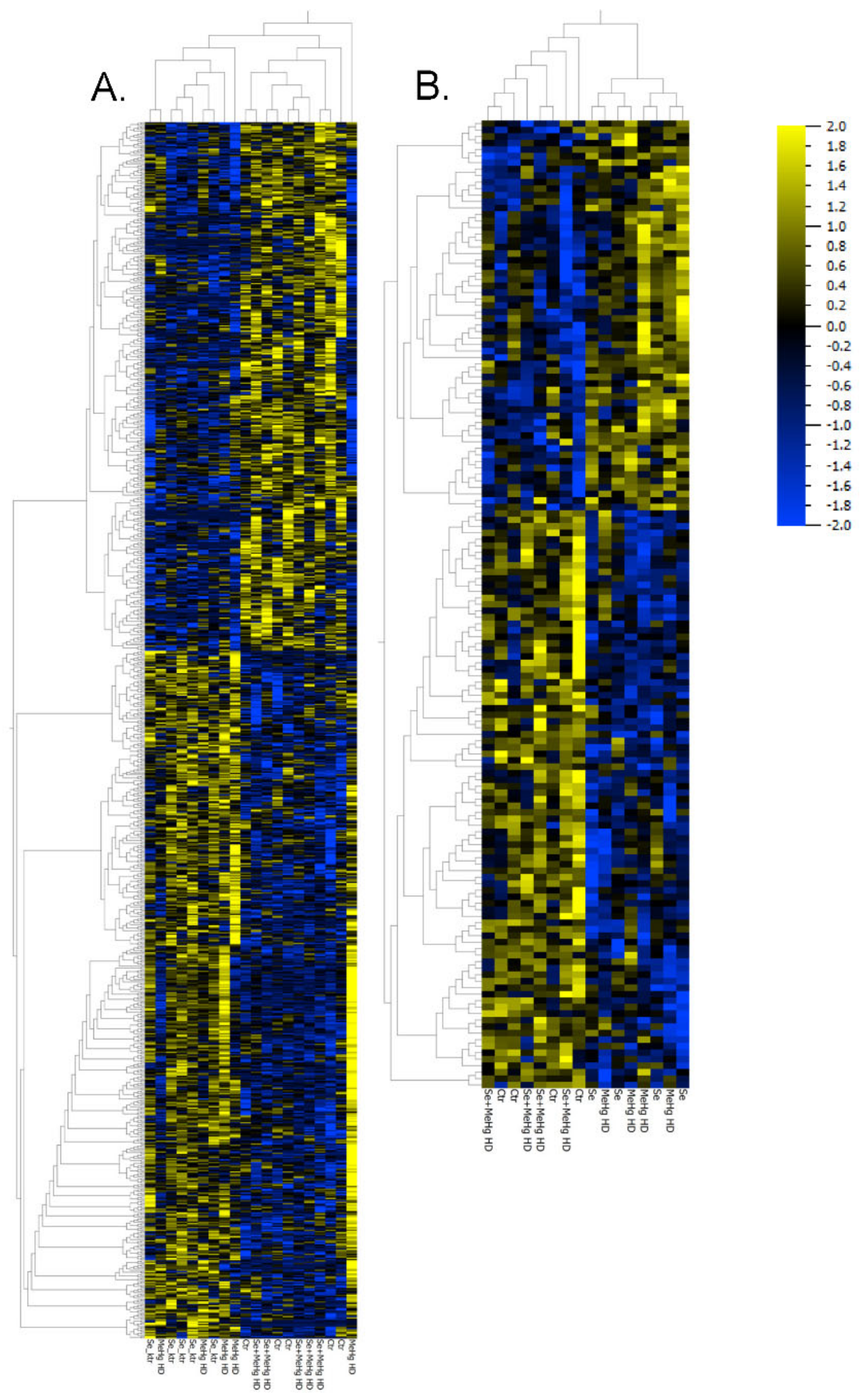
2.3. Proteomics, Transcriptomics and Pathway Analyses
3. Discussion
Limitations
4. Material and Methods
4.1. Experimental Design and Sampling
4.2. Mercury and Selenium Determination
4.3. Proteomics Analysis and RNA Sequencing
4.4. Statistics and Bioinformatics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Crowe, W.; Allsopp, P.J.; Watson, G.E.; Magee, P.J.; Strain, J.J.; Armstrong, D.J.; Ball, E.; McSorley, E.M. Mercury as an environmental stimulus in the development of autoimmunity—A systematic review. Autoimmun. Rev. 2017, 16, 72–80. [Google Scholar] [CrossRef]
- Chen, C.Y.; Stemberger, R.S.; Klaue, B.; Blum, J.D.; Pickhardt, P.C.; Folt, C.L. Accumulation of heavy metals in food web components across a gradient of lakes. Limnol. Oceanogr. 2000, 45, 1525–1536. [Google Scholar] [CrossRef]
- CTM. Toxicological Effects of Methylmercury; National Academies Press: Washington, DC, USA, 2001; p. 344. [Google Scholar]
- EFSA. Scientific Opinion on the risk for public health related to the presence of mercury and methylmercury in food. EFSA J. 2012, 10, 2985. [Google Scholar] [CrossRef]
- Kirkpatrick, M.; Benoit, J.; Everett, W.; Gibson, J.; Rist, M.; Fredette, N.J.N. The effects of methylmercury exposure on behavior and biomarkers of oxidative stress in adult mice. Neurotoxicology 2015, 50, 170–178. [Google Scholar] [CrossRef]
- Bellum, S.; Thuett, K.A.; Grajeda, R.; Abbott, L.C. Coordination deficits induced in young adult mice treated with methylmercury. Int. J. Toxicol. 2007, 26, 115–121. [Google Scholar] [CrossRef]
- Bittencourt, L.O.; Dionizio, A.; Nascimento, P.C.; Puty, B.; Leao, L.K.R.; Luz, D.A.; Silva, M.C.F.; Amado, L.L.; Leite, A.; Buzalaf, M.R.; et al. Proteomic approach underlying the hippocampal neurodegeneration caused by low doses of methylmercury after long-term exposure in adult rats. Met. Integr. Biometal Sci. 2019, 11, 390–403. [Google Scholar] [CrossRef]
- Santana, L.; Bittencourt, L.O.; Nascimento, P.C.; Fernandes, R.M.; Teixeira, F.B.; Fernandes, L.M.P.; Freitas Silva, M.C.; Nogueira, L.S.; Amado, L.L.; Crespo-Lopez, M.E.; et al. Low doses of methylmercury exposure during adulthood in rats display oxidative stress, neurodegeneration in the motor cortex and lead to impairment of motor skills. J. Trace Elem. Med. Biol. 2019, 51, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Myers, G.J.; Davidson, P.W. Prenatal methylmercury exposure and children: Neurologic, developmental, and behavioral research. Environ. Health Perspect. 1998, 106 (Suppl. 3), 841–847. [Google Scholar]
- Castoldi, A.F.; Coccini, T.; Manzo, L. Neurotoxic and molecular effects of methylmercury in humans. Rev. Environ. Health 2003, 18, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Sanfeliu, C.; Sebastia, J.; Cristofol, R.; Rodriguez-Farre, E. Neurotoxicity of organomercurial compounds. Neurotox. Res. 2003, 5, 283–305. [Google Scholar] [CrossRef] [PubMed]
- Chapman, L.; Chan, H.M. The influence of nutrition on methyl mercury intoxication. Environ. Health Perspect. 2000, 108 (Suppl. 1), 29–56. [Google Scholar]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef]
- Papp, L.V.; Lu, J.; Holmgren, A.; Khanna, K.K. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid. Redox Signal. 2007, 9, 775–806. [Google Scholar] [CrossRef] [PubMed]
- Aakre, I.; Naess, S.; Kjellevold, M.; Markhus, M.W.; Alvheim, A.R.; Dalane, J.O.; Kielland, E.; Dahl, L. New data on nutrient composition in large selection of commercially available seafood products and its impact on micronutrient intake. Food Nutr. Res. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.M.; Frantzen, S.; Bank, M.S.; Nilsen, B.M.; Duinker, A.; Madsen, L.; Maage, A. Effects of geography and species variation on selenium and mercury molar ratios in Northeast Atlantic marine fish communities. Sci. Total Environ. 2019, 652, 1482–1496. [Google Scholar] [CrossRef]
- Gropper, S.S. Advanced Nutrition and Human Metabolism, 6th ed.; Wadsworth: Belmont, MA, USA, 2013. [Google Scholar]
- Parizek, J.; Ostadalova, I. The protective effect of small amounts of selenite in sublimate intoxication. Experientia 1967, 23, 142–143. [Google Scholar] [CrossRef] [PubMed]
- Dyrssen, D.; Wedborg, M. The sulphur-mercury(II) system in natural waters. Water Air Soil Pollut. 1991, 56, 507–519. [Google Scholar] [CrossRef]
- Ralston, N.V.C.; Raymond, L.J. Dietary selenium’s protective effects against methylmercury toxicity. Toxicology 2010, 278, 112–123. [Google Scholar] [CrossRef]
- Spiller, H.A. Rethinking mercury: The role of selenium in the pathophysiology of mercury toxicity. Clin. Toxicol. 2018, 56, 313–326. [Google Scholar] [CrossRef]
- Korbas, M.; O’Donoghue, J.L.; Watson, G.E.; Pickering, I.J.; Singh, S.P.; Myers, G.J.; Clarkson, T.W.; George, G.N. The Chemical Nature of Mercury in Human Brain Following Poisoning or Environmental Exposure. ACS Chem. Neurosci. 2010, 1, 810–818. [Google Scholar] [CrossRef]
- Ralston, N.V.C.; Raymond, L.J. Mercury’s neurotoxicity is characterized by its disruption of selenium biochemistry. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2018, 1862, 2405–2416. [Google Scholar] [CrossRef] [PubMed]
- Prohaska, J.R.; Ganther, H.E. Interactions between selenium and methylmercury in rat brain. Chem.-Biol. Interact. 1977, 16, 155–167. [Google Scholar] [CrossRef]
- Ralston, N.V. Selenium health benefit values as seafood safety criteria. EcoHealth 2008, 5, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.A.; Ralston, N.V.; Peck, D.V.; Van Sickle, J.; Robertson, J.D.; Spate, V.L.; Morris, J.S. How might selenium moderate the toxic effects of mercury in stream fish of the western U.S.? Environ. Sci. Technol. 2009, 43, 3919–3925. [Google Scholar] [CrossRef]
- Sele, V.; Ornsrud, R.; Sloth, J.J.; Berntssen, M.H.G.; Amlund, H. Selenium and selenium species in feeds and muscle tissue of Atlantic salmon. J. Trace Elem. Med. Biol. 2018, 47, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Kristan, U.; Arribere, M.A.; Stibilj, V. Selenium species and their distribution in freshwater fish from Argentina. Biol. Trace Elem. Res. 2013, 151, 240–246. [Google Scholar] [CrossRef]
- Amlund, H.; Lundebye, A.K.; Boyle, D.; Ellingsen, S. Dietary selenomethionine influences the accumulation and depuration of dietary methylmercury in zebrafish (Danio rerio). Aquat. Toxicol. 2015, 158, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Rasinger, J.D.; Lundebye, A.K.; Penglase, S.J.; Ellingsen, S.; Amlund, H. Methylmercury Induced Neurotoxicity and the Influence of Selenium in the Brains of Adult Zebrafish (Danio rerio). Int. J. Mol. Sci. 2017, 18, 725. [Google Scholar] [CrossRef]
- European Food Safety Authority; Aguilera, J.; Aguilera-Gomez, M.; Barrucci, F.; Cocconcelli, P.S.; Davies, H.; Denslow, N.; Lou Dorne, J.; Grohmann, L.; Herman, L.J.E.S.P. EFSA Scientific Colloquium 24–’omics in risk assessment: State of the art and next steps. EFSA Support. Publ. 2018, 15, 1512E. [Google Scholar]
- Waters, M.D.; Fostel, J.M. Toxicogenomics and systems toxicology: Aims and prospects. Nat. Rev. Genet. 2004, 5, 936. [Google Scholar] [CrossRef] [PubMed]
- Krewski, D.; Andersen, M.E.; Tyshenko, M.G.; Krishnan, K.; Hartung, T.; Boekelheide, K.; Wambaugh, J.F.; Jones, D.; Whelan, M.; Thomas, R. Toxicity testing in the 21st century: Progress in the past decade and future perspectives. Arch. Toxicol. 2020, 94, 1–58. [Google Scholar] [CrossRef]
- Canzler, S.; Schor, J.; Busch, W.; Schubert, K.; Rolle-Kampczyk, U.E.; Seitz, H.; Kamp, H.; von Bergen, M.; Buesen, R.; Hackermüller, J. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 2020, 94, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.; Barone, S., Jr. Critical periods of vulnerability for the developing nervous system: Evidence from humans and animal models. Environ. Health Perspect. 2000, 108, 511–533. [Google Scholar]
- Rasinger, J.; Carroll, T.; Lundebye, A.; Hogstrand, C.J.T. Cross-omics gene and protein expression profiling in juvenile female mice highlights disruption of calcium and zinc signalling in the brain following dietary exposure to CB-153, BDE-47, HBCD or TCDD. Toxicology 2014, 321, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rasinger, J.; Carroll, T.; Maranghi, F.; Tassinari, R.; Moracci, G.; Altieri, I.; Mantovani, A.; Lundebye, A.-K.; Hogstrand, C.J.R.T. Low dose exposure to HBCD, CB-153 or TCDD induces histopathological and hormonal effects and changes in brain protein and gene expression in juvenile female BALB/c mice. Reprod. Toxicol. 2018, 80, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Doi, R.; Kobayashi, T. Organ distribution and biological half-time of methylmercury in four strains of mice. Jpn. J. Exp. Med. 1982, 52, 307–314. [Google Scholar]
- Yasutake, A.; Hirayama, K. Strain difference in mercury excretion in methylmercury-treated mice. Arch. Toxicol. 1986, 59, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.Y.; Nakai, K.; Kasanuma, Y.; Satoh, H. Comparison of neurobehavioral changes in three inbred strains of mice prenatally exposed to methylmercury. Neurotoxicol. Teratol. 2000, 22, 397–403. [Google Scholar] [CrossRef]
- Mellingen, R.M.; Myrmel, L.S.; Lie, K.K.; Rasinger, J.D.; Madsen, L.; Nøstbakken, O.J.J.M. RNA sequencing and proteomic profiling reveal different alterations by dietary methylmercury in the hippocampal transcriptome and proteome in BALB/c mice. Metallomics 2021, 13, mfab022. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.X. Selenium induces the demethylation of mercury in marine fish. Environ. Pollut. 2017, 231, 1543–1551. [Google Scholar] [CrossRef]
- Li, X.; Yin, D.; Li, J.; Wang, R. Protective effects of selenium on mercury induced immunotoxic effects in mice by way of concurrent drinking water exposure. Arch. Environ. Contam. Toxicol. 2014, 67, 104–114. [Google Scholar] [CrossRef]
- Garcia-Sevillano, M.A.; Rodriguez-Moro, G.; Garcia-Barrera, T.; Navarro, F.; Gomez-Ariza, J.L. Biological interactions between mercury and selenium in distribution and detoxification processes in mice under controlled exposure. Effects on selenoprotein. Chem. Biol. Interact. 2015, 229, 82–90. [Google Scholar] [CrossRef]
- Li, Y.; Fan, Y.; Zhao, J.; Xu, X.; Jing, H.; Shang, L.; Gao, Y.; Li, B.; Li, Y.F. Elevated mercury bound to serum proteins in methylmercury poisoned rats after selenium treatment. Biometals 2016, 29, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Yasutake, A.; Kakita, A.; Ryufuku, M.; Chan, H.M.; Yamamoto, M.; Oumi, S.; Kobayashi, S.; Watanabe, C. Selenomethionine protects against neuronal degeneration by methylmercury in the developing rat cerebrum. Environ. Sci. Technol. 2013, 47, 2862–2868. [Google Scholar] [CrossRef]
- Berry, M.J.; Ralston, N. Mercury Toxicity and the Mitigating Role of Selenium. EcoHealth 2008, 5, 456–459. [Google Scholar] [CrossRef]
- Bjerregaard, P.; Christensen, A. Selenium reduces the retention of methyl mercury in the brown shrimp Crangon crangon. Environ. Sci. Technol. 2012, 46, 6324–6329. [Google Scholar] [CrossRef] [PubMed]
- Bjerregaard, P.; Christensen, L. Accumulation of organic and inorganic mercury from food in the tissues of Carcinus maenas: Effect of waterborne selenium. Mar. Ecol. Prog. Ser. 1993, 99, 271–281. [Google Scholar] [CrossRef]
- Bjerregaard, P.; Andersen, B.W.; Rankin, J. Retention of methyl mercury and inorganic mercury in rainbow trout Oncorhynchus mykiss (W): Effect of dietary selenium. Aquat. Toxicol. 1999, 45, 171–180. [Google Scholar] [CrossRef]
- Bjerregaard, P.; Fjordside, S.; Hansen, M.G.; Petrova, M.B. Dietary selenium reduces retention of methyl mercury in freshwater fish. Environ. Sci. Technol. 2011, 45, 9793–9798. [Google Scholar] [CrossRef]
- Storelli, M.M.; Ceci, E.; Marcotrigiano, G.O. Comparison of total mercury, methylmercury, and selenium in muscle tissues and in the liver of Stenella coeruleoalba (Meyen) and Caretta caretta (Linnaeus). Bull. Environ. Contam. Toxicol. 1998, 61, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.W.; Whanger, P.D.; Fang, S.C. Diversion of mercury binding in rat tissues by selenium: A possible mechanism of protection. Pharmacol. Res. Commun. 1974, 6, 571–579. [Google Scholar] [CrossRef]
- Ralston, N.V.C.; Blackwell, J.L.; Raymond, L.J. Importance of Molar Ratios in Selenium-Dependent Protection Against Methylmercury Toxicity. Biol. Trace Element Res. 2007, 119, 255–268. [Google Scholar] [CrossRef]
- Shi, C.Y.; Lane, A.T.; Clarkson, T.W. Uptake of mercury by the hair of methylmercury-treated newborn mice. Environ. Res. 1990, 51, 170–181. [Google Scholar] [CrossRef]
- Evans, R.D.; Grochowina, N.M.; Basu, N.; O’Connor, E.M.; Hickie, B.E.; Rouvinen-Watt, K.; Evans, H.E.; Chan, H.M. Uptake of selenium and mercury by captive mink: Results of a controlled feeding experiment. Chemosphere 2016, 144, 1582–1588. [Google Scholar] [CrossRef]
- Fujimura, M.; Usuki, F. Methylmercury-Mediated Oxidative Stress and Activation of the Cellular Protective System. Antioxidants 2020, 9, 1004. [Google Scholar] [CrossRef]
- Farina, M.; Aschner, M. Methylmercury-Induced Neurotoxicity: Focus on Pro-oxidative Events and Related Consequences. Adv. Neurobiol. 2017, 18, 267–286. [Google Scholar] [CrossRef]
- Branco, V.; Godinho-Santos, A.; Gonçalves, J.; Lu, J.; Holmgren, A.; Carvalho, C. Mitochondrial thioredoxin reductase inhibition, selenium status, and Nrf-2 activation are determinant factors modulating the toxicity of mercury compounds. Free. Radic. Biol. Med. 2014, 73, 95–105. [Google Scholar] [CrossRef]
- Zemolin, A.; Meinerz, D.; De Paula, M.; Mariano, D.; Rocha, J.; Pereira, A.; Posser, T.; Franco, J. Evidences for a role of glutathione peroxidase 4 (GPx4) in methylmercury induced neurotoxicity in vivo. Toxicology 2012, 302, 60–67. [Google Scholar] [CrossRef]
- Franco, J.L.; Posser, T.; Dunkley, P.R.; Dickson, P.W.; Mattos, J.J.; Martins, R.; Bainy, A.C.; Marques, M.R.; Dafre, A.L.; Farina, M. Methylmercury neurotoxicity is associated with inhibition of the antioxidant enzyme glutathione peroxidase. Free Radic. Biol. Med. 2009, 47, 449–457. [Google Scholar] [CrossRef]
- Penglase, S.; Hamre, K.; Ellingsen, S. Selenium prevents downregulation of antioxidant selenoprotein genes by methylmercury. Free Radic. Biol. Med. 2014, 75, 95–104. [Google Scholar] [CrossRef]
- Pollard, K.M.; Cauvi, D.M.; Toomey, C.B.; Hultman, P.; Kono, D.H. Mercury-induced inflammation and autoimmunity. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2019, 1863, 129299. [Google Scholar] [CrossRef] [PubMed]
- Turksoy, V.A.; Tutkun, L.; Gunduzoz, M.; Oztan, O.; Deniz, S.; Iritas, S.B. Changing levels of selenium and zinc in cadmium-exposed workers: Probable association with the intensity of inflammation. Mol. Biol. Rep. 2019, 46, 5455–5464. [Google Scholar] [CrossRef] [PubMed]
- Duntas, L. Selenium and inflammation: Underlying anti-inflammatory mechanisms. J. Horm. Metab. Res. 2009, 41, 443–447. [Google Scholar] [CrossRef]
- Li, X.; Yin, D.; Yin, J.; Chen, Q.; Wang, R.J.F.; Toxicology, C. Dietary selenium protect against redox-mediated immune suppression induced by methylmercury exposure. Food Chem. Toxicol. 2014, 72, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Deshimaru, M.; Sumiyoshi, S.; Teraoka, A.; Udo, N.; Hattori, E.; Tatetsu, S. Experimental organic mercury poisoning—Pathological changes in peripheral nerves. Acta Neuropathol. 1970, 15, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Imura, N. Mechanism of cytotoxicity of methylmercury. With special reference to microtubule disruption. Biol. Trace Elem. Res. 1989, 21, 313–316. [Google Scholar] [CrossRef]
- Chan, M.C.; Bautista, E.; Alvarado-Cruz, I.; Quintanilla-Vega, B.; Segovia, J. Inorganic mercury prevents the differentiation of SH-SY5Y cells: Amyloid precursor protein, microtubule associated proteins and ROS as potential targets. J. Trace Elem. Med. Biol. 2017, 41, 119–128. [Google Scholar] [CrossRef]
- Xu, F.; Farkas, S.; Kortbeek, S.; Zhang, F.-X.; Chen, L.; Zamponi, G.W.; Syed, N.I. Mercury-induced toxicity of rat cortical neurons is mediated through N-methyl-D-Aspartate receptors. Mol. Brain 2012, 5, 30. [Google Scholar] [CrossRef]
- Miura, K.; Himeno, S.; Koide, N.; Imura, N. Effects of methylmercury and inorganic mercury on the growth of nerve fibers in cultured chick dorsal root ganglia. Tohoku J. Exp. Med. 2000, 192, 195–210. [Google Scholar] [CrossRef][Green Version]
- Rainville, A. Axon growth in the presence of mercury: The effects on cell adhesion. In Neurobiology Short Report; Wheaton College: Norton, MA, USA, 2013. [Google Scholar]
- Cariccio, V.L.; Samà, A.; Bramanti, P.; Mazzon, E. Mercury involvement in neuronal damage and in neurodegenerative diseases. Biol. Trace Elem. Res. 2019, 187, 341–356. [Google Scholar] [CrossRef]
- Shao, Y.; Yamamoto, M.; Figeys, D.; Ning, Z.; Chan, H.M.J.T. Proteome profiling reveals regional protein alteration in cerebrum of common marmoset (Callithrix jacchus) exposed to methylmercury. Toxicology 2016, 347, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Qiu, S.; Wang, Z.; Wang, R.; Wang, C.; Tian, J.; Liu, Q. Direct interaction between selenoprotein P and tubulin. Int. J. Mol. Sci. 2014, 15, 10199–10214. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, N.; Drobyshev, E.; Bjorklund, G.; Dubrovskii, Y.; Lysiuk, R.; Rayman, M.P. Selenium, selenoprotein P, and Alzheimer’s disease: Is there a link? Free Radic. Biol. Med. 2018, 127, 124–133. [Google Scholar] [CrossRef]
- Drouet, B.; Pincon-Raymond, M.; Chambaz, J.; Pillot, T. Molecular basis of Alzheimer’s disease. Cell. Mol. Life Sci. CMLS 2000, 57, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Maccioni, R.B.; Munoz, J.P.; Barbeito, L. The molecular bases of Alzheimer’s disease and other neurodegenerative disorders. Arch. Med. Res. 2001, 32, 367–381. [Google Scholar] [CrossRef]
- Long, J.; Pan, G.; Ifeachor, E.; Belshaw, R.; Li, X. Discovery of Novel Biomarkers for Alzheimer’s Disease from Blood. Dis. Mrk. 2016, 2016, 4250480. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S. The role of mTOR signaling in Alzheimer disease. Front. Biosci. (Sch. Ed.) 2012, 4, 941. [Google Scholar] [CrossRef]
- Ferrer, I.; Andrés-Benito, P.J.N.; Neurobiology, A. White matter alterations in Alzheimer’s disease without concomitant pathologies. Neuropathol. Appl. Neurobiol. 2020, 46, 654–672. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72, 3–12. [Google Scholar] [CrossRef]
- Pereira, J.B.; Janelidze, S.; Smith, R.; Mattsson-Carlgren, N.; Palmqvist, S.; Teunissen, C.E.; Zetterberg, H.; Stomrud, E.; Ashton, N.J.; Blennow, K.J.B. Plasma GFAP is an early marker of amyloid-β but not tau pathology in Alzheimer’s disease. Brain 2021, 144, 3505–3516. [Google Scholar] [CrossRef]
- Hol, E.; Roelofs, R.; Moraal, E.; Sonnemans, M.; Sluijs, J.; Proper, E.; De Graan, P.; Fischer, D.; Van Leeuwen, F. Neuronal expression of GFAP in patients with Alzheimer pathology and identification of novel GFAP splice forms. Mol. Psychiatr. 2003, 8, 786–796. [Google Scholar] [CrossRef]
- Kamphuis, W.; Middeldorp, J.; Kooijman, L.; Sluijs, J.A.; Kooi, E.-J.; Moeton, M.; Freriks, M.; Mizee, M.R.; Hol, E.M. Glial fibrillary acidic protein isoform expression in plaque related astrogliosis in Alzheimer’s disease. Neurobiol. Aging 2014, 35, 492–510. [Google Scholar] [CrossRef] [PubMed]
- Bjorklund, G.; Tinkov, A.A.; Dadar, M.; Rahman, M.M.; Chirumbolo, S.; Skalny, A.V.; Skalnaya, M.G.; Haley, B.E.; Ajsuvakova, O.P.; Aaseth, J. Insights into the Potential Role of Mercury in Alzheimer’s Disease. J. Mol. Neurosci. 2019, 67, 511–533. [Google Scholar] [CrossRef] [PubMed]
- Mutter, J.; Naumann, J.; Sadaghiani, C.; Schneider, R.; Walach, H. Alzheimer disease: Mercury as pathogenetic factor and apolipoprotein E as a moderator. Neuro Endocrinol. Lett. 2004, 25, 331–339. [Google Scholar] [PubMed]
- Haley, B.E. The relationship of the toxic effects of mercury to exacerbation of the medical condition classified as Alzheimer’s disease. Med. Veritas 2007, 4, 1484–1498. [Google Scholar]
- Loef, M.; Schrauzer, G.N.; Walach, H. Selenium and Alzheimer’s disease: A systematic review. J. Alzheimer’s Dis. JAD 2011, 26, 81–104. [Google Scholar] [CrossRef]
- Song, G.; Zhang, Z.; Wen, L.; Chen, C.; Shi, Q.; Zhang, Y.; Ni, J.; Liu, Q. Selenomethionine ameliorates cognitive decline, reduces tau hyperphosphorylation, and reverses synaptic deficit in the triple transgenic mouse model of Alzheimer’s disease. J. Alzheimer’s Dis. JAD 2014, 41, 85–99. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, W.; Liu, X.; Zhang, C.; Wang, P.; Zhao, X. Circulatory Levels of Toxic Metals (Aluminum, Cadmium, Mercury, Lead) in Patients with Alzheimer’s Disease: A Quantitative Meta-Analysis and Systematic Review. J. Alzheimer’s Dis. JAD 2018, 62, 361–372. [Google Scholar] [CrossRef]
- Siblerud, R.; Mutter, J.; Moore, E.; Naumann, J.; Walach, H. A hypothesis and evidence that mercury may be an etiological factor in Alzheimer’s disease. Int. J. Environ. Res. Public Health 2019, 16, 5152. [Google Scholar] [CrossRef]
- Du, X.; Wang, C.; Liu, Q. Potential roles of selenium and selenoproteins in the prevention of Alzheimer’s disease. Curr. Top. Med. Chem. 2016, 16, 835–848. [Google Scholar] [CrossRef]
- Kryscio, R.J.; Abner, E.L.; Caban-Holt, A.; Lovell, M.; Goodman, P.; Darke, A.K.; Yee, M.; Crowley, J.; Schmitt, F.A. Association of antioxidant supplement use and dementia in the prevention of Alzheimer’s disease by vitamin E and selenium trial (PREADViSE). JAMA Neurol. 2017, 74, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Brockman, J.; Schneider, J.A.; Wang, Y.; Bennett, D.A.; Tangney, C.C.; van de Rest, O. Association of Seafood Consumption, Brain Mercury Level, and APOE epsilon4 Status With Brain Neuropathology in Older Adults. Jama 2016, 315, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; Hampel, H.; Feldman, H.H.; Scheltens, P.; Aisen, P.; Andrieu, S.; Bakardjian, H.; Benali, H.; Bertram, L.; Blennow, K.; et al. Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria. Alzheimer’s Dement. 2016, 12, 292–323. [Google Scholar] [CrossRef] [PubMed]
- Paczkowska, M.; Barenboim, J.; Sintupisut, N.; Fox, N.S.; Zhu, H.; Abd-Rabbo, D.; Mee, M.W.; Boutros, P.C.; Reimand, J. Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun. 2020, 11, 735. [Google Scholar] [CrossRef] [PubMed]
- Boomhower, S.R.; Newland, M.C. Adolescent methylmercury exposure affects choice and delay discounting in mice. Neurotoxicology 2016, 57, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Bourdineaud, J.P.; Fujimura, M.; Laclau, M.; Sawada, M.; Yasutake, A. Deleterious effects in mice of fish-associated methylmercury contained in a diet mimicking the Western populations’ average fish consumption. Environ. Int. 2011, 37, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Julshamn, K.; Maage, A.; Norli, H.S.; Grobecker, K.H.; Jorhem, L.; Fecher, P. Determination of arsenic, cadmium, mercury, and lead by inductively coupled plasma/mass spectrometry in foods after pressure digestion: NMKL interlaboratory study. J. AOAC Int. 2007, 90, 844–856. [Google Scholar] [CrossRef]
- Epa, U. Method 7473 (SW-846). In Mercury in Solids and Solutions by Thermal Decomposition, Amalgamation, and Atomic Absorption Spectrophotometry; Environmental Protection Agency: Washington, DC, USA, 2007. [Google Scholar]
- Obiorah, M.; McCandlish, E.; Buckley, B.; DiCicco-Bloom, E. Hippocampal developmental vulnerability to methylmercury extends into prepubescence. Front. Neurosci. 2015, 9, 150. [Google Scholar] [CrossRef]
- Sokolowski, K.; Falluel-Morel, A.; Zhou, X.; DiCicco-Bloom, E. Methylmercury (MeHg) elicits mitochondrial-dependent apoptosis in developing hippocampus and acts at low exposures. Neurotoxicology 2011, 32, 535–544. [Google Scholar] [CrossRef]
- Bernhard, A.; Rasinger, J.D.; Wisløff, H.; Kolbjørnsen, Ø.; Myrmel, L.S.; Berntssen, M.H.; Lundebye, A.-K.; Ørnsrud, R.; Madsen, L.J.F.; Toxicology, C. Subchronic dietary exposure to ethoxyquin dimer induces microvesicular steatosis in male BALB/c mice. Food Chem. Toxicol. 2018, 118, 608–625. [Google Scholar] [CrossRef]
- Mair, P.; Wilcox, R.J.B.R.M. Robust statistical methods in R using the WRS2 package. Behav. Res. Methods 2020, 52, 464–488. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.R-project.org/ (accessed on 1 May 2022).
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015; Available online: http://www.rstudio.com/ (accessed on 1 May 2022).
- Ralston, N.V.C.; Kaneko, J.J.; Raymond, L.J. Selenium health benefit values provide a reliable index of seafood benefits vs. risks. J. Trace Elem. Med. Biol. 2019, 55, 50–57. [Google Scholar] [CrossRef]
| No SeMet | SeMet 3 | |
|---|---|---|
| No MeHg | Ctr (n = 6) | Se (n = 6) |
| Low dose (LD) 1 | MeHg LD (n = 6) | Se + MeHg LD (n = 6) |
| High dose (HD) 2 | MeHg HD (n = 6) | Se + MeHg HD (n = 6) |
| Two Group Comparison | Proteomics | RNA Sequencing | ||
| Group | Vs. | Group | Proteins (n) | Transcripts (n) |
| p = 0.05 | p = 0.05 | |||
| Ctr | MeHg HD | 120 | 1411 | |
| Ctr | Se | 79 | 856 | |
| Ctr | Se+MeHg HD | 137 | 1043 | |
| Se | MeHg HD | 66 | 1927 | |
| Se | Se+MeHg HD | 121 | 938 | |
| HD | Se+MeHg HD | 113 | 1349 | |
| Multi Group Comparison (ANOVA) | Proteomics | RNA sequencing | ||
| Effect | Groups included | Proteins (n) | Transcripts (n) | |
| p = 0.05 | p = 0.05 | |||
| MeHg HD main effect | Ctr, MeHg HD, Se, Se+MeHg HD | 128 | 1775 | |
| SeMet main effect | Ctr, MeHg HD, Se, Se+MeHg HD | 95 | 1572 | |
| Interaction effect HD | Ctr, MeHg HD, Se, Se+MeHg HD | 149 | 916 | |
| Proteomics | Transcriptomics | ||||
|---|---|---|---|---|---|
| Diseases or functions annotation | p-value | Molecules from dataset | Diseases or functions annotation | p-value | Molecules from dataset |
| Neuromuscular disease | *** | ACP1, ATP2B2, CTNNA2, HSBP1, MBP, NRCAM, AP1S1, ATP5MG, FABP7, HSPA5, MOG, TPT1, APOE, ATP6V1A, GAP43, IDH3A, NDUFB5, VAMP1, ATP2B1, COX5A, GFAP, KCNAB2, NDUFS6 | Quantity of neuroglia | *** | Csf1r, Galns, Mmp12, Ptprc, Trem2, Cxcl2, Hdac2, Myd88, Sox2, Tyrobp, Daam2, Il1b, Neil3, Sox9, Fgf2, Kcnj10, Notch1, St8sia4 |
| Progressive neurological disorder | *** | APOE, GAP43, MBP, PRDX6, WDR7, CST3, GFAP, MOBP, SLC2A1, CTNNA2, HSPA5, MOG, SORL1, FAAH, MAG, NRCAM, TPT1 | Quantity of leukocytes | *** | Alcam, Csf1r, Neil3, C1qa, Il1b, Pycard, C4a/C4b, Il27ra, Trem2, Cnr2, Myd88, Tyrobp |
| Disorder of the basal ganglia | *** | ACP1, ATP2B2, CTNNA2, GRIN2B, MBP, TPT1, AP1S1, ATP5MG, FABP7, HSBP1, MOG, VAMP1, APOE, ATP6V1A, GAP43, IDH3A, NDUFB5, ATP2B1, COX5A, GFAP, KCNAB2, NDUFS6 | Recruitment of phagocytes | *** | Cxcl2, Il1b, Myd88, Tlr2 |
| Morphology of the nervous system | *** | ADAM22, BRSK2, CST3, GAP43, KIF5C, MOG, PTK2, SLC44A2, VAMP1, APOE, CHMP4B, CTNNA2, GFAP, MAG, NFASC, PTPRS, SORL1, ASPA, CNTNAP2, DHRS7B, GRIN2B, MAOA, NRCAM, RHEB, UBQLN2, ATP2B2, CSNK2A1, FKBP8, HSPA5, MBP, PRMT8, SLC2A1, UCHL1 | Loss of neuronal progenitor cells | *** | Foxo3, Neil3, Notch1 |
| Alzheimer disease | *** | APOE, GAP43, MOG, WDR7, CST3, GFAP, PRDX6, CTNNA2, MAG, SLC2A1, FAAH, MOBP, SORL1 | Damage of central nervous system | *** | Cnr2, Gpr34, Lepr, Sparc, Cx3cr1, Il1b, Mt3, Thbs1, Entpd1, Kcnk2, Olfml3, Tlr2, Fgf2, Lancl1, Pycard |
| Canonical pathways | Canonical pathways | ||||
| Remodeling of Epithelial Adherens Junctions | *** | CTNNA2, EXOC2, MAPRE1, MAPRE2, TUBA8, TUBA4A | Complement System | *** | C1qa, C1qb, C1qc, C3ar1, C4a/C4b, Itgam, Itgax |
| Phagosome Maturation | *** | ATP6AP1, ATP6V1A, ATP6V1F, CALR, PRDX6, TUBA8, TUBA4A | Inflammasome Pathway | *** | Il1b, Myd88, Naip, P2rx7, Pycard |
| Glutathione Redox Reactions | ** | GPX4, MGST3, PRDX6 | Pyroptosis Signaling Pathway | ** | Casp4, Foxo3, Gsdmd, Il1b, Naip, Nol3, P2rx7, Pycard, Tlr2 |
| Semaphorin Signaling in Neurons | ** | FNBP1, PLXNA1, PLXNB1, PTK2 | Urea Cycle | ** | Ass1, Cps1 |
| LPS/IL-1 Mediated Inhibition of RXR Function | ** | ACSL1, ALDH18A1, APOE, FABP3, FABP7, MAOA, MGST3 | Phospholipases | ** | Gpld1, Hmox1, Plaat1, Plb1, Plcz1, Prdx6 |
| Upstream regulators | Upstream regulators | ||||
| MAPT | *** | UCHL1, MBP, ATP6V1A, TUBA8, NDUFS6, TPT1, HSPH1, GRIN2B, GAP43, MAPRE2, PRDX6, MOG, GFAP, SEC31A, HSPA5, CIQB, TUBA4A, WDR7 | KDM1A | *** | Tlr2, Tent5c, Slc43a3, Phf11, Il27ra, Apobec1, C4a/C4b, Trim21, C3ar1, Sox2, Cd22, Hmox1, Clec7a, Ctss, LCp1, Dock2, Itgax, Ptprc, Ccl4, Cst7, Gfap, Il21r, Ccl3l3, C1qb, Tyrobp, Glycam1 |
| PSEN1 | *** | UCHL1, MBP, ATP6V1A, ACSL1, TUBA8, NDUFS6, TPT1, APOE, GRIN2B, PRDX6, GFAP, HSPA5, KCNAB2, C1QB, TUBA4A | MAPT | *** | Prdx6, Tubb2a, Tlr2, Tent5c, Mt3, Slc43a3, Phf11, Abcg1, Ctsz, Tubb2b, Apobec1, C4a/C4b, Trim21, Map6, Thbs1, Pea15, C3ar1, Dbi, Camk2g, Cd63, CD22, Hmox1, Ctsd, Clec7a, Ctss, Lcp1, Dock2, Gucy1b1, Itgax, Ptprc, Ccl4, Cst7, Gfap, Il21r, Ccl3l3, Il1b, C1qb, Tyrobp, Gad1, Glycam1 |
| mTOR | *** | MBP, SERPINB1, ATP2B1, GAP43, MAG, UBE2O, MOG | ST8SIA1 | *** | C4a/C4b, C3ar1, Sox2, C1qc, C1qa, Il1b, C1qb |
| APP | *** | UCHL1, MBP, ATP6V1A, TUBA8, NDUFS6, TPT1, APOE, GRIN2B, FABP3, GAP43, PRDX6, GFAP, HSPA5, TUBA4A | B4GALNT1 | *** | C4a/C4b, C3ar1, C1qc, C1qa, Il1b, C1qb |
| MYRF | *** | MBP, MAG, MOG | L2HGDH | *** | Clec7a, Itgax, Ccl4, Cd68, Ccl3l3 |
| Selected Features (abb.) | Level of Regulation | p-Value Interaction Effect | MeHg HD | Se | Se + MeHg HD | |
|---|---|---|---|---|---|---|
| Cell plasticity | TUBA8 | Protein | * | |||
| TUBA4A | Protein | ** | * | * | ||
| MAPRE1 | Protein | ** | ||||
| MAPRE2 | Protein | * | ||||
| PLXNA1 | Protein | * | * | |||
| PLXNB1 | Protein | ** | ||||
| Oxidative stress | GPX4 | Protein | * | |||
| PRDX6 | Protein | * | * | |||
| Prdx6 | RNA | * | ||||
| MGST3 | Protein | * | * | |||
| TRXR1 | Protein | *** | *** | |||
| Immune response | Il1b | RNA | ** | ** | ||
| C1qa | RNA | ** | *** | |||
| C1QB | Protein | ** | ** | |||
| C1qb | RNA | ** | *** | |||
| C1qc | RNA | ** | ** | |||
| Alzheimer’s disease | APOE | Protein | * | * | ||
| GFAP | Protein | * | *** | |||
| Gfap | RNA | ** | *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mellingen, R.M.; Myrmel, L.S.; Rasinger, J.D.; Lie, K.K.; Bernhard, A.; Madsen, L.; Nøstbakken, O.J. Dietary Selenomethionine Reduce Mercury Tissue Levels and Modulate Methylmercury Induced Proteomic and Transcriptomic Alterations in Hippocampi of Adolescent BALB/c Mice. Int. J. Mol. Sci. 2022, 23, 12242. https://doi.org/10.3390/ijms232012242
Mellingen RM, Myrmel LS, Rasinger JD, Lie KK, Bernhard A, Madsen L, Nøstbakken OJ. Dietary Selenomethionine Reduce Mercury Tissue Levels and Modulate Methylmercury Induced Proteomic and Transcriptomic Alterations in Hippocampi of Adolescent BALB/c Mice. International Journal of Molecular Sciences. 2022; 23(20):12242. https://doi.org/10.3390/ijms232012242
Chicago/Turabian StyleMellingen, Ragnhild Marie, Lene Secher Myrmel, Josef Daniel Rasinger, Kai Kristoffer Lie, Annette Bernhard, Lise Madsen, and Ole Jakob Nøstbakken. 2022. "Dietary Selenomethionine Reduce Mercury Tissue Levels and Modulate Methylmercury Induced Proteomic and Transcriptomic Alterations in Hippocampi of Adolescent BALB/c Mice" International Journal of Molecular Sciences 23, no. 20: 12242. https://doi.org/10.3390/ijms232012242
APA StyleMellingen, R. M., Myrmel, L. S., Rasinger, J. D., Lie, K. K., Bernhard, A., Madsen, L., & Nøstbakken, O. J. (2022). Dietary Selenomethionine Reduce Mercury Tissue Levels and Modulate Methylmercury Induced Proteomic and Transcriptomic Alterations in Hippocampi of Adolescent BALB/c Mice. International Journal of Molecular Sciences, 23(20), 12242. https://doi.org/10.3390/ijms232012242







