Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant
Abstract
1. Introduction
2. Results
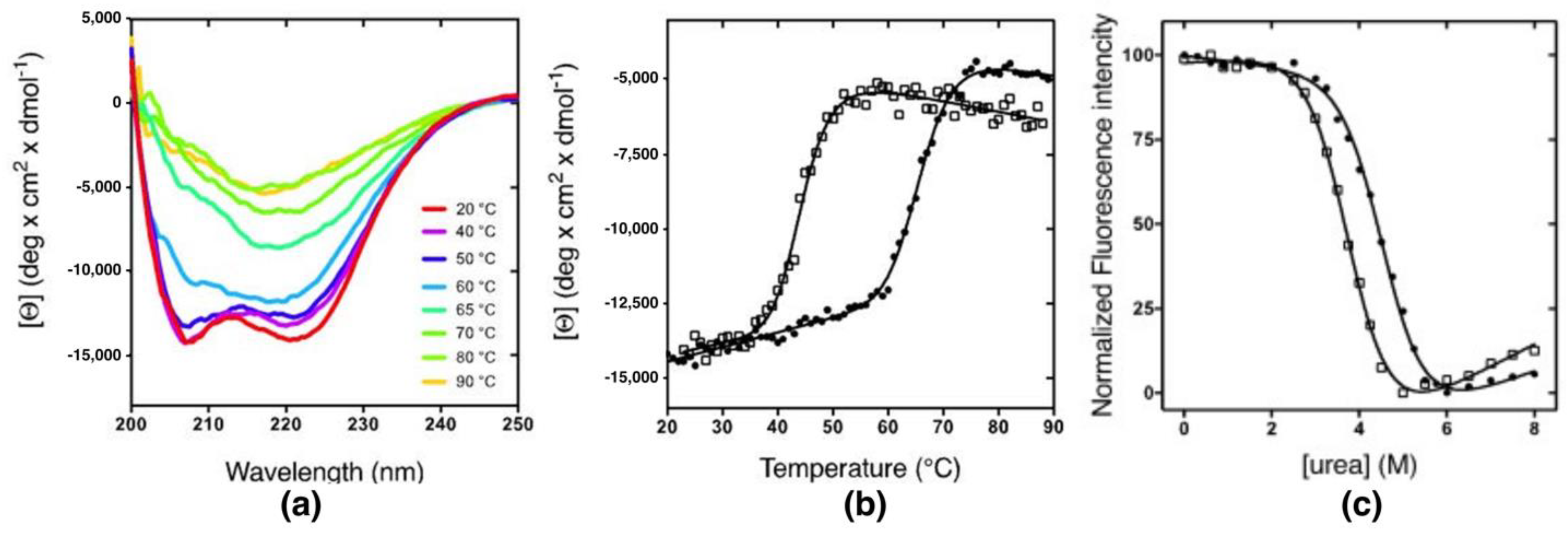
2.1. Equilibrium Experiments
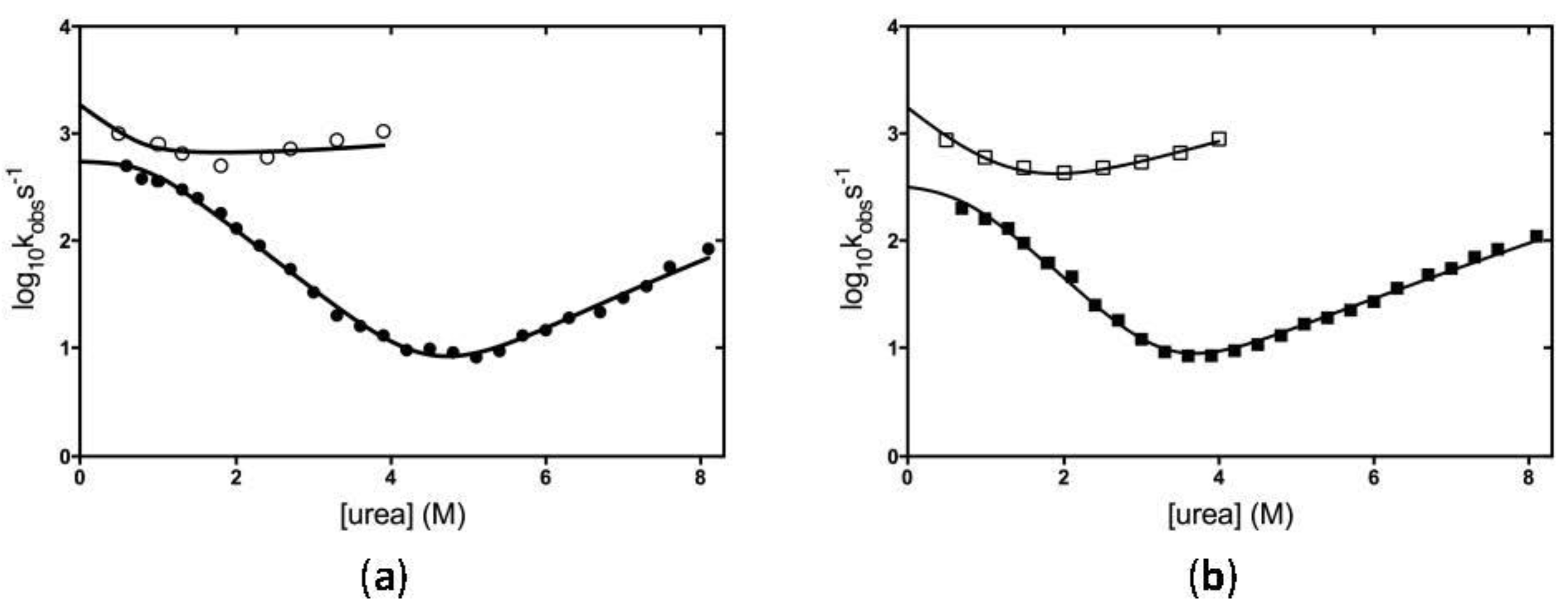
2.2. The Folding Mechanism of KH0 and KH0-R138Q
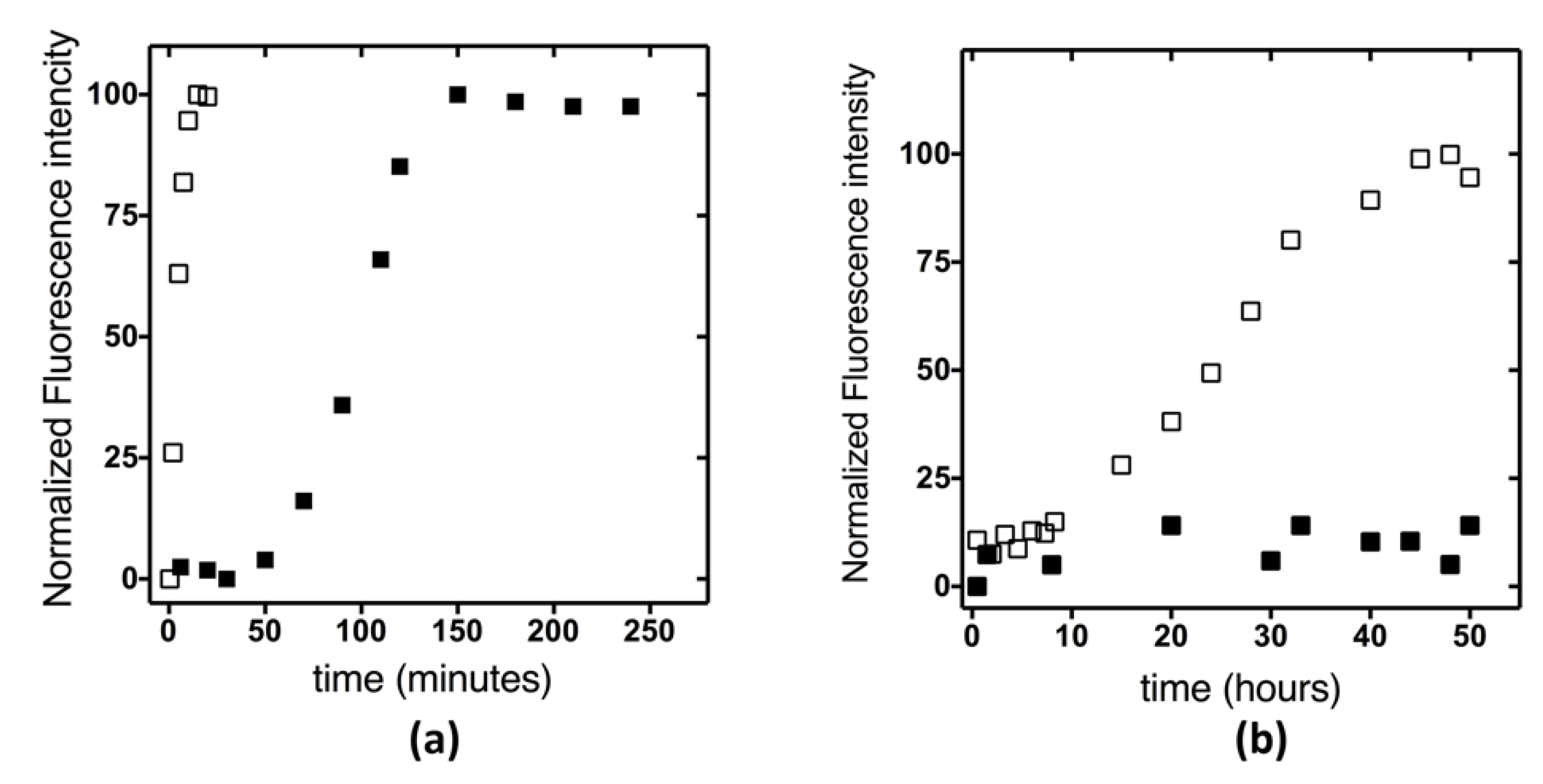
2.3. Aggregation and Fibrillogenesis Propensity of KH0 and KH0-R138Q
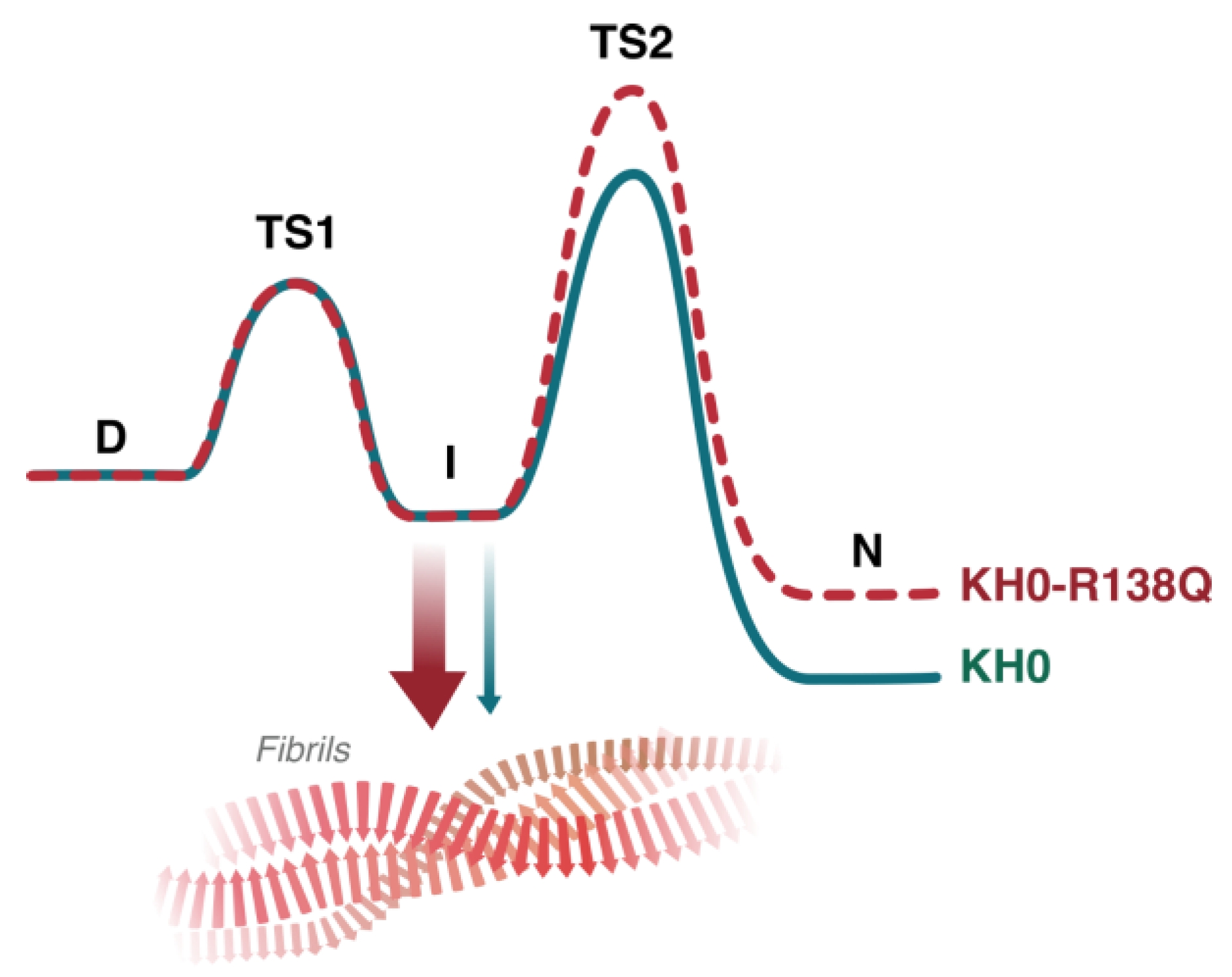
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Equilibrium Experiments
4.3. Kinetic (Un)Folding Experiments
4.4. Thioflavin T Fluorescence Assay
4.5. TEM Experiments
4.6. X-ray Diffraction
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siomi, H.; Matunis, M.J.; Michael, W.M.; Dreyfuss, G. The pre-mRNA binding K protein contains a novel evolutionary conserved motif. Nucleic Acids Res. 1993, 21, 1193–1198. [Google Scholar] [CrossRef]
- Valverde, R.; Edwards, L.; Regan, L. Structure and function of KH domains. FEBS J. 2008, 275, 2712–2726. [Google Scholar] [CrossRef]
- Nicastro, G.; Taylor, I.A.; Ramos, A. KH-RNA interactions: Back in the groove. Curr. Opin. Struct. Biol. 2015, 30, 63–70. [Google Scholar] [CrossRef]
- Hollingworth, D.; Candel, A.M.; Nicastro, G.; Martin, S.R.; Briata, P.; Gherzi, R.; Ramos, A. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 2012, 40, 6873–6886. [Google Scholar] [CrossRef]
- Pereira, J.; Lupas, A.N. The ancestral KH peptide at the root of a domain family with three different folds. Bioinformatics 2018, 34, 3961–3965. [Google Scholar] [CrossRef]
- Santorelli, D.; Rocchio, S.; Fata, F.; Silvestri, I.; Angelucci, F.; Imperi, F.; Marasco, D.; Diaferia, C.; Gigli, L.; Demitri, N.; et al. The folding and aggregation properties of a single KH-domain protein: Ribosome binding factor A (RbfA) from Pseudomonas aeruginosa. Biochim. Biophys. Acta—Gen. Subj. 2021, 1865, 129780. [Google Scholar] [CrossRef]
- Silva, A.P.G.; Chechik, M.; Byrne, R.T.; Waterman, D.G.; Ng, C.L.; Dodson, E.J.; Koonin, E.V.; Antson, A.A.; Smits, C. Structure and activity of a novel archaeal β-CASP protein with N-terminal KH domains. Structure 2011, 19, 622–632. [Google Scholar] [CrossRef]
- Chmiel, N.H.; Rio, D.C.; Doudna, J.A. Distinct contributions of KH domains to substrate binding affinity of Drosophila P-element somatic inhibitor protein. RNA 2006, 12, 283–291. [Google Scholar] [CrossRef]
- Cheng, M.H.K.; Jansen, R.P. A jack of all trades: The RNA-binding protein vigilin. RNA 2017, 8, e1448. [Google Scholar] [CrossRef]
- Oddone, A.; Lorentzen, E.; Basquin, J.; Gasch, A.; Rybin, V.; Conti, E.; Sattler, M. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep. 2007, 8, 63–69. [Google Scholar] [CrossRef]
- Myrick, L.K.; Hashimoto, H.; Cheng, X.; Warren, S.T. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 2015, 24, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- D’Annessa, I.; Cicconardi, F.; di Marino, D. Handling FMRP and its molecular partners: Structural insights into Fragile X Syndrome. Prog. Biophys. Mol. Biol. 2019, 141, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.K.; Broadie, K. Multifarious Functions of the Fragile X Mental Retardation Protein. Trends Genet. 2017, 33, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef]
- Starke, E.L.; Zius, K.; Barbee, S.A. FXS causing missense mutations disrupt FMRP granule formation, dynamics, and function. PloS Genet. 2022, 18, e1010084. [Google Scholar] [CrossRef]
- Lai, A.; Valdez-Sinon, A.N.; Bassell, G.J.; Gary Bassell, C.J. Regulation of RNA granules by FMRP and implications for neurological diseases. Traffic 2020, 2, 454–462. [Google Scholar] [CrossRef]
- Gareau, C.; Martel, D.; Coudert, L.; Mellaoui, S.; Mazroui, R. Characterization of fragile X mental retardation protein granules formation and dynamics in Drosophila. Biol. Open 2013, 2, 68–81. [Google Scholar] [CrossRef][Green Version]
- Adinolfi, S.; Bagni, C.; Musco, G.; Gibson, T.; Mazzarella, L.; Pastore, A. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA 1999, 5, 1248–1258. [Google Scholar] [CrossRef]
- Musco, G.; Kharrat, A.; Fraternali, F.; Gibson, T.J.; Nilgen, M.; Pastore, A. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat. Struct. Biol. 1997, 4, 712–716. [Google Scholar] [CrossRef]
- Musco, G.; Stier, G.; Joseph, C.; Morelli, M.A.C.; Nilges, M.; Gibson, T.J.; Pastore, A. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 1996, 85, 237–245. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Prim. 2017, 3, 17065. [Google Scholar] [CrossRef] [PubMed]
- Sitzmann, A.F.; Hagelstrom, R.T.; Tassone, F.; Hagerman, R.J.; Butler, M.G. Rare FMR1 gene mutations causing fragile X syndrome: A review. Am. J. Med. Genet. Part A 2018, 176, 11–18. [Google Scholar] [CrossRef]
- Quartier, A.; Poquet, H.; Gilbert-Dussardier, B.; Rossi, M.; Casteleyn, A.S.; Portes, V.d.; Feger, C.; Nourisson, E.; Kuentz, P.; Redin, C.; et al. Intragenic FMR1 disease-causing variants: A significant mutational mechanism leading to Fragile-X syndrome. Eur. J. Hum. Genet. 2017, 25, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.C.; Bray, S.M.; Suhl, J.A.; Cutler, D.J.; Coffee, B.; Zwick, M.E.; Warren, S.T. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am. J. Med. Genet. A 2010, 152, 2512–2520. [Google Scholar] [CrossRef]
- Diaz, J.; Scheiner, C. Leon E Presentation of a recurrent FMR1 missense mutation (R138Q) in an affected female. Transl. Sci. Rare Dis. 2018, 3, 139–144. [Google Scholar]
- Myrick, L.K.; Deng, P.Y.; Hashimoto, H.; Oh, Y.M.; Cho, Y.; Poidevin, M.J.; Suhl, J.A.; Visootsak, J.; Cavalli, V.; Jin, P.; et al. Independent role for presynaptic FMRP revealed by an FMR1 missense mutation associated with intellectual disability and seizures. Proc. Natl. Acad. Sci. USA 2015, 112, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Suhl, J.A.; Warren, S.T. Single-Nucleotide mutations in FMR1 reveal novel functions and regulatory mechanisms of the fragile X syndrome protein FMRP. J. Exp. Neurosci. 2015, 9, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Prieto, M.; Folci, A.; Poupon, G.; Schiavi, S.; Buzzelli, V.; Pronot, M.; François, U.; Pousinha, P.; Lattuada, N.; Abelanet, S.; et al. Missense mutation of Fmr1 results in impaired AMPAR-mediated plasticity and socio-cognitive deficits in mice. Nat. Commun. 2021, 12, 1557. [Google Scholar] [CrossRef] [PubMed]
- Sjekloća, L.; Pauwels, K.; Pastore, A. On the aggregation properties of FMRP—A link with the FXTAS syndrome? FEBS J. 2011, 278, 1912–1921. [Google Scholar] [CrossRef]
- Parker, M.J.; Spencer, J.; Clarke, A.R. An Integrated Kinetic Analysis of Intermediates and Transition States in Protein Folding Reactions. J. Mol. Biol. 1995, 253, 771–786. [Google Scholar] [CrossRef]
- Gianni, S.; Ivarsson, Y.; Jemth, P.; Brunori, M.; Travaglini-Allocatelli, C. Identification and characterization of protein folding intermediates. Biophys. Chem. 2007, 128, 105–113. [Google Scholar] [CrossRef]
- Malgieri, G.; D’Abrosca, G.; Pirone, L.; Toto, A.; Palmieri, M.; Russo, L.; Sciacca, M.F.M.; Tatè, R.; Sivo, V.; Baglivo, I.; et al. Folding mechanisms steer the amyloid fibril formation propensity of highly homologous proteins. Chem. Sci. 2018, 9, 3290–3298. [Google Scholar] [CrossRef]
- del Poggetto, E.; Chiti, F.; Bemporad, F. The Folding process of Human Profilin-1, a novel protein associated with familial amyotrophic lateral sclerosis. Sci. Rep. 2015, 5, 12332. [Google Scholar] [CrossRef]
- Neudecker, P.; Robustelli, P.; Cavalli, A.; Walsh, P.; Lundström, P.; Zarrine-Afsar, A.; Sharpe, S.; Vendruscolo, M.; Kay, L.E. Structure of an Intermediate State in Protein Folding and Aggregation. Science 2012, 336, 362–366. [Google Scholar] [CrossRef]
- Chakraborty, I.; Kar, R.K.; Sarkar, D.; Kumar, S.; Maiti, N.C.; Mandal, A.K.; Bhunia, A. Solvent Relaxation NMR: A Tool for Real-Time Monitoring Water Dynamics in Protein Aggregation Landscape. ACS Chem. Neurosci. 2021, 12, 2903–2916. [Google Scholar] [CrossRef]
- Nilsson, M.R. Techniques to study amyloid fibril formation in vitro. Methods 2004, 34, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Sulatskaya, A.I.; Maskevich, A.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Fluorescence Quantum Yield of Thioflavin T in Rigid Isotropic Solution and Incorporated into the Amyloid Fibrils. PLoS ONE 2010, 5, e15385. [Google Scholar] [CrossRef] [PubMed]
- Goldsbury, C.; Baxa, U.; Simon, M.N.; Steven, A.C.; Engel, A.; Wall, J.S.; Aebi, U.; Müller, S.A. Amyloid structure and assembly: Insights from scanning transmission electron microscopy. J. Struct. Biol. 2011, 173, 1–13. [Google Scholar] [CrossRef]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Novak, E.; Petrosino, M.; Santorelli, D.; Chiaraluce, R.; Consalvi, V.; Pasquo, A.; Travaglini-Allocatelli, C. A Glimpse into the Structural Properties of the Intermediate and Transition State in the Folding of Bromodomain 2 Domain 2 by Φ Value Analysis. Int. J. Mol. Sci. 2021, 22, 5953. [Google Scholar] [CrossRef]
- Fersht, A.R.; Sato, S. Value analysis and the nature of protein-folding transition states. Proc. Natl. Acad. Sci. USA 2004, 101, 7976–7981. [Google Scholar] [CrossRef]
- Zheng, W.; Tsai, M.Y.; Chen, M.; Wolynes, P.G. Exploring the aggregation free energy landscape of the amyloid-β protein (1–40). Proc. Natl. Acad. Sci. USA 2016, 113, 11835–11840. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wolynes, P.G. Aggregation landscapes of Huntingtin exon 1 protein fragments and the critical repeat length for the onset of Huntington’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, 4406–4411. [Google Scholar] [CrossRef]
- Grishin, N.V. Fold change in evolution of protein structures. J. Struct. Biol. 2001, 134, 167–185. [Google Scholar] [CrossRef]
- Grishin, N.V. KH domain: One motif, two folds. Nucleic Acids Res. 2001, 29, 638–643. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Lausi, A.; Polentarutti, M.; Onesti, S.; Plaisier, J.R.; Busetto, E.; Bais, G.; Barba, L.; Cassetta, A.; Campi, G.; Lamba, D.; et al. Status of the crystallography beamlines at Elettra. Eur. Phys. J. Plus 2015, 130, 43. [Google Scholar] [CrossRef]
- Hammersley, A.P. FIT2D: A multi-purpose data reduction, analysis and visualization program. J. Appl. Crystallogr. 2016, 49, 646–652. [Google Scholar] [CrossRef]
- Huang, T.C.; Toraya, H.; Blanton, T.N.; Wu, Y. X-ray Powder Diffraction Analysis of Silver Behenate, a Possible Low-Angle Diffraction Standard. J. Appl. Cryst. 1993, 26, 180–184. [Google Scholar] [CrossRef]
- Santoro, M.; Bolen, D.W. Unfolding Free Energy Changes Determined by the Linear Extrapolation Method. 1. Unfolding Phenylmethanesulfonyl a-Chymotrypsin Using Different Denaturants. Biochemistry 1988, 27, 8063–8068. [Google Scholar] [CrossRef] [PubMed]


| Circular Dichroism | Fluorescence | |||
|---|---|---|---|---|
| KH0 | KH0-R138Q | W-KH0 | W-KH0 R138Q | |
| [Urea]1/2 | 4.30 ± 0.04 | 3.46 ± 0.06 | 4.56 ± 0.04 | 3.77 ± 0.04 |
| m | 1.29 ± 0.06 | 1.29 ± 0.06 | 1.25 ± 0.06 | 1.25 ± 0.06 |
| DGD-N | 5.50 ± 0.20 | 4.46 ± 0.23 | 5.70 ± 0.32 | 4.70 ± 0.24 |
| W-KH0 | W-KH0 R138Q | |
|---|---|---|
| kDI | 1860 ± 220 | 1723 ± 123 |
| mDI | 0.75 ± 0.04 | 0.87 ± 0.02 |
| kID | 29 ± 20 | 54 ± 6 |
| mID | 0.22 ± 0.10 | 0.33 ± 0.03 |
| kIN | 584 ± 58 | 243 ± 12 |
| mIN | 10−12 ± 10−3 | 10−12 ± 10−3 |
| kNI | 1.62 ± 1.1 | 1.20 ± 0.04 |
| mNI | 0.32 ± 0.06 | 0.32 (fixed) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santorelli, D.; Troilo, F.; Fata, F.; Angelucci, F.; Demitri, N.; Giardina, G.; Federici, L.; Catalano, F.; Di Matteo, A.; Travaglini-Allocatelli, C. Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant. Int. J. Mol. Sci. 2022, 23, 12178. https://doi.org/10.3390/ijms232012178
Santorelli D, Troilo F, Fata F, Angelucci F, Demitri N, Giardina G, Federici L, Catalano F, Di Matteo A, Travaglini-Allocatelli C. Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant. International Journal of Molecular Sciences. 2022; 23(20):12178. https://doi.org/10.3390/ijms232012178
Chicago/Turabian StyleSantorelli, Daniele, Francesca Troilo, Francesca Fata, Francesco Angelucci, Nicola Demitri, Giorgio Giardina, Luca Federici, Flavia Catalano, Adele Di Matteo, and Carlo Travaglini-Allocatelli. 2022. "Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant" International Journal of Molecular Sciences 23, no. 20: 12178. https://doi.org/10.3390/ijms232012178
APA StyleSantorelli, D., Troilo, F., Fata, F., Angelucci, F., Demitri, N., Giardina, G., Federici, L., Catalano, F., Di Matteo, A., & Travaglini-Allocatelli, C. (2022). Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant. International Journal of Molecular Sciences, 23(20), 12178. https://doi.org/10.3390/ijms232012178







