Methylome Profiling in Fabry Disease in Clinical Practice: A Proof of Concept
Abstract
1. Introduction
2. Results and Discussion
2.1. Study Cohort
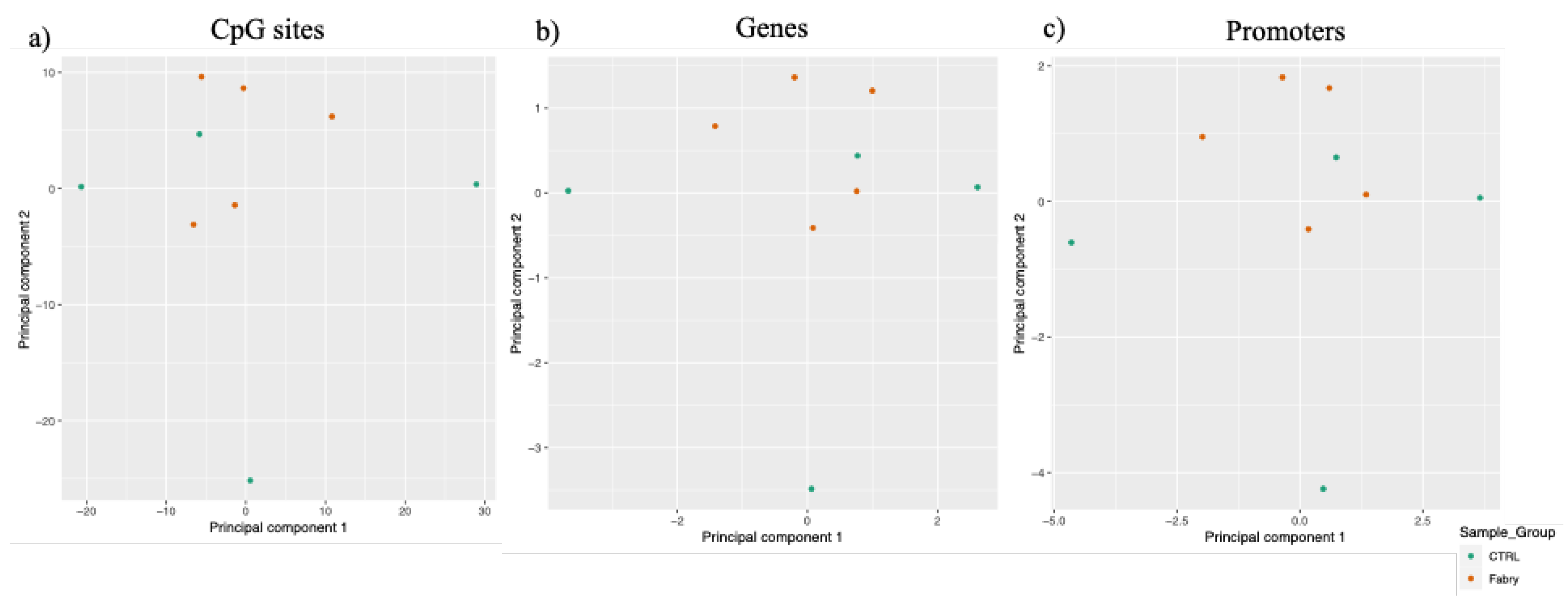
2.2. Genome-Wide DNA Methylation Analyses in FD Patients and in Healthy Controls
2.3. Identification of Differentially Methylated CpG Sites and Differentially Methylated Regions
3. Materials and Methods
3.1. Samples Processing
3.2. DNA Methylome Analyses
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Desnick, R.; Ioannou, Y.; Eng, C. α-Galactosidase A deficiency: Fabry disease. In The Metabolic and Molecular Bases of Inherited Disease, 8th ed.; Scriver, C.R.B.A., Sly, W.S., Valle, D., Eds.; McGraw-Hill: New York, NY, USA, 2001; pp. 3733–3774. [Google Scholar]
- Chien, Y.H.; Lee, N.C.; Chiang, S.C.; Desnick, R.J.; Hwu, W.L. Fabry disease: Incidence of the common later-onset α-galactosidase A IVS4+919G→A mutation in Taiwanese newborns—Superiority of DNA-based to enzyme-based newborn screening for common mutations. Mol. Med. 2012, 18, 780–784. [Google Scholar] [CrossRef] [PubMed]
- Crutchfield, K.E.; Patronas, N.J.; Dambrosia, J.M.; Frei, K.P.; Banerjee, T.K.; Barton, N.W.; Schiffmann, R. Quantitative analysis of cerebral vasculopathy in patients with Fabry disease. Neurology 1998, 50, 1746–1749. [Google Scholar] [CrossRef] [PubMed]
- Germain, D.P. Fabry disease. Orphanet. J. Rare Dis. 2010, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Hopwood, J.J.; Clague, A.E.; Carey, W.F. Prevalence of lysosomal storage disorders. JAMA 1999, 281, 249–254. [Google Scholar] [CrossRef]
- Van der Tol, L.; Smid, B.E.; Poorthuis, B.J.; Biegstraaten, M.; Deprez, R.H.; Linthorst, G.E.; Hollak, C.E. A systematic review on screening for Fabry disease: Prevalence of individuals with genetic variants of unknown significance. J. Med. Genet. 2014, 51, 1–9. [Google Scholar] [CrossRef]
- Limongelli, G.; Adorisio, R.; Baggio, C.; Bauce, B.; Biagini, E.; Castelletti, S.; Favilli, S.; Imazio, M.; Lioncino, M.; Merlo, M.; et al. Diagnosis and Management of Rare Cardiomyopathies in Adult and Paediatric Patients. A Position Paper of the Italian Society of Cardiology (SIC) and Italian Society of Paediatric Cardiology (SICP). Int. J. Cardiol. 2022, 357, 55–71. [Google Scholar] [CrossRef]
- Rubino, M.; Monda, E.; Lioncino, M.; Caiazza, M.; Palmiero, G.; Dongiglio, F.; Fusco, A.; Cirillo, A.; Cesaro, A.; Capodicasa, L.; et al. Diagnosis and Management of Cardiovascular Involvement in Fabry Disease. Heart Fail Clin. 2022, 18, 39–49. [Google Scholar] [CrossRef]
- Sadikovic, B.; Aref-Eshghi, E.; Levy, M.A.; Rodenhiser, D. DNA methylation signatures in mendelian developmental disorders as a diagnostic bridge between genotype and phenotype. Epigenomics 2019, 11, 563–575. [Google Scholar] [CrossRef]
- Haghshenas, S.; Bhai, P.; Aref-Eshghi, E.; Sadikovic, B. Diagnostic Utility of Genome-Wide DNA Methylation Analysis in Mendelian Neurodevelopmental Disorders. Int. J. Mol. Sci. 2020, 21, 9303. [Google Scholar] [CrossRef]
- Schenkel, L.C.; Kernohan, K.D.; McBride, A.; Reina, D.; Hodge, A.; Ainsworth, P.J.; Rodenhiser, D.I.; Pare, G.; Bérubé, N.G.; Skinner, C.; et al. Identification of epigenetic signature associated with alpha thalassemia/mental retardation X-linked syndrome. Epigenetics Chromatin 2017, 10, 10. [Google Scholar] [CrossRef]
- Guastafierro, T.; Bacalini, M.G.; Marcoccia, A.; Gentilini, D.; Pisoni, S.; Di Blasio, A.M.; Corsi, A.; Franceschi, C.; Raimondo, D.; Spanò, A.; et al. Genome-wide DNA methylation analysis in blood cells from patients with Werner syndrome. Clin. Epigenetics 2017, 9, 92. [Google Scholar] [CrossRef]
- Eisenberger, T.; Neuhaus, C.; Khan, A.O.; Decker, C.; Preising, M.N.; Friedburg, C.; Bieg, A.; Gliem, M.; Charbel Issa, P.; Holz, F.G.; et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: The example of retinal dystrophies. PLoS ONE 2013, 8, e78496. [Google Scholar] [CrossRef]
- Chater-Diehl, E.; Goodman, S.J.; Cytrynbaum, C.; Turinsky, A.L.; Choufani, S.; Weksberg, R. Anatomy of DNA methylation signatures: Emerging insights and applications. Am. J. Hum. Genet. 2021, 108, 1359–1366. [Google Scholar] [CrossRef]
- Butcher, D.T.; Cytrynbaum, C.; Turinsky, A.L.; Siu, M.T.; Inbar-Feigenberg, M.; Mendoza-Londono, R.; Chitayat, D.; Walker, S.; Machado, J.; Caluseriu, O.; et al. CHARGE and Kabuki Syndromes: Gene-Specific DNA Methylation Signatures Identify Epigenetic Mechanisms Linking These Clinically Overlapping Conditions. Am. J. Hum. Genet. 2017, 100, 773–788. [Google Scholar] [CrossRef]
- Choufani, S.; Cytrynbaum, C.; Chung, B.H.; Turinsky, A.L.; Grafodatskaya, D.; Chen, Y.A.; Cohen, A.S.; Dupuis, L.; Butcher, D.T.; Siu, M.T.; et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015, 6, 10207. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; Kerkhof, J.; Pedro, V.P.; Groupe, D.I.F.; Barat-Houari, M.; Ruiz-Pallares, N.; Andrau, J.C.; Lacombe, D.; Van-Gils, J.; Fergelot, P.; et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020, 106, 356–370. [Google Scholar] [CrossRef]
- Krzyzewska, I.M.; Maas, S.M.; Henneman, P.; Lip, K.V.D.; Venema, A.; Baranano, K.; Chassevent, A.; Aref-Eshghi, E.; van Essen, A.J.; Fukuda, T.; et al. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin. Epigenetics 2019, 11, 156. [Google Scholar] [CrossRef]
- Schenkel, L.C.; Rodenhiser, D.I.; Ainsworth, P.J.; Paré, G.; Sadikovic, B. DNA methylation analysis in constitutional disorders: Clinical implications of the epigenome. Crit. Rev. Clin. Lab. Sci. 2016, 53, 147–165. [Google Scholar] [CrossRef]
- De Riso, G.; Cuomo, M.; Di Risi, T.; Della Monica, R.; Buonaiuto, M.; Costabile, D.; Pisani, A.; Cocozza, S.; Chiariotti, L. Ultra-Deep DNA Methylation Analysis of X-Linked Genes: GLA and AR as Model Genes. Genes 2020, 11, 620. [Google Scholar] [CrossRef]
- Di Risi, T.; Vinciguerra, R.; Cuomo, M.; Della Monica, R.; Riccio, E.; Cocozza, S.; Imbriaco, M.; Duro, G.; Pisani, A.; Chiariotti, L. DNA methylation impact on Fabry disease. Clin. Epigenetics 2021, 13, 24. [Google Scholar] [CrossRef]
- Müller, F.; Scherer, M.; Assenov, Y.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. RnBeads 2.0: Comprehensive analysis of DNA methylation data. Genome Biol. 2019, 20, 55. [Google Scholar] [CrossRef]
- Assenov, Y.; Müller, F.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods. 2014, 11, 1138–1140. [Google Scholar] [CrossRef]
- Shi, H.; Strogantsev, R.; Takahashi, N.; Kazachenka, A.; Lorincz, M.C.; Hemberger, M.; Ferguson-Smith, A.C. ZFP57 regulation of transposable elements and gene expression within and beyond imprinted domains. Epigenetics Chromatin 2019, 12, 49. [Google Scholar] [CrossRef]
- Bak, M.; Boonen, S.E.; Dahl, C.; Hahnemann, J.M.; Mackay, D.J.; Tümer, Z.; Grønskov, K.; Temple, I.K.; Guldberg, P.; Tommerup, N. Genome-wide DNA methylation analysis of transient neonatal diabetes type 1 patients with mutations in ZFP57. BMC Med. Genet. 2016, 17, 29. [Google Scholar] [CrossRef]


| Sample | Age | Sex | Gene Variant | Genomic Position (GRCh38) | Type and Classification of Variant | Interpretation of Variant (ClinVar-NCBI) | α-gal A Activity | Clinical Involvement | Therapy |
|---|---|---|---|---|---|---|---|---|---|
| FD1 | 56 | M | c.901C>G p.Arg301Gly (EXON 6) | chrX: 101398468 | SNV Missense | Pathogenic | Very Low | Renal, Cardiac and Neuropathic | Agalasidase alfa |
| FD2 | 26 | M | c.901C>G p.Arg301Gly (EXON 6) | chrX: 101398468 | SNV Missense | Pathogenic | Very Low | Renal and Cardiac | Agalasidase alfa |
| FD3 | 31 | M | c.901C>G p.Arg301Gly (EXON 6) | chrX: 101398468 | SNV Missense | Pathogenic | Undetected | Renal | Agalasidase alfa |
| FD4 | 43 | M | c.901C>G p.Arg301Gly (EXON 6) | chrX: 101398468 | SNV Missense | Pathogenic | Very Low | Renal, Cardiac and Neurological | Agalasidase alfa |
| FD5 | 38 | M | c.901C>G p.Arg301Gly (EXON 6) | chrX: 101398468 | SNV Missense | Pathogenic | Low | Renal, Cardiac and Neurological | Agalasidase alfa |
| CTRL1 | 38 | M | - | - | - | - | - | - | - |
| CTRL2 | 42 | M | - | - | - | - | - | - | - |
| CTRL3 | 32 | M | - | - | - | - | - | - | - |
| CTRL4 | 48 | M | - | - | - | - | - | - | - |
| Gene Name | Average Methylation in FD (%) | Average Methylation in CTRL (%) | Number of Differentially Methylated CpGs within the Gene | Description |
|---|---|---|---|---|
| ZFP57 | 78.1 | 65.9 | 19 | Zinc finger protein containing a KRAB domain. Diseases associated: diabetes mellitus, transient neonatal, 1 and transient neonatal diabetes mellitus. |
| NUDT12 | 50.7 | 43.8 | 7 | Nudix hydrolase 12. Related pathways: NAD metabolism and nicotinate metabolism. |
| AS3MT | 52 | 45.4 | 5 | Arsenite methyltransferase. Catalyzes the transfer of a methyl group from S-adenosyl-L-methionine (AdoMet) to trivalent arsenical and may play a role in arsenic metabolism. Diseases associated: Borst–Jadassohn intraepidermal carcinoma and schizophrenia. Related pathways: metabolism and metapathway biotransformation phase I and II. |
| RARRES2 | 59.9 | 53.6 | 6 | Retinoic acid receptor responder 2. Diseases associated: monckeberg arteriosclerosis and diabetes. Related pathways: Response to elevated platelet cytosolic Ca2+. |
| PRSS41 | 83.7 | 78.7 | 6 | Serine protease 41. Predicted to enable sodium channel regulator activity. Predicted to be involved in proteolysis. |
| ALOX15B | 60.3 | 63.2 | 10 | Arachidonate 15-lipoxygenase type B. Encodes a member of the lipoxygenase family of structurally related nonheme iron dioxygenases involved in the production of fatty acid hydroperoxides. Diseases associated: autosomal recessive congenital ichthyosis and embryoma. Related pathways: arachidonic acid metabolism and prostaglandin and leukotriene metabolism in senescence. |
| TOMM5 | 58.9 | 56.4 | 5 | Translocase of outer mitochondrial membrane 5. Located in mitochondrion, predicted to be involved in protein targeting to mitochondria. Diseases associated: Sengers syndrome. Related pathways: mitophagy and selective autophagy. |
| CCNT1 | 60.6 | 59.2 | 7 | Cyclin T1. Encodes a member of the highly conserved cyclin C subfamily. The encoded protein associated with cyclin-dependent kinase 9 acts as a cofactor of human immunodeficiency virus type 1 (HIV-1) Tat protein and is necessary for full activation of viral transcription. Overexpression of this gene is implicated in tumor growth. Diseases associated: human immunodeficiency virus type 1 and human cytomegalovirus infection. Related pathways: male infertility and GPCR pathway. |
| Gene Name | Average Methylation in FD (%) | Average Methylation in CTRL (%) | CpG Sites | Description |
|---|---|---|---|---|
| FAM163B | 66 | 67.3 | 5 | Family with sequence similarity 163 member B. Forecasted to be integral component of membrane. |
| CREB3 | 8.4 | 9.4 | 6 | CAMP-responsive element-binding protein 3. Diseases associated: herpes simplex and ventricular tachycardia, catecholaminergic polymorphic, 2. related pathways: MIF mediated glucocorticoid regulation and development beta-adrenergic receptors regulation of ERK. |
| FAM167B | 26 | 28 | 6 | Family with sequence similarity 167 member B. |
| PAPOLB | 57.5 | 59.5 | 6 | Poly(A) polymerase beta. Allows polynucleotide adenylyltransferase activity, involved in mRNA polyadenylation. Forecasted to be located in endoplasmic reticulum and to be active in nucleus. |
| LRRC24 | 50.6 | 52.8 | 9 | Leucine-rich repeat-containing 24. Predicted to act upstream of or within positive regulation of synapse assembly, to be integral component of membrane and to be active in extracellular matrix and extracellular space. Diseases associated: retroperitoneum carcinoma and atrial septal defect 2. |
| BDH2 | 60.1 | 62.4 | 5 | 3-Hydroxybutyrate dehydrogenase 2. Located in cytosol, allows 3-hydroxybutyrate dehydrogenase activity and NAD binding activity. Involved in epithelial cell differentiation and fatty acid beta-oxidation. Diseases associated: alpha-methylacetoacetic aciduria. Related pathways: ketone body metabolism and metabolism. |
| ZNF429 | 46.6 | 49 | 10 | Zinc finger protein 429. It could allow DNA-binding transcription factor activity, RNA polymerase II-specific and RNA polymerase II cis-regulatory region sequence-specific DNA-binding activity. May be involved in regulation of transcription by RNA polymerase II and active in nucleus. Diseases associated: reflex sympathetic dystrophy. Related pathways: gene expression (transcription). |
| KLK7 | 54.3 | 58.3 | 13 | Kallikrein related peptidase 7. The encoded protein may be involved in cancer invasion and metastasis, and increased expression of this gene is associated with unfavorable prognosis and progression of several types of cancer; has chymotrypsin-like activity and plays a role in the proteolysis of intercellular cohesive structures that precedes desquamation, the shedding of the outermost layer of the epidermis. Could be involved in the activation of precursors to inflammatory cytokines. Diseases associated: Netherton syndrome and dermatitis. Related pathways: extracellular matrix organization and collagen chain trimerization. |
| ADRB1 | 20.9 | 25 | 8 | Adrenoceptor beta 1. The adrenergic receptors (subtypes alpha 1, alpha 2, beta 1 and beta 2) are a prototypic family of guanine nucleotide-binding regulatory protein-coupled receptors that mediate the physiological effects of the hormone epinephrine and the neurotransmitter norepinephrine. They are located primarily in the CNS, heart, coronary artery, kidney and muscle. Involved in the regulation of sleep/wake behaviors. Diseases associated: resting heart rate, variation in and short sleep, familial natural, 2. Related pathways: development ligand-independent activation of ESR1 and ESR2 and ADORA2B mediated anti-inflammatory cytokines production. |
| PROB1 | 22.2 | 26.8 | 10 | Proline-rich basic protein 1. Located in nucleoplasm. Diseases associated: Hyperphenylalaninemia, Bh4-Deficient, A. |
| FTH1P22 | 81.6 | 87 | 5 | Ferritin heavy chain 1 pseudogene 22. |
| EGLN1P1 | 35.6 | 41.6 | 5 | Egl-9 family hypoxia-inducible factor 1 pseudogene 1. |
| PLEKHA4 | 54.1 | 60.4 | 12 | Pleckstrin homology domain-containing A4. The encoded protein is a pleckstrin homology (PH) domain-containing protein. Elevated expression of this gene has been observed in some melanomas. Related pathways: PI metabolism and glycerophospholipid biosynthesis. |
| TNNI3 | 51 | 57.9 | 9 | Troponin I3, cardiac type. This gene encodes the TnI-cardiac protein and is exclusively expressed in cardiac muscle tissues. Diseases associated: cardiomyopathy, dilated, 2A and cardiomyopathy, familial Hypertrophic, 7. Related pathways: beta-2 adrenergic-dependent CFTR expression and cytoskeletal Signaling. |
| NKX1-1 | 22.6 | 29.5 | 10 | Encoded protein is a transcription factor of NKX family of homeodomain-containing proteins which are critical regulators of organ development. Diseases associated: orofaciodigital syndrome VIII and twin-to-twin transfusion syndrome. |
| TNNT1 | 31.3 | 40.4 | 7 | Troponin T1, slow skeletal type. Encoded protein is a subunit of troponin, which is a regulatory complex located on the thin filament of the sarcomere. This complex regulates striated muscle contraction in response to fluctuations in intracellular calcium concentration. Diseases associated: nemaline myopathy 5 and nemaline myopathy. Related pathways: malignant pleural mesothelioma and striated muscle contraction pathway. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Risi, T.; Cuomo, M.; Vinciguerra, R.; Ferraro, S.; Della Monica, R.; Costabile, D.; Buonaiuto, M.; Trio, F.; Capoluongo, E.; Visconti, R.; et al. Methylome Profiling in Fabry Disease in Clinical Practice: A Proof of Concept. Int. J. Mol. Sci. 2022, 23, 12110. https://doi.org/10.3390/ijms232012110
Di Risi T, Cuomo M, Vinciguerra R, Ferraro S, Della Monica R, Costabile D, Buonaiuto M, Trio F, Capoluongo E, Visconti R, et al. Methylome Profiling in Fabry Disease in Clinical Practice: A Proof of Concept. International Journal of Molecular Sciences. 2022; 23(20):12110. https://doi.org/10.3390/ijms232012110
Chicago/Turabian StyleDi Risi, Teodolinda, Mariella Cuomo, Roberta Vinciguerra, Sara Ferraro, Rosa Della Monica, Davide Costabile, Michela Buonaiuto, Federica Trio, Ettore Capoluongo, Roberta Visconti, and et al. 2022. "Methylome Profiling in Fabry Disease in Clinical Practice: A Proof of Concept" International Journal of Molecular Sciences 23, no. 20: 12110. https://doi.org/10.3390/ijms232012110
APA StyleDi Risi, T., Cuomo, M., Vinciguerra, R., Ferraro, S., Della Monica, R., Costabile, D., Buonaiuto, M., Trio, F., Capoluongo, E., Visconti, R., Riccio, E., Pisani, A., & Chiariotti, L. (2022). Methylome Profiling in Fabry Disease in Clinical Practice: A Proof of Concept. International Journal of Molecular Sciences, 23(20), 12110. https://doi.org/10.3390/ijms232012110









